Abstract
Salt-induced genes in the cyanobacterium Anabaena sp. strain PCC 7120 were identified by use of a Tn5-based transposon bearing luxAB as a reporter. The genomic sequence adjacent to one site of insertion of the transposon was identical in part to the sequence of the lti2 gene, which was previously identified in a differential screen for cold-induced transcripts in Anabaena variabilis. The lti2-like gene was induced by sucrose and other osmotica and by low temperature, in addition to salt. Regulatory components necessary for the induction of this gene by osmotica were sought by a further round of transposon mutagenesis. One mutant that displayed reduced transcriptional activity of the lti2-like gene in response to exposure to osmotica had an insertion in an open reading frame, which was denoted orrA, whose predicted product showed sequence similarity to response regulators from two-component regulatory systems. The corresponding mutation was reconstructed and was shown, like the second-site transposon mutation, to result in reduced response to osmotic stress. Induction of the lux reporter gene by osmotica was restored by complementation with a genomic fragment containing the entire open reading frame for the presumptive response regulator, whereas a fragment containing a truncated copy of the open reading frame for the response regulator did not complement the mutation.
High concentrations of salt, incipient freezing, and desiccation stress organisms by decreasing the availability of free water (35). So-called compatible solutes, which have the capacity to substitute for water in the stabilization of proteins and membranes, protect against the effects of water stress (36, 37). Numerous prokaryotes, including cyanobacteria, possess uptake systems for compatible solutes (24, 32) and/or synthesize a variety of compatible solutes, including glucosylglycerol (25), sucrose, trehalose, and betaines (36, 37). As many as 100 genes, some of which may function in the synthesis or uptake of compatible solutes, have been reported to be induced by salt stress in Anabaena torulosa (reference 4; see also references 3 and 19). However, the functions of most of these genes are unknown. Salt-induced genes may have homologs among the desiccation-induced genes of higher plants (12, 13), but the functions of the latter genes are also largely unknown, as are the mechanisms of perception of osmotic stress and the subsequent signal transduction pathways that lead to altered genetic expression.
Two-component regulatory systems mediate physiological responses to the environment in a wide range of prokaryotes. Most two-component systems consist of a sensor and a response regulator (21, 41). The sensor is often a transmembrane protein that perceives changes in the extracellular environment and phosphorylates a histidine in its own cytoplasmically localized transmitter domain. The phosphate is subsequently transferred to an aspartate in the amino terminus of the corresponding response regulator, which may then alter transcription or other cellular processes. Two-component regulatory systems are often identified on the basis of sequence similarity. The cytoplasmic domains of the sensory kinases are highly conserved, and the amino termini of response regulators share 20 to 30% amino acid identity on average (21). Although the carboxy termini of response regulators are not conserved, the secondary structure often has a helix-turn-helix DNA binding motif (34).
In cyanobacteria, several functions have been attributed to two-component regulatory systems, including the regulation of developmental processes (10, 31, 46), responses to variations in the spectra of incident light (11, 28), responses to deprivation of phosphate (1, 33, 44), and, in the cyanobacterium Synechocystis sp. strain PCC 6803, responses to salt stress (22, 23). Completion of the chromosomal sequence of Synechocystis sp. has led to the finding that the predicted products of close to 1% of the genes bear extensive similarity to response regulators of two-component systems (27); however, the functions of few of them are known.
We report the identification of a mutant of the filamentous cyanobacterium Anabaena sp. strain PCC 7120, which displays reduced expression of several salt stress-induced genes. The corresponding mutation results from the disruption of an open reading frame (ORF) whose product bears sequence similarity to response regulators of two-component signal transduction systems.
MATERIALS AND METHODS
Culture conditions and transformation of Anabaena sp.
Anabaena sp. strain PCC 7120 was grown on solid (AAN) and in liquid (AA/8 nitrate) nitrate-containing media at 30°C (when indicated, at 15°C) as described by Hu et al. (26). Plasmids (all plasmids used are described in Table 1) were introduced into Anabaena sp. strain PCC 7120 by triparental matings on Nuclepore REC-85 membrane filters with Escherichia coli J53 (RP4) and a second E. coli strain containing both the plasmid to be transferred and helper plasmid pRL528 (14).
TABLE 1.
Plasmids used in this study
| Plasmid | Characteristics and (or) derivationa | Source or reference |
|---|---|---|
| pAHS1 | Apr; clone, in the SalI site of pRL171 (15), of the 15-kb insert (derived from a partial Sau3AI fragment of Anabaena sp. DNA) of a λEMBL3 phage whose insert hybridizes with pRL871 | This study |
| pAHS2 | Apr; 1.7-kb fragment of Anabaena sp. genomic DNA, excised from pAHS1 with SalI (from the vector) and DraI, subcloned between the SalI and EcoRV sites of 2.67-kb plasmid pBluescript SK(+) (Stratagene) | This study |
| pAHS3 and pAHS4 | Apr; 4.2- and 3.9-kb plasmids, respectively, generated by complete digestion of pAHS2 with SalI and partial digestion with StyI, blunting with the Klenow fragment, and religation | This study |
| pAHS5, pAHS6, and pAHS7 | Smr Spr; Cmr cassette C.C1, isolated from pRL178 (15) by digestion with SalI, was inserted into the unique XhoI sites of pAHS2, pAHS3, and pAHS4, respectively, followed by transfer of a BamHI fragment containing the Anabaena sp. DNA into the BamHI site of pRL1049 (6) | This study |
| pRL386a (see Fig. 1) | Cmr Emr; to exchange Tn5-1063 for a lux cassette with a different marker, permitting second-site mutagenesis with transposon Tn5-1058 | This study |
| pRL487 | Cmr Kmr; S.K4/L.XSX1/C.C1 (nomenclature from reference 15) | 17 |
| pRL528 | Cmr; encodes methylases that modify sites targeted by the restriction endonucleases AvaI and AvaII | 14, 18 |
| pRL864 | Bmr Kmr Smr; EcoRI transposon recovery of mutant AB5 | This study |
| pRL871 | Bmr Kmr Smr; DraI transposon recovery of second-site mutant SA6 | This study |
| pRL1058 | Bmr Kmr Smr; bears transposon Tn5-1058 | 45 |
| pRL1063a | Bmr Kmr Smr; bears transposon Tn5-1063, a luxAB-containing derivative of Tn5-1058 | 45 |
| pRL1130a | Smr Spr; bears a cassette that contains sacB and aadA, whose product confers Smr and Spr | 9 |
| pRL1380 | Bmr Kmr Smr Spr; pRL871, linearized with DraI, ligated to the FspI fragment of pRL1130a that contains genes aadA and sacB | This study |
| pRL2270 | Bmr Kmr Smr; ClaI transposon recovery of mutant AB5 | This study |
| RP4 | Apr Kmr Tcr; conjugative plasmid | 42 |
Ap, ampicillin; Bm, bleomycin; Cm, chloramphenicol; Em, erythromycin; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Tc, tetracycline.
Identification of salt-induced genes and quantitation of luminescence.
Strain PCC 7120 was mutagenized with the transposon-bearing plasmid pRL1063a. Insertions into salt-induced genes were identified (see reference 45) by comparing images of luminescence of filter-grown colonies prior to and 5 h following a shift to hyperosmotic conditions, i.e., from AAN medium to AAN medium supplemented with 0.1 M NaCl. Luminescence of spots of cells was measured with a Hamamatsu Photonics System (model C1966-20 [7]); measurements were corrected for instrumental background. Luciferase activities of suspensions were measured with an ATP photometer (Turner Designs, Sunnyvale, Calif. [16]).
Cloning genomic DNA adjacent to transposon insertions.
DNA purified (8) from AB5, a mutant derivative of Anabaena sp. strain PCC 7120, was digested with EcoRI and ligated in a volume of 100 μl. The products of ligation were precipitated and used to transform E. coli DH10B (20) by electroporation. A kanamycin-resistant colony bore a plasmid, designated pRL864, that had resulted from intramolecular ligation and that contained the inserted Tn5-1063 (which carries an oriV) and adjacent cyanobacterial genomic DNA. A genomic fragment cloned similarly by digestion with ClaI was designated pRL2270, and a genomic fragment bearing transposon Tn5-1058 was cloned similarly from mutant strain SA6 and was designated pRL871. We shall refer to this procedure as transposon rescue. A PCR clone of wild-type PCC 7120 DNA bracketing the transposon in mutant AB5 was generated with the primers 5′-GGGGGCAAAACCATTCTAC-3′ and 5′-CTTCGGGATTTTCGTGGATG-3′, whose sequences are based on the sequence of gene lti2 (40).
Restructuring the AB5 mutant and second-site mutagenesis.
In order to perform second-site mutagenesis by insertion of a transposon, plasmid pRL386a (Fig. 1b) was transferred to AB5 by conjugation. Several single recombinants were selected on erythromycin, grown in liquid culture, and plated on medium containing 5% sucrose to select for double recombinants lacking the sacB gene. Survivors that were presumptively identified as double recombinants by their sucrose-resistant, erythromycin-resistant, neomycin-sensitive, and spectinomycin-sensitive phenotype were confirmed as double recombinants by Southern analysis and showed an induction of luciferase by salt that was equivalent to the induction shown by AB5 (data not shown). One double recombinant, designated AB5DR386a, was subjected to second-site mutagenesis by use of the transposon-bearing plasmid pRL1058 (45).
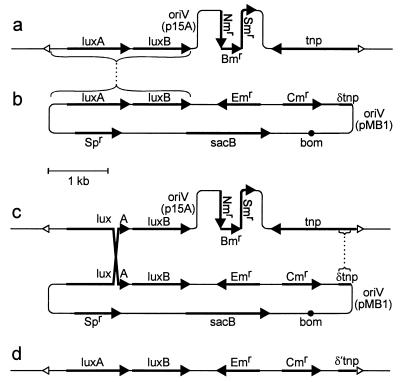
FIG. 1.
Marker exchange. (a) Transposon Tn5-1063 (45), bounded by open triangles, inserted in the genome (thin lines extending to the left and right) of Anabaena sp. (b) pRL386a, which contains genes luxA and luxB for recombination with inserted transposon Tn5-1063 and for transcriptional reporting. Shown are an erythromycin resistance gene (Emr) and a gene (Spr) conferring resistance to spectinomycin and streptomycin for selection of recombinants in Anabaena sp., with Spr also providing a simple screen to distinguish loss of the Spr-sacB portion from mutation of sacB; a chloramphenicol resistance gene (Cmr) for selection in E. coli; a portion (δtnp) of the transposase gene (tnp) to provide a second site for homologous recombination; a sacB gene to provide positive selection for double reciprocal recombination (8); an oriV (from plasmid pMB1) to allow replication in E. coli; and a bom site to allow conjugal mobilization. pRL386a is a derivative of pRL277 (7), which has unique BglII, dam-unmethylated XbaI, and NruI sites. V. fischeri luxAB as a BamHI fragment from pRL1063a was placed in the BglII site, cassette C.CE2 from pRL598a (6) was inserted in the dam-unmethylated XbaI site, and a 199-bp BalI-RsaI fragment from IS50R was transferred first to the EcoRV site of positive selection vector pRL487 and then moved as an NruI fragment to the pRL277-derived NruI site. The dotted lines link regions of sequence identity (brackets) that provide loci for homologous recombination. (c) Product of a recombinational event, obtained upon selection for resistance to erythromycin. (d) Product of a second recombinational event, obtained upon subsequent selection for resistance to erythromycin and sucrose. δ′tnp extends to the end of tnp.
Following mutagenesis, Nuclepore membranes bearing colonies derived from transposition were transferred from AAN medium to AAN medium supplemented with 0.1 M NaCl. Colonies without increased luciferase activity were identified by overlaying images of luminescence 5 h after transfer to salt-containing medium, with images portraying the positions of colonies. One derivative thus identified, which was designated SA6, was selected for further experimentation.
Isolation of a genomic clone of SA6.
An EMBL3 lambda library (6) was screened with pRL871, which had been labeled in a random primer labeling reaction (Promega, Madison, Wis.). Phage DNA from positive plaques was isolated from plate lysates (39); digested with SalI, sites for which bracket the cloned insert; and ligated into the SalI site of pRL171 (15). One resulting plasmid was designated pAHS1. Restriction fragments subcloned into pBluescript SK(+) (Stratagene, La Jolla, Calif.) were subjected to automated sequencing (Applied Biosystems Inc., Foster City, Calif.) on both strands of the DNA. Database comparisons and alignments of the translated sequences were performed by using the default settings of the algorithm developed by Altschul et al. (2) at the National Center for Biotechnology Information with the BLAST network service. The molecular weight of OrrA was calculated by the ExPASy webserver at the Swiss Institute of Bioinformatics, Geneva, Switzerland.
Reconstruction of mutant SA6.
Plasmid pRL1380 was transferred to AB5DR386a, and recombinants were selected on medium containing 400 μg of neomycin ml−1. Several recombinants were grown in liquid culture and were then plated on agar medium containing 5% (wt/vol) sucrose.
Protein analysis.
Cultures of wild-type PCC 7120, AB5DR386a, and SA6 were induced for 8 h by exposure to AAN medium supplemented with 75 mM NaCl and 75 mM KCl. The cells were then harvested by centrifugation, frozen in liquid N2, resuspended in 1× Laemmli loading buffer (30), and heated to 100°C for 5 min. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (gradients of 7.5 to 15% acrylamide), stained with silver, and sized with low-range standards from Bio-Rad Laboratories (Hercules, Calif.).
Complementation.
pAHS5, pAHS6, and pAHS7 were introduced into SA6 by conjugation, with selection on AAN containing 10 μg of spectinomycin ml−1.
Nucleotide sequence accession number.
The sequence of orrA reported in this paper has been submitted to GenBank under accession no. AF056042.
RESULTS
Selection of mutants with insertions in salt-induced genes.
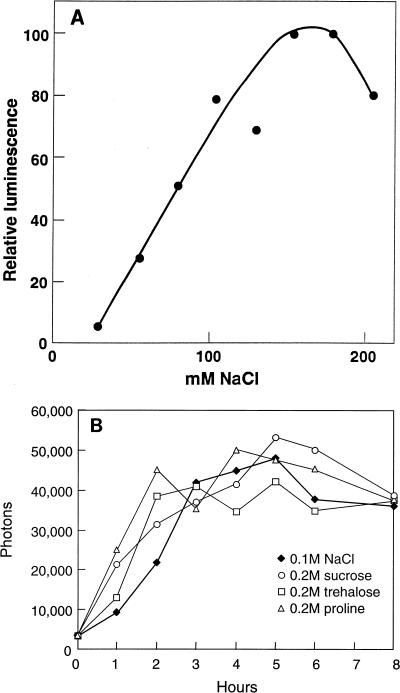
Filters bearing transposition-derived colonies were transferred from solid AAN medium without NaCl to solid AAN medium supplemented with 0.1 M NaCl. Insertions of luxAB-bearing transposons into salt-inducible genes were identified by comparison of images of luminescence prior and subsequent to transfer. Thirty-four mutants of Anabaena sp. strain PCC 7120 that showed salt-induced elevation of luciferase activity were isolated. The response of one of these mutants, AB5, to different concentrations of NaCl is depicted in Fig. 2A. The transposon in mutant AB5 is present within a sequence that shows extensive nucleotide identity to gene lti2 of Anabaena variabilis (40). That is, (i) a short (103-bp) manual sequence outward from IS50R in pRL864 showed near identity to the sequence of lti2 reported by Sato (40); (ii) the PCR product from PCC 7120 that bracketed the site of transposition in AB5 showed complete nucleotide identity with bp 1733 to 1986 of the sequence from within lti2 (40); (iii) based on sequence from single strands, that PCR product also showed probable nucleotide identity with bp 1599 to 2127 of lti2 (the PCC 7120 equivalent of bp 1978 was adjacent to IS50R upon transposition); and (iv) based on automated sequencing of one strand of pRL2270 and derivatives of it, the sequence between the KpnI site in the gene 5′ from the lti2-like gene and the transposon showed probable nucleotide identity with the entire corresponding sequence reported by Sato (40).
FIG. 2.
Relative luminescence of suspensions of mutant AB5 as a function of the concentration of NaCl, after 2 h of exposure, as measured with an ATP photometer (A) and of AB5DR386a as a function of the time of exposure to 0.1 M NaCl or 0.2 M sucrose, trehalose, or proline (B). Readings in panel B are mean values of captured photons minute−1 spot−1 (3 ng of chlorophyll a per spot; average standard error of the mean/mean, 0.085).
Response of AB5DR386a to various osmotica and to temperature.
Whereas lti2 was identified by its strong induction by a decrease in temperature, the lti2-like gene of PCC 7120 was identified by its strong transcriptional responsiveness to NaCl and other osmotica (Fig. 2A and B). Because possible variations in RNA stability (e.g., see reference 38) would not necessarily be reflected in the activity of luciferase, we compared the responsiveness of the lti2-like gene to salt and low temperature by Northern blotting. In several experiments, Northern blots of PCC 7120 grown at 26 to 27°C and either not treated further, shifted to 15°C for 2 h, or supplemented with 0.1 M NaCl for 2 h were probed with a PCR fragment of the lti2-like gene. The increase in transcript abundance in response to the downshift in temperature approximated the increase in transcript abundance occasioned by exposure to salt (data not shown).
Second-site mutagenesis to identify regulatory components.
To identify genes that control expression of the lti2-like gene, much of Tn5-1063 in mutant AB5 was first replaced with a portion of pRL386a. This replacement served several purposes. It (i) eliminated interference with transposition by a preexisting copy of Tn5, (ii) nearly completely prevented the competing reaction of homologous recombination with preexisting transposon sequences, and (iii) permitted repeated use of the powerful, Tn5-derived neomycin, bleomycin, and streptomycin resistance genes. AB5DR386a was then mutagenized with transposon Tn5-1058. Mutants with reduced luciferase expression in response to salt induction were isolated. This phenotype could result from a transposon insertion in a regulatory locus but might also result from a spontaneous mutation, a transposon insertion in the lux reporter, or a severe metabolic impairment.
AB5DR386a from AA/8 nitrate typically showed 0.025 to 0.07 as much luminescence as that after 2 h of exposure to AA/8 nitrate supplemented with 0.1 M NaCl. One mutant, which was designated SA6, expressed a normal basal level of luciferase activity (and was therefore not due to transposition within luxAB) that was not increased by salt. SA6 from AA/8 nitrate with or without 2 h of exposure to 0.1 M NaCl showed only about as much luciferase activity as uninduced AB5DR386a. Growth experiments (data not shown) indicated that SA6 had no substantial metabolic impairment relative to the wild-type strain or AB5, even at elevated concentrations (0.1 to 0.4 M) of NaCl. To examine the relationship between the observed phenotype and the insertion of Tn5-1058, DNA proximal to the site of the Tn5-1058 insertion was isolated from SA6 by transposon rescue as plasmid pRL871. This plasmid was used to construct pRL1380 and to isolate a genomic clone from which pAHS1 was constructed. All tested sucrose-resistant, neomycin-resistant recombinants of AB5DR386a with pRL1380, with the exception of several that also proved resistant to spectinomycin, showed no induction of luciferase activity in response to salt stress after 2 h, i.e., the original phenotype of SA6 had been reproduced (data not shown).
Other, unpublished experiments made use of plasmid pRL386b, which differs from pRL386a by the opposite orientation of its fragment of IS50R. In those experiments, sucrose-resistant, erythromycin-resistant colonies were also neomycin and spectinomycin resistant, with sucrose resistance evidently due to a mutation in the sacB gene, as described earlier (8). These experiments illustrated that for a second homologous recombination to take place, permitting replacement of the Tn5-derived resistance genes, the δtnp fragment of IS50R (Fig. 1b) must be in the same orientation relative to luxAB as that in pRL1063a (i.e., as that in pRL386a).
The transposon in SA6 was inserted within a 720-bp ORF whose predicted product (molecular weight, 26,234) is similar in sequence to response regulators from two-component regulatory systems. We designate this ORF orrA (for osmoticum response regulator A). The closest match from the sequence databases is a hypothetical ORF from the genome of Synechocystis sp. (27), and a known response regulator from Bacillus subtilis is also a close match (29) (Fig. 3).
FIG. 3.
Similarity of OrrA to the predicted products of translation of ORFs Ssp1 and Ssp2 of Synechocystis sp. strain PCC 6803 (Ssp1, GenBank accession no. D90914, PID:d1019125; data are from reference 27; Ssp2 [in lowercase] corresponds to the N-terminal third of a sequence from PCC 6803 that may regulate osmoprotection [23]), and of an ORF from B. subtilis (Bsu; GenBank accession no. Z99122, PID:e1184455 [data are from reference 29]) whose product regulates an extracellular proteinase. Identical (∗) and similar (+) residues are indicated, with those at the top of series of sequences referring to comparison (Gapped BLAST [2]) of the predicted sequence of OrrA with Ssp1 and Bsu, and those below comparing OrrA with Ssp2. The presumptive phosphorylation site is shown in boldface, and a potential helix-turn-helix motif in OrrA is underlined.
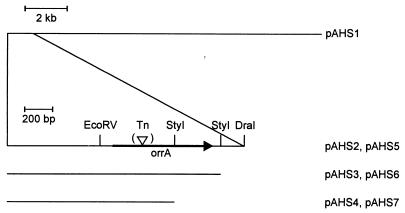
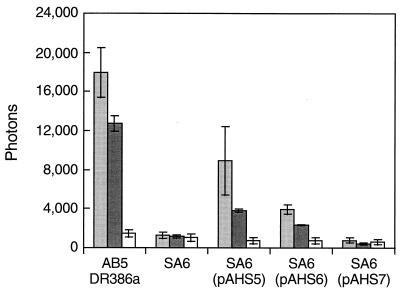
Plasmids pAHS5, pAHS6, and pAHS7 (Fig. 4) were transferred to SA6; exconjugants were resistant to spectinomycin, erythromycin, and neomycin. pAHS5 and pAHS6 both contain the entire ORF of the putative response regulator and complemented the SA6 mutation (Fig. 5); in contrast, pAHS7, which lacks the probable DNA-binding region of the product of that ORF, failed to complement the SA6 mutant (Fig. 5).
FIG. 4.
Map of the region of the genome of PCC 7120 in the vicinity of orrA and origin of the DNA fragments carried by the indicated plasmids. ▿, site of transposon Tn5-1058 in mutant SA6.
FIG. 5.
Complementation of SA6 (a mutant derivative of osmotic stress-responsive strain AB5DR386a) to osmoticum responsiveness by pAHS5 and pAHS6, but not by pAHS7, after 2 h of exposure to 0.1 M NaCl (light gray), 0.2 M sucrose (dark gray), or no supplemental osmoticum (open bar). Values of luminescence are presented as described in the legend to Fig. 2B.
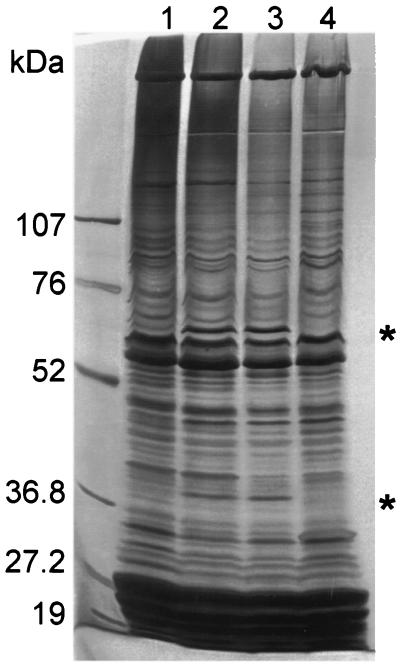
Protein extracts were prepared from an unstressed culture of wild-type PCC 7120 and from salt-induced cultures of PCC 7120, AB5, and SA6. Two proteins with molecular masses of approximately 40 and 67 kDa (asterisks in Fig. 6) were induced in PCC 7120 and in AB5, but not in SA6, in response to salt stress. Because these two bands correspond to proteins that are much larger than OrrA, they cannot represent OrrA or products of its degradation. No banding pattern that could correspond to what was expected for a product of the lti2-like gene was observed (perhaps a protein of the size of Lti2 from A. variabilis [63.8 kDa] [40]). That is, no band was produced much more abundantly in the presence than in the absence of salt, was replaced in AB5 by a truncated product and by proteins of the size of Vibrio fischeri LuxA and LuxB (i.e., 40.3 and 37.3 kDa), and was absent from SA6. Corresponding proteins were presumably too limited in abundance to permit visualization by silver staining. Nonetheless, it appears clear that two or more proteins, and presumably at least three proteins (the third being the product of the lti2-like gene), whose production is blocked in SA6 are produced by wild-type PCC 7120 in response to salt.
FIG. 6.
Proteins produced in response to salt induction in several strains. Lanes: 1, PCC 7120; 2, 3, and 4, salt-treated PCC 7120, AB5, and SA6, respectively. ∗, proteins of ca. 40 and 67 kDa induced in PCC 7120 in response to salinization and whose production in salt-treated strain AB5 is blocked by the orrA mutation in SA6.
DISCUSSION
We have described a system based on successive rounds of transposon mutagenesis for identification of regulatory genes that mediate responses of Anabaena sp. to environmental perturbations. As presented earlier (9, 45), one first makes use of luxAB, in transposon Tn5-1063, as a reporter gene to identify environmentally regulated genes. After exchange of most of Tn5-1063 for part of pRL386a, a second round of transposon mutagenesis then identifies genes that regulate the activity of the luxAB-tagged gene. We do not know of earlier marker exchange experiments having been performed with Anabaena sp. strain PCC 7120.
A screen for salt responsiveness identified a gene that showed extensive similarity to a previously identified gene designated lti2. lti2, whose function is unknown, encodes a protein with similarity to α-glucanotransferases and was induced by approximately 40-fold within 1 h after a downshift of A. variabilis from 38 to 22°C (40). A similar gene in PCC 7120 mutant AB5 was induced by elevated levels of salt and other osmotica, as well as by low temperature. The responsiveness of AB5 to high salt concentration was blocked in transposon mutant SA6. Reproduction of the phenotype of SA6 by double recombinants with pRL1380 indicated that the transposon is closely linked to the mutation that results in that phenotype. Complementation of the mutation by plasmids pAHS5 and pAHS6, but not by pAHS7, confirmed that interpretation and further implicated the gene orrA, which was truncated in pAHS7, in the regulation of the lti2-like gene.
Two-component regulatory systems are the predominant signal transduction mechanism by which prokaryotes sense and respond to their environment. Environmental responses in cyanobacteria, as in other prokaryotes, are evidently often mediated by two-component regulatory systems (see the introduction). On the basis of alignments of the sequences of many response regulators and solution of the crystal structure of CheY from E. coli, numerous residues that have extensive but various degrees of conservation have been identified (43). The amino terminus, which interacts with the corresponding sensory kinase, is highly conserved (21). The most highly conserved residues are aspartates at positions 12 and 13 (here perhaps replaced by glutamate-10 and aspartate-11 [Fig. 3]), which bind Mg2+; an aspartate at position 57 (here, 56), which is phosphorylated; a threonine or serine at position 87 (here, threonine-89), which donates a proton at the active site; and a lysine at position 109 (here, position 111), which is required for activation. Evidence of predicted α helices and β strands (43) is also observed (http://www.cmpharm.ucsf.edu/cgi-bin/nnpredict.pl), as is a potential γ-turn loop, which is a secondary structure near the phosphorylation region in many response regulators and which may interact with corresponding sensory kinases (43). Additional conserved residues found in the amino terminus may maintain the structural integrity of the protein. The carboxy terminus of the response regulators is normally much more variable.
Many of the deviations of the sequence of OrrA from the consensus sequence are conservative substitutions that are found in a number of putative response regulators from PCC 6803. Its predicted protein (Fig. 3) shares extensive sequence similarity with the deduced amino acid sequence of ORF sll0921 (GenBank accession no. D90914, PID:d1019125), whose function is unknown, from Synechocystis sp. (Ssp1 in Fig. 3). The BLAST score of 200 bits corresponds to an E value of 5e-51, i.e., is statistically highly significant. The similarity through the carboxy terminus suggests that this gene product may serve the same function in Synechocystis sp. as does OrrA in PCC 7120. In contrast, the BLAST score of 30.1 bits for the salt-regulatory protein encoded by the gene identified by Hagemann et al. (23) (Ssp2 in Fig. 3) corresponds to an E value of 11 (2), i.e., shows a degree of similarity that is not statistically significant.
A motif database (http://www.genome.ad.jp/SIT/MOTIF.html) indicates that a sequence near the carboxy terminus of OrrA contains a helix-turn-helix motif (Fig. 3) which is characteristic of the LuxR family of bacterial regulatory proteins. Because helix-turn-helix motifs are found in many DNA-binding proteins (5), OrrA, like other response regulators, may modulate genetic expression. Such an interpretation is consistent with the observation that the mutation in SA6 affects the production of several proteins that normally increase in amount in response to high salt concentration. That observation also suggests that the lti2-like gene acts in concert with other genes to respond to that stimulus. The second-site mutagenesis strategy described here should be useful in identifying additional components, such as a sensory kinase, of the osmoticum-induced signal transduction pathway.
ACKNOWLEDGMENTS
We thank Jeff Elhai for a program that facilitates the comparison of images of luminescence and bright-field images.
This work was supported by the U.S. Department of Energy under grant DOE-FG02-91ER20021 and by NSF grant MCB 9723193.
REFERENCES
- 1.Aiba H, Nagaya M, Mizuno T. Sensor and regulator proteins from the cyanobacterium Synechococcus species PCC7942 that belong to the bacterial signal-transduction protein families: implication in the adaptive response to phosphate limitation. Mol Microbiol. 1993;8:81–91. doi: 10.1111/j.1365-2958.1993.tb01205.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apte S K, Bhagwat A A. Salinity-stress-induced proteins in two nitrogen-fixing Anabaena strains differentially tolerant to salt. J Bacteriol. 1989;171:909–915. doi: 10.1128/jb.171.2.909-915.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apte S K, Haselkorn R. Cloning of salinity stress-induced genes from the salt-tolerant nitrogen-fixing cyanobacterium Anabaena torulosa. Plant Mol Biol. 1990;15:723–733. doi: 10.1007/BF00016122. [DOI] [PubMed] [Google Scholar]
- 5.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Gunsalas R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 6.Black T A, Wolk C P. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J Bacteriol. 1994;176:2282–2292. doi: 10.1128/jb.176.8.2282-2292.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black T A, Cai Y, Wolk C P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 8.Cai Y, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Y, Wolk C P. Anabaena sp. strain PCC 7120 responds to nitrogen deprivation with a cascade-like sequence of transcriptional activations. J Bacteriol. 1997;179:267–271. doi: 10.1128/jb.179.1.267-271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell E L, Hagen K D, Cohen M F, Summers M L, Meeks J C. The devR gene product is characteristic of receivers of two-component regulatory systems and is essential for heterocyst development in the filamentous cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol. 1996;178:2037–2043. doi: 10.1128/jb.178.7.2037-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang G G, Schaefer M R, Grossman A R. Complementation of a red-light-indifferent cyanobacterial mutant. Proc Natl Acad Sci USA. 1992;89:9415–9419. doi: 10.1073/pnas.89.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Close T J, Lammers P J. An osmotic stress protein of cyanobacteria is immunologically related to plant dehydrins. Plant Physiol. 1993;101:773–779. doi: 10.1104/pp.101.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curry J, Walker-Simmons M K. Sequence analysis of wheat cDNAs for abscisic acid-responsive genes expressed in dehydrated wheat seedlings and the cyanobacterium, Anabaena. In: Close T J, Bray E A, editors. Plant responses to cellular dehydration during environmental stress. Rockville, Md: American Society for Plant Physiology; 1993. pp. 128–136. [Google Scholar]
- 14.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 15.Elhai J, Wolk C P. A versatile class of positive selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 16.Elhai J, Wolk C P. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 1990;9:3379–3388. doi: 10.1002/j.1460-2075.1990.tb07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elhai, J., and C. P. Wolk. Unpublished data.
- 18.Elhai J, Vepritskiy A, Muro-Pastor A M, Flores E, Wolk C P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes T A, Iyer V, Apte S K. Differential responses of nitrogen-fixing cyanobacteria to salinity and osmotic stresses. Appl Environ Microbiol. 1993;59:899–904. doi: 10.1128/aem.59.3.899-904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross R, Aricó B, Rappuoli R. Families of bacterial signal transducing proteins. Mol Microbiol. 1989;3:1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 22.Hagemann M, Golldack D, Biggins J, Erdmann N. Salt-dependent protein phosphorylation in the cyanobacterium Synechocystis PCC 6803. FEMS Microbiol Lett. 1993;113:205–210. [Google Scholar]
- 23.Hagemann M, Richter S, Zuther E, Schoor A. Characterization of a glucosylglycerol-phosphate-accumulating, salt-sensitive mutant of the cyanobacterium Synechocystis sp. strain PCC 6803. Arch Microbiol. 1996;166:83–91. doi: 10.1007/s002030050360. [DOI] [PubMed] [Google Scholar]
- 24.Hagemann M, Richter S, Mikkat S. The ggtA gene encodes a subunit of the transport system for the osmoprotective compound glucosylglycerol in Synechocystis sp. strain PCC 6803. J Bacteriol. 1997;179:714–720. doi: 10.1128/jb.179.3.714-720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagemann M, Schoor A, Jeanjean R, Zuther E, Joset F. The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J Bacteriol. 1997;179:1727–1733. doi: 10.1128/jb.179.5.1727-1733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu N-T, Thiel T, Giddings T H, Wolk C P. New Anabaena and Nostoc cyanophages from sewage settling ponds. Virology. 1981;114:236–246. doi: 10.1016/0042-6822(81)90269-5. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 28.Kehoe D M, Grossman A R. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J Bacteriol. 1997;179:3914–3921. doi: 10.1128/jb.179.12.3914-3921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Liang J, Scappino L, Haselkorn R. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc Natl Acad Sci USA. 1992;89:5655–5659. doi: 10.1073/pnas.89.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkat S, Hagemann M, Schoor A. Active transport of glucosylglycerol is involved in salt adaptation of the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 1996;142:1725–1732. doi: 10.1099/13500872-142-7-1725. [DOI] [PubMed] [Google Scholar]
- 33.Nagaya M, Aiba H, Mizuno T. The sphR product, a two-component system response regulator protein, regulates phosphate assimilation in Synechococcus sp. strain PCC 7942 by binding to two sites upstream from the phoA promoter. J Bacteriol. 1994;176:2210–2215. doi: 10.1128/jb.176.8.2210-2215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 35.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805. doi: 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed R H, Richardson D L, Warr S R C, Stewart W D P. Carbohydrate accumulation and osmotic stress in cyanobacteria. J Gen Microbiol. 1984;130:1–4. [Google Scholar]
- 37.Reed R H. Osmotic adjustment: organic solutes. Methods Enzymol. 1988;167:528–534. [Google Scholar]
- 38.Sakamoto T, Bryant D A. Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. strain PCC 7002. Mol Microbiol. 1997;23:1281–1292. doi: 10.1046/j.1365-2958.1997.3071676.x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sato N. Cloning of a low-temperature induced gene lti2 from the cyanobacterium Anabaena variabilis M3 that is homologous to α-amylases. Plant Mol Biol. 1992;18:165–170. doi: 10.1007/BF00018474. [DOI] [PubMed] [Google Scholar]
- 41.Swanson R V, Alex L A, Simon M I. Histidine and aspartate phosphorylation: two-component systems and the limits of homology. Trends Biochem Sci. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 42.Thomas C M, Smith C A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 43.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 44.Watson G M F, Scanlan D J, Mann N H. Characterization of the genes encoding a phosphate-regulated two component sensory system in the marine cyanobacterium Synechococcus sp. WH7803. FEMS Microbiol Lett. 1996;142:105–109. doi: 10.1111/j.1574-6968.1996.tb08415.x. [DOI] [PubMed] [Google Scholar]
- 45.Wolk C P, Cai Y, Panoff J-M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Kong R, Wolk C P. Regulation of hepA of Anabaena sp. strain PCC 7120 by elements 5′ from the gene and by hepK. J Bacteriol. 1998;180:4233–4242. doi: 10.1128/jb.180.16.4233-4242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]