Abstract
A two-component signal transduction system encoded by the yycF and yycG genes is part of an operon containing three genes, yycH, yycI, and yycJ, with no known function and a gene, yycK, coding for an HtrA-like protease. This operon was transcribed during growth, and its transcription shut down as the cells approached stationary phase. This decreased transcription was not Spo0A dependent. The HtrA protease gene was separately controlled during sporulation from a ςG promoter. Studies using insertional inactivation plasmids revealed that neither yycF nor yycG could be inactivated, whereas the other genes were inactivated without loss of viability. A temperature-sensitive YycF response regulator mutant was isolated and shown to have an H215P mutation in a putative DNA-binding domain which is closely related to the OmpR family of response regulators. At the nonpermissive temperature, cultures of the mutant strain stopped growth within 30 min, and this was followed by a decrease in optical density. Microscopically, many of the cells appeared to retain their structure while being empty of their contents. The essential processes regulated by this two-component system remain unknown. A search of the genome databases revealed YycF, YycG, and YycJ homologues encoded by three linked genes in Streptococcus pyogenes. The high level of identity of these proteins (71% for YycF) suggests that this system may play a similar role in gram-positive pathogens.
In order for bacteria to effectively compete and survive, they have to sense environmental conditions and respond accordingly. Two-component systems are a predominant mode of environmental sensing and signal transduction in bacteria. This system of adaptive responses occurs through sensor kinases that mediate reversible phosphorylation events controlling downstream effectors (20, 27). Sensor kinases are generally integral membrane proteins that respond to specific environmental signals, and response regulators are often transcription factors whose affinity for DNA is modulated by phosphorylation. Sensor histidine kinases autophosphorylate on a conserved histidine residue and serve as phosphodonors to an invariant aspartic acid residue on a response regulator protein to which it is paired. The phosphorylated response regulator is thus able to mediate changes in gene expression, leading to the appropriate cellular response.
It is generally believed that two-component systems are used to sense environmental levels of essential substances, such as nitrogen, phosphate, and carbon sources, as well as a variety of nonessential molecules that signal adaptive responses. The discovery of two-component signal transduction involvement in the cell cycle and division of Caulobacter crescentus showed for the first time that this mechanism of genetic control had been adopted to regulate vital functions in the cell (7, 22). Because of the therapeutic implications of the discovery, it was important to determine if this adaptation was widespread in bacteria (5). The complete sequence of the Bacillus subtilis genome (10) provided a resource to inactivate all of the two-component systems of a single organism by insertional inactivation and determine the phenotype of such mutations. In a study of this kind, a two-component signal transduction system was found that could not be inactivated, and this communication describes the consequences to the cell of inactivation of this system.
MATERIALS AND METHODS
Bacterial strains and plasmids.
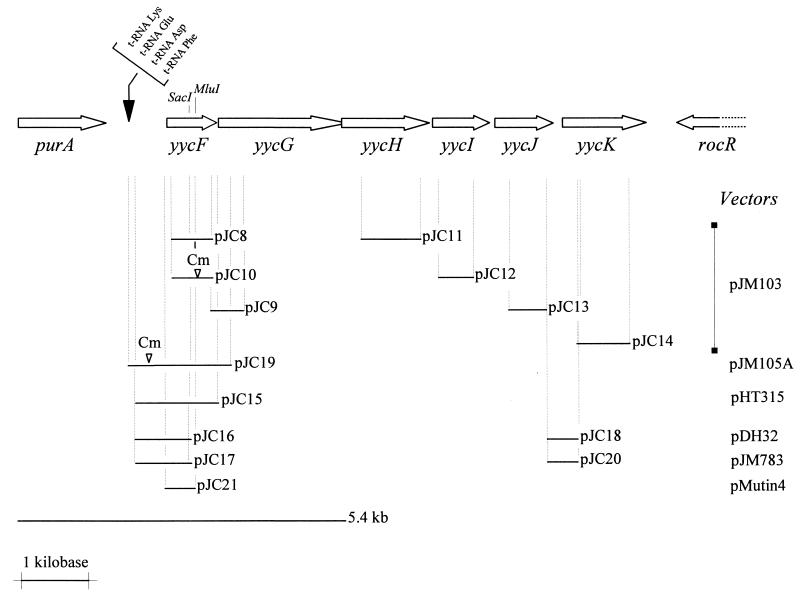
The bacterial strains and plasmids used in this study are listed in Table 1. Plasmids were constructed as follows. DNA fragments from the yycFIJK region were identified by the base pair numbering in the sequence submitted to GenBank under accession no. D78193 (18). For gene disruptions, the PCR was used to amplify regions encoding an internal part of each gene, with SphI and BamHI sites added at the ends. After digestion with appropriate enzymes, PCR fragments were ligated to SphI-BamHI-digested pJM103 integrative vectors (21). pJC8 contains a 594-bp internal DNA fragment of the yycF gene (bp 35285 to 35879). To obtain pJC10, the chloramphenicol resistance gene from pJC8 was first inactivated by removing the 400-bp MunI-NcoI 3′-end fragment; the Klenow-treated plasmid was self-ligated, and then the chloramphenicol resistance gene, without a terminator, on a 955-bp HincII-SmaI fragment from pJM105B (21) was inserted into the MluI-blunted site within the coding region of yycF. pJC9 contains the 3′ end of yycF and the 5′ end of yycG on a 475-bp DNA fragment (bp 34867 to 35342). pJC11 to pJC14 contain DNA fragments internal to yycH (bp 32233 to 33107), yycI (bp 31434 to 31924), yycJ (bp 30423 to 30994), and yycK (bp 29383 to 30167), respectively. For construction of multicopy plasmid pJC15, containing the yycF promoter region and the entire yycF gene, a 1,152-bp fragment (bp 1651 to 2803 in the nucleotide sequence with GenBank accession no. D26185 [18]) was generated by PCR with EcoRI and BamHI sites on each end and cloned into the EcoRI-BamHI sites of pHT315. The same PCR fragment, amplified from the chromosomal DNA of thermosensitive strain JH17041, was cloned in a similar way into pJM103. The lacZ transcriptional fusions were made from the EcoRI-SacI PCR fragment of pJC15 which contains the yycF promoter region and the 5′ terminus of yycF. The vectors used were pJM783 for integration at the original locus of the fused promoter and pDH32 for double-crossover integration at the amyE locus (21). To construct pJC16, the 750-bp EcoRI-SacI DNA fragment was blunted by using the Klenow fragment and T4DNA polymerase and then subcloned into the EcoRI site of Klenow-treated pDH32. For pJC17, the 750-bp DNA fragment, blunted only at the SacI site, was ligated to the EcoRI-SmaI-digested pJM783 vector. A 292-bp HaeIII fragment carrying the DNA region upstream of the yycK gene (bp 30125 to 30417 in D78193) was first obtained from a PCR-amplified fragment (bp 29383 to 30994), Klenow blunted, and cloned into the SmaI site of pJM103. The EcoRI-BamHI fragment from this plasmid was then subcloned into EcoRI-BamHI-digested pDH32 and pJM783 to produce the pJC18 and pJC20 vectors, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| B. subtilis straina or plasmid | Genotype or descriptionb | Sourcec |
|---|---|---|
| JH642 | pheA1 trpC2 | Laboratory stock |
| JH646 | pheA1 trpC2 spo0A12 | Laboratory stock |
| JH17022 | amyE::(yycF′-lacZ cat) | pJC16→JH642 |
| JH17023 | yycF::(yycF′-lacZ cat) | pJC17→JH642 |
| JH17024 | spo0A12 amyE::(yycF′-lacZ cat) | JH17022 DNA→JH646 |
| JH17025 | spo0A12 yycF::(yycF′-lacZ cat) | JH17023 DNA→JH646 |
| JH17026 | yycH::pJC11 (Cmr) | pJC11→JH642 |
| JH17027 | yycI::pJC12 (Cmr) | pJC12→JH642 |
| JH17028 | yycJ::pJC13 (Cmr) | pJC13→JH642 |
| JH17029 | yycK::pJC14 (Cmr) | pJC14→JH642 |
| JH17040 | yycF::(yycF′-lacZ Err)-PspacyycFGHIJK | pJC21→JH642 |
| Mu8u5u16 | ade-16 leu-8 met-5 | Laboratory stock |
| JH17041 | leu-8 met-5 yycF (H215P) (temperature-sensitive strain) | 5.4-kb PCR→Mu8u5u16 |
All JH strains were derived from JH642.
cat is the chloramphenicol acetyltransferase gene from pC194. Plasmid-encoded antibiotic resistance: Cmr, chloramphenicol resistance; Err, erythromycin resistance.
Arrows indicate construction by transformation.
To create a conditional mutation, a 474-bp HindIII-BamHI fragment containing the ribosome-binding site and the N-terminal part of yycF (bp 35526 to 36000) was amplified by PCR and ligated with the HindIII-BamHI-digested integration vector pMutin4 (30), generating plasmid pJC21. Upon transformation into B. subtilis JH642 and integration into the chromosome, pJC21 should disrupt the yycF gene and place a second copy of yycF, as well as yycGHIJK, under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Pspac. At the same time, a transcriptional fusion between the natural yycF promoter and the lacZ gene is created. The resulting strain was designated JH17040.
The 5.4-kb DNA fragment corresponding to the region from purA to yycG (bp 5011 in the sequence with GenBank accession no. D26185 to bp 33234 in the sequence with GenBank accession no. D78193) was amplified by using the Expand PCR kit (Boehringer Mannheim). Conditions recommended by the manufacturer were used, except for the PCR program, which was 10 s at 94°C, 20 cycles of 30 s at 55°C and 4 min at 68°C, and finally 10 min at 68°C.
To sequence the yycF gene from the thermosensitive strain, the primers previously used to construct pJC15 were utilized in PCRs carried out with purified chromosomal DNA. After EcoRI-BamHI digestion, the amplified DNA fragment (1,152 bp) was cloned into the pJM103 vector and sequenced. For the yycG gene, the upstream primer used in the case of pJC9 and the downstream primer from the 5.4-kb amplified DNA fragment were combined (see Fig. 1). The PCR product (2,139 bp) was digested by SphI, cloned into pJM103, and sequenced.
FIG. 1.
Genetic organization at the B. subtilis yycFGHIJK operon. purA codes for adenylosuccinate synthetase and rocR for a positive regulator controlling arginine utilization. The arrows identify the coding region of the genes. The restriction sites (SacI and MluI) used in some constructions are indicated. DNA inserts cloned into plasmids are represented by solid lines, with the corresponding names. The original vector for each construction is indicated at the extreme right. ▿, position of insertion of a terminatorless Cmr-encoding gene from pJM105B into the cloned DNA. The 5.4-kb fragment shows the extent of the PCRs used to generate the temperature-sensitive mutant.
Media, growth conditions, and genetic techniques.
The plasmids were transformed first into Escherichia coli DH5α and later into B. subtilis. Transformation of E. coli was performed by electroporation with a Bio-Rad apparatus in accordance with the manufacturer’s recommendations. Selection was done on Luria-Bertani (LB) broth supplemented, when appropriate, with 50-μg/ml ampicillin or 10-μg/ml chloramphenicol. Transformation of B. subtilis with chromosomal or plasmid DNA was carried out as previously described (2). Strains were maintained on Schaeffer’s sporulation medium (SM) (23) and selected with 5 μg of chloramphenicol per ml or (pMutin4) 1 μg of erythromycin per ml.
β-Galactosidase assay.
β-Galactosidase activity was determined as previously described (6) on cells grown in LB or SM. Specific activity was expressed in Miller units (16).
Sequence analyses.
DNA and protein sequences were analyzed and manipulated by using programs in the University of Wisconsin Genetics Computer Group software. The predicted product of each open reading frame was compared to protein sequence databases by using the BLAST network service (1). The hydropathy profiles were obtained with the TMpred program provided by the EMBL worldwide web servers (8).
RESULTS
In a survey of the 35 two-component signal transduction systems in B. subtilis (5a), the yycF and yycG genes encoding a two-component system with no known function could not be inactivated, suggesting that this system was essential for growth. Sequence analysis of the yycFG region revealed six open reading frames (18) (Fig. 1). A putative −10 promoter sequence (TATAAT) was identified in the DNA region upstream of the yycF open reading frame, but no sequence substantially similar to the consensus −35 sequence of ςA promoters was found in this region. Downstream from the yycF and yycG genes, four open reading frames (yycH to yycK) are closely spaced (<22 bp for the three first open reading frames and 69 bp between yycJ and yycK) with no obvious internal transcription terminators. Furthermore, the overlap between open reading frames yycG and yycH (Fig. 1) is consistent with a translational coupling strategy. These data suggested that the six open reading frames may comprise an operon.
Similarity among yycF, YycG, and two-component proteins.
The deduced sequences of the putative YycG and YycF proteins exhibit structural similarity to histidine kinases and response regulators, respectively, of two-component regulatory systems. The yycG gene encodes a protein of 611 amino acids with a calculated molecular mass of 70 kDa. The C-terminal portion of the protein contains the five blocks of conserved amino acids characteristic of the histidine protein kinase family (Fig. 2); i.e., a conserved histidine, the site of phosphorylation, and the N, G1, F, and G2 boxes, which presumably form a nucleotide-binding surface within the active site (20). The predicted sequence shows the highest similarity to the B. subtilis ResE protein (26) (30% identity) and slightly less similarity to the PhoR protein (25) (28 to 29% identity) (Fig. 2). The hydropathy profile of the N-terminal putative sensory domain suggests the presence of two stretches of hydrophobic amino acids sufficiently long to span the membrane (data not shown). Amino acids 35 to 182 could be oriented toward the environment in the periplasm, perhaps to sense a specific signal.
FIG. 2.
Amino acid sequence alignment of B. subtilis YycG with B. subtilis ResE (SwissProt accession no. P35164) and PhoR (SwissProt accession no. P23545). The predicted transmembrane helix domains are underlined. Conserved motifs present in the histidine kinase family, designated H, N, G1, F, and G2, are indicated by lines above the corresponding sequences.
The yycF gene encodes a putative protein of 235 amino acids with a calculated molecular mass of 27 kDa. The deduced amino acid sequence shows significant similarity to those of numerous response regulator proteins, such as B. subtilis PhoP (24) (54% identity) and the vancomycin response regulator of Enterococcus faecium (VanR) (3) (43% identity). The YycF response regulator is characterized by a conserved N-terminal domain of approximately 125 amino acids, containing the aspartates and lysine that form the active site.
Response regulators have been classified into subfamilies, according to sequence homologies in their DNA-binding domains (27, 28). On the basis of sequence similarity, the YycF protein should be assigned to the OmpR-PhoB subgroup, to which B. subtilis PhoP belongs (11). Proteins from this subclass are thought to bind to promoter sequences that are recognized by the major form of RNA polymerase holoenzyme, corresponding to B. subtilis EςA (27). It thus seems likely that YycF is a DNA-binding transcriptional regulator.
Sequence analysis of the predicted YycH, YycI, YycJ, and YycK products.
The predicted products of yycH, yycI, and yycJ do not show any significant matches when compared to the protein sequence in databases. The calculated molecular masses of these putative proteins are 52.5 kDa for YycH, 32.5 kDa for YycI, and 30 kDa for YycJ. Based on their hydrophobicity profiles (data not shown), YycH and YycI could be exported proteins or could be anchored to the membrane by a segment close to the N terminus. YycJ is predicted to be a cytoplasmic protein.
The putative YycK protein displays similarity to members of the HtrA serine protease family (34 to 35% identity with HtrA from Synechocystis sp. [9] and E. coli [12]). HtrA is involved in the proteolysis of abnormal proteins and is required for resistance to oxidative and heat stress in enteric bacteria (13, 19). YycK contains the motif Gly-Asp-Ser-Gly-Gly-Ala-Lys, which is very similar to the consensus sequence surrounding the active serine residues of the catalytic domains of known serine proteases. The sequence analysis of YycK suggests a 42.8-kDa membrane-anchored protein located outside the cell.
yycF and yycG are essential genes.
In an effort to determine the functions of YycF and YycG, we used an integrative plasmid to interrupt their genes and analyze the mutant phenotypes. An internal region of each gene was cloned into the pJM103 integrative vector, and the plasmids obtained (pJC8 and pJC9) were used to transform B. subtilis JH642. Numerous attempts to integrate these circular plasmids into these genes by a single-crossover event did not yield transformants, suggesting that null mutations in yycF or yycG may not be constructed.
The same strategy was used for the other open reading frames, yycH, yycI, yycJ, and yycK, to determine if mutation of one of them was lethal. By using plasmids pJC11, -12, -13, and -14, transformants were obtained in each case (strains JH17026 to JH17029, respectively), indicating that the deleterious effect observed was due to yycF and yycG inactivation and not to polar effects on the downstream genes.
In addition, attempts to interrupt yycF with a chloramphenicol resistance cassette, designed to produce nonpolar mutations (21), were also unsuccessful. Transformation of B. subtilis with linearized pJC10 plasmid DNA (derived from pJC8) should have produced recombinants carrying a terminatorless chloramphenicol resistance gene integrated within the yycF gene by double crossing over, allowing transcription of downstream genes in the putative operon. Thus, both yycF and yycG appear to encode products essential for growth.
Transcriptional activity of the yycFG regulatory region.
To study expression from the yycFG promoter, the 400 nucleotides preceding the start codon of yycF were cloned in front of the promoterless lacZ gene in pDH32. The resulting plasmid (pJC16) was linearized and integrated at the nonessential amyE locus of B. subtilis JH642, giving strain JH17022. To eliminate any effects due to the chromosomal location, the same transcriptional fusion was also introduced directly into the original yycF locus. To construct this strain, B. subtilis JH642 was transformed with circular plasmid pJC17, derived from pJM783, giving strain JH17023.
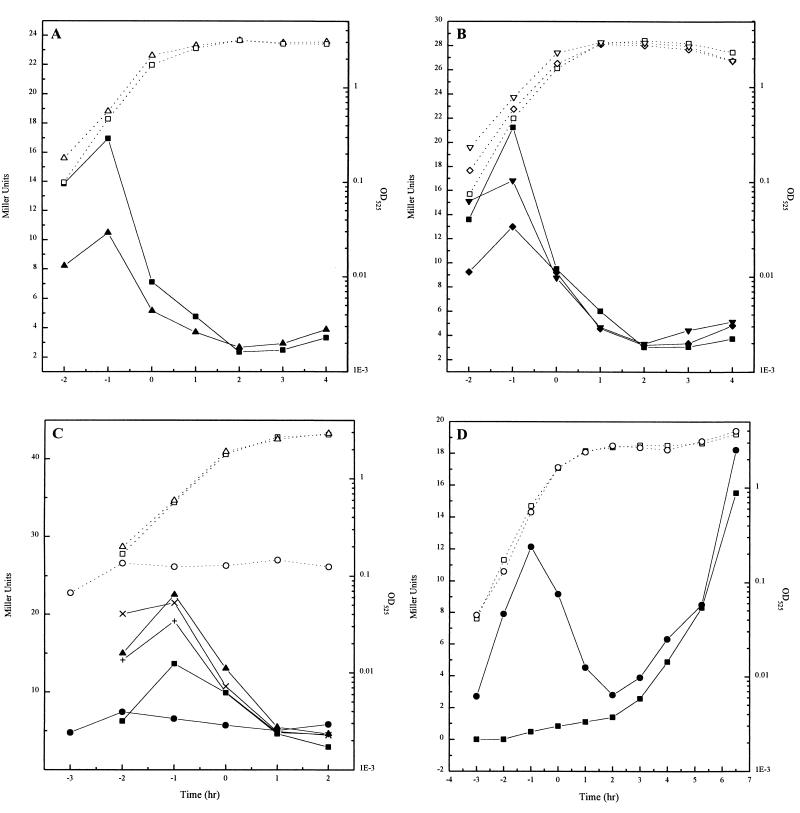
The strains were grown in SM or LB broth and assayed for β-galactosidase activity. The results obtained were generally identical for both constructions, indicating a promoter that functions primarily during vegetative growth. As shown in Fig. 3A, the yycF promoter in cells grown in SM was active during exponential growth, reaching a peak of 17 U 1 h before the onset of sporulation, whereupon it dramatically decreased toward zero as the cells entered stationary phase. β-Galactosidase production in cells grown in LB broth (where the culture sporulates poorly) was similar throughout exponential growth, but relative to growth in SM, the rate of decrease was delayed and levels remained higher in the later stages. The activity was maximum at the onset of stationary phase and slowly decreased during the following 2 h (data not shown). These results are consistent with expression from the promoter upstream of yycF being turned off by some regulatory protein related to the end of growth or the onset of sporulation.
FIG. 3.
Expression of the yycFG and yycK promoters. (A) Growth in SM (open symbols) of JH17022 (▵) or JH17023 (□) and β-galactosidase specific activity of the yycF-lacZ promoter fusions (solid symbols). (B) Growth in SM (open symbols) of JH17023 (□), JH17024 (spo0A strain with the yycF-lacZ fusion integrated into the amyE locus) (◊), JH17025 (spo0A mutant with the fusion in the chromosomal yycF locus) (▿), and β-galactosidase specific activity of the promoter fusion (solid symbols). (C) Effect of variations in the level of the YycF and YycG proteins on the growth in SM (open symbols) and expression of the operon (solid symbols) of JH17040 without IPTG (○), with 50 μM IPTG (▵), with 0.5 mM IPTG (X), and with 1 mM IPTG (+). Growth of JH17023 (□) is plotted as a control. (D) Growth in SM (open symbols) and β-galactosidase specific activity (solid symbols) of JH642 with the yycK-lacZ fusion integrated into the amyE locus (pJC18) (□) or into the yycK locus (pJC20) (○).
To investigate the possibility that Spo0A, the major transcription factor required for sporulation, might repress the expression of the yycF and yycG genes upon entry into stationary phase, the transcriptional fusions were introduced into a spo0A mutant strain (JH646). For construction of strains JH17024 (fusion integrated into the amyE locus) and JH17025 (fusion integrated into the yycF locus), JH646 (spo0A12) was transformed with chromosomal DNAs from JH17022 and JH17023, respectively. The β-galactosidase activities, assayed on cells grown in SM, were similar to those in the wild-type strains (Fig. 3B). Therefore, Spo0A does not appear to play a role in the regulation of the expression of the yycF and yycG genes.
In order to determine if the yycF and yycG gene products autoregulate their own synthesis, β-galactosidase activity was assayed in a strain in which the level of YycF and YycG was controlled by the Pspac promoter of the pMutin 4 vector, pJC21, and the yycF promoter was coupled to lacZ to determine its level of expression. In this strain, JH17040 containing integrated pJC21, the level of proteins YycF and YycG was regulated by the amount of IPTG added. As seen in Fig. 3C, modulation of yycFG expression by varying the concentration of IPTG had no obvious effect on the transcription of yycF and lacZ. β-Galactosidase activities were similar, regardless of the IPTG concentration used. Moreover, the strain required IPTG for growth. These data suggest that this two-component system does not regulate its own expression.
The noncoding DNA region (69 bp) upstream of the yycK gene was checked for the presence of a potential promoter. The pJC18 vector was integrated into the amyE locus of JH642, and β-galactosidase activity was assayed on cells grown in SM or LB broth. No activity was detected when cells were grown in LB broth (data not shown), but in SM, β-galactosidase expression started between 3 and 4 h (Fig. 3D). A putative ςG-dependent promoter was identified in this DNA fragment with the −10 sequence CATATTA [consensus, CAT(AT)(AC)TA] (17) and the −35 sequence GAGTT [consensus, G(AC)AT(AG)] (17). The possibility that yycK was transcribed from the yycFG promoter was analyzed by using the pJC20 vector to transform JH642 and to obtain a transcriptional fusion with lacZ in the original yycK locus. β-Galactosidase production measured in cells grown in SM showed expression during exponential phase, followed by a decay as the cells neared stationary phase and renewed expression during sporulation (Fig. 3D). Thus, the yycK gene belongs to the yycFGHIJ operon and is under the control of two promoters: one for expression during exponential growth and one for expression starting at stage III of sporulation, probably in the forespore.
Isolation of a thermosensitive mutation in the yycFG operon.
Upstream of the yycFG operon, four genes are located for tRNA-Phe, tRNA-Asp, tRNA-Glu, and tRNA-Lys, preceded by the purA gene encoding the adenylosuccinate synthetase involved in adenine biosynthesis (Fig. 1). The region encompassing all of these genes plus the yycF and yycG genes (5.4 kb) was amplified by PCR, and the resulting DNA fragment was used to transform strain Mu8u5u16 to Ade+. About 3,200 transformants were transferred to replicate plates and incubated at 30 or 47°C. One strain did not grow at 47°C on either minimal medium or SM plates.
In order to locate the mutation giving the phenotype, chromosomal DNA was prepared from the thermosensitive mutant strain (JH17041) and used as template DNA in PCRs to amplify the entire yycF gene (the fragment in pJC15 [Fig. 1]) and the entire yycG gene (see Materials and Methods). The amplified DNA fragments were then cloned and sequenced. Only one alteration was found, and that was localized in the yycF gene. Mutation of A to C at position 35303 (sequence with GenBank accession no. D78193) changed histidine 215 to a proline.
To determine if this mutation was responsible for the thermosensitive phenotype, the PCR-amplified fragment containing the yycF mutant gene was cloned into the integrative pJM103 vector and used to transform JH642. The fragment was identical to that cloned into pJC15 (Fig. 1). Among the transformants, 35% displayed a thermosensitive phenotype. The same experiment, when done with the identical PCR-amplified fragment from the parental strain, did not result in thermosensitivity.
A second experiment was done to confirm that the yycF mutation was responsible for the thermosensitive phenotype. The fragment in plasmid pJC19 (Fig. 1) was obtained in two pieces by two PCRs from the DNA of thermosensitive strain JH17041, and they were ligated to the chloramphenicol resistance gene and cloned into pJM105A. This placed the Cmr cassette upstream of the promoter and allowed the insertion of the entire fragment into the chromosome as a double-crossover event. Sequence analyses confirmed the presence of only a single mutation in yycF, as previously described. Transformation of this plasmid in JH642 gave 30% thermosensitive transformants.
Growth and phenotypic characteristics of the thermosensitive strain.
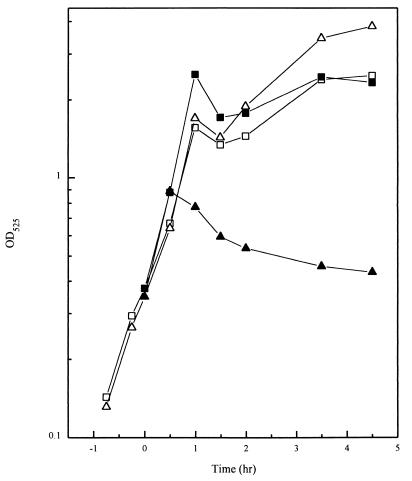
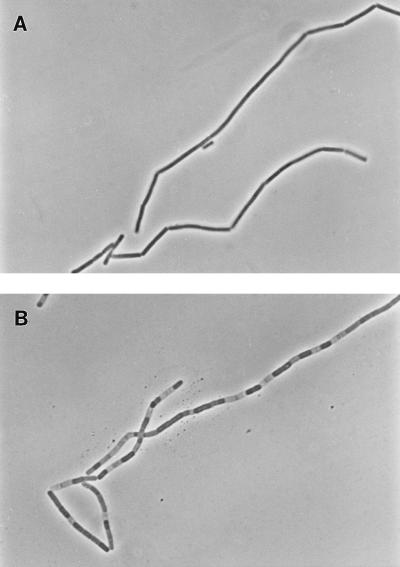
The thermosensitive and parental strains were grown in LB broth at 30°C until an optical density at 525 nm (OD525) of 0.4 was reached; when half of each culture was transferred to 47°C. At 30°C, both strains grew similarly (Fig. 4). Upon a shift to 47°C, the thermosensitive strain stopped growing after approximately 30 min and the OD525 decreased. When cultures were observed with a microscope, the thermosensitive strain at the permissive temperature produced chains of cells characteristic of early exponential-phase cultures that persisted longer than parental strains under the same conditions (Fig. 5A). Within an hour from when the cultures were shifted to the nonpermissive temperature, the chains of the thermosensitive mutant cells were interspersed with empty sections, as if some of the cells had lost their cytoplasmic contents and only the cell wall remained (Fig. 5B). The parental strain showed none of this behavior and had separated into single or double cells after 1 h at 47°C (data not shown).
FIG. 4.
Growth characteristics of the temperature-sensitive strain. Growth curves of the Mu8u5u16 (□) and JH17041 (temperature-sensitive) (▵) strains in LB broth at 30°C (open symbols) or at 47°C (solid symbols). Time zero is when the culture was shifted to 47°C from 30°C.
FIG. 5.
Cellular effect of the thermosensitive mutation. Strain JH17041 was grown in LB broth at 30°C to an OD525 of 0.35, when half the culture was shifted at 47°C. Phase-contrast images of cells taken 1.5 h after the shift are shown. A, 30°C (OD525 = 1.7); B, 47°C (OD525 = 0.6).
DISCUSSION
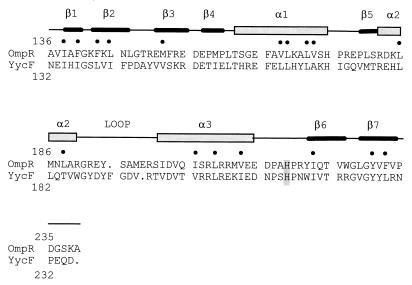
On the basis of the sequence similarity of its C-terminal domain, the YycF response regulator has been assigned to the OmpR subfamily. The crystal structure of the E. coli OmpR C-terminal domain revealed that members of the subfamily belong to the winged helix-turn-helix family of DNA-binding proteins (Fig. 6) (15). Of the 17 residues known to make up the hydrophobic core of OmpR, all are either identical or conserved hydrophobic in YycF. Analogy with other winged helix-turn-helix proteins and correlation with mutagenesis data have allowed the identification of important structural features: a recognition helix, a wing involved in DNA binding, and an extensive loop preceding the recognition helix involved in interaction with the α subunit of RNA polymerase (15). The residues thought to be important for DNA binding in OmpR, such as Arg182, Val203, and Thr224, are identical in YycF, and Ser200 is replaced with a similar amino acid, threonine. The thermosensitive mutation of YycF was localized to the loop connecting the α3 helix to the β6 and β7 C-terminal strands. Substitution of a proline for a histidine at this site may perturb the DNA-binding properties of this region by destabilizing the interaction of β6 and β7 with the rest of the molecule at elevated temperature.
FIG. 6.
Sequence alignment of the DNA-binding domains of B. subtilis YycF and E. coli OmpR (SwissProt accession no. P03025). A schematic diagram of the secondary structure of OmpR is shown (15). The dots mark residues corresponding to the OmpR hydrophobic core. The position of thermosensitive mutation is shaded.
The YycG kinase is related to the ResE and PhoR kinases of B. subtilis. All of these kinases appear to have two membrane-spanning regions which form a putative periplasmic loop of about 140 amino acids. The active-site histidine of the cytoplasmic catalytic domain is preceded by about 160 amino acids forming a domain with no known function in all cases. The highest homology among these proteins is found in the catalytic domain, with less in the cytoplasmic domain. Very little amino acid sequence conservation was found among all three kinases in the putative periplasmic domain, but YycF and ResE have 34% identical and similar residues in this region. If all of these proteins evolved from an ancestral protein, there appears to be more pressure to conserve the histidine- and ATP-binding regions.
The YycF response regulator is likely to regulate several genes which remain unidentified. Clearly, loss of this activity in the thermosensitive mutant results in rapid cessation of growth. The cellular effect of this loss as revealed by microscopy was the generation of empty cells which maintained their structural rigidity. This was especially apparent in septated chains of cells characteristic of low-density cultures when transferred to the nonpermissive temperature. The molecular basis of this phenomenon is unknown. YycF may control some genes whose products are essential for normal cell growth and are likely to be integrated with other cell cycle-dependent events, since transcription of the operon containing the yycF and yycG genes occurs only during growth and is shut off as the cells approach stationary phase. This shutoff is not mediated by the sporulation regulator Spo0A but may be due to loss of an inducing signal from, or dependent on, cell growth. Clearly, there are many unknown features of this system, including what signals regulate the YycG kinase.
Essential two-component signal transduction systems have been found in C. crescentus. In C. crescentus, a system of histidine kinases and response regulators controls cell division and motility (7, 22). The growth signals that this system interprets to affect cell division remain obscure. Similarly, YycF and YycG are likely to respond to growth signals in B. subtilis. In fact, very little is known about how cells coordinate processes such as cell wall and membrane growth with DNA replication to precisely control cell division. Certainly, bacteria do this well and very quickly. It seems possible that YycF regulates one aspect of these processes and that YycG responds to vital growth signals involved in the coordination.
A yycFG-like operon was found in the Streptococcus pyogenes genome (Fig. 7). This gram-positive bacterium is a member of the group A streptococci and is known to be responsible for a wide variety of human diseases (29). Homologies were found with the response regulator YycF (71% identity) and the histidine kinase YycG (44% identity), as well as with the predicted YycJ protein (55% identity) (Fig. 7). No open reading frames corresponding to yycH, yycI, or yycK were identified.
FIG. 7.
Organization of the yycFG-like genes in S. pyogenes M1 GAS (sequenced at Oklahoma University [http://www.genome.ou.edu/strep.html]) and amino acid sequence alignment of the predicted products (YycFGJSt) with the homologs in B. subtilis (YycFGJBs). Conserved motifs in the histidine kinase proteins are indicated by lines above the corresponding sequences.
This very high identity between the B. subtilis and S. pyogenes YycF proteins suggests that they carry out the same function in the two organisms. If the consequences of loss of YycF activity are the same in both organisms, inhibition of YycF in S. pyogenes should effectively curtail growth and result in rapid cell death. This system may be a target for a series of two-component signal transduction inhibitors with bactericidal properties that have recently been described. These inhibitors were especially effective on gram-positive pathogens, such as methicillin-resistant Staphylococcus aureus. Since the genome sequence of S. aureus is proprietary, it cannot be determined by us whether a YycF equivalent exists in this organism, but the S. pyogenes results suggest that such a system is common to gram-positive microorganisms (4). Resistance to these inhibitors was multifactorial, indicating that more than one target was being affected by the inhibitors (4). This may indicate that more than one kinase is essential for growth or that general kinase inhibition is a lethal event. The ResE-ResD signal transduction system to which YycG and YycF are closely related is essential for growth in the absence of glucose. How many other systems of their type, when mutated singly or in combination, would have this phenotype has yet to be determined. The presence of similar two-component systems in gram-positive pathogens suggests that kinase or response regulator inhibitors would be effective bactericidal anti-infective agents (4, 5, 14).
ACKNOWLEDGMENTS
This research was supported in part by grant GM19416 from the National Institute of General Medical Sciences, National Institutes of Health, USPHS. We thank B. A. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, M. McShan, and J. Ferretti for the S. pyogenes data made available through the Streptococcal Genome Sequencing Project funded by AI38406 from the USPHS.
Footnotes
Publication 11729-MEM from the Department of Molecular and Experimental Medicine at The Scripps Research Institute.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett J F, Goldschmidt R M, Lawrence L E, Foleno B, Chen R, Demers J P, Johnson S, Kanojia R, Fernandez J, Bernstein J, Licata L, Donetz A, Huang S, Hlasta D J, Macielag M J, Ohemeng K, Frechette R, Frosco M B, Klaubert D H, Whiteley J M, Wang L, Hoch J A. Antibacterial agents that inhibit two-component signal transduction systems. Proc Natl Acad Sci USA. 1998;95:5317–5322. doi: 10.1073/pnas.95.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett J F, Hoch J A. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob Agents Chemother. 1998;42:1529–1536. doi: 10.1128/aac.42.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Fabret, C., and J. A. Hoch. Unpublished data.
- 6.Ferrari E, Henner D J, Perego M, Hoch J A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988;170:289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecht G B, Lane T, Ohta N, Sommer J, Newton A. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166–169. [Google Scholar]
- 9.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 10.Kunst F, et al. The complete genome sequence of the gram-positive model organism Bacillus subtilis (strain 168) Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 11.Lee J W, Hulett F M. Nucleotide sequence of the phoP gene encoding PhoP, the response regulator of the phosphate regulon of Bacillus subtilis. Nucleic Acids Res. 1992;20:5848. doi: 10.1093/nar/20.21.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macielag M, Demers J P, Fraga-Spano S A, Hlasta D J, Johnson S G, Kanojia R M, Russell R, Sui Z, Weidner-Wells M A, Werblood H, Foleno B D, Goldschmidt R M, Loeloff M, Webb G C, Barrett J F. Substituted salicylanilides as inhibitors of two-component regulatory systems in bacteria. J Med Chem. 1998;41:2939–2945. doi: 10.1021/jm9803572. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 17.Nicholson W L, Sun D, Setlow B, Setlow P. Promoter specificity of ςG-containing RNA polymerase from sporulating cells of Bacillus subtilis: identification of a group of forespore-specific promoters. J Bacteriol. 1989;171:2708–2718. doi: 10.1128/jb.171.5.2708-2718.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Pallen M J, Wren B. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 21.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 22.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 23.Schaeffer P, Millet J, Aubert J P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:701–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seki T, Yoshikawa H, Takahashi H, Saito H. Cloning and nucleotide sequence of phoP, the regulatory gene for alkaline phosphatase and phosphodiesterase in Bacillus subtilis. J Bacteriol. 1987;169:2913–2916. doi: 10.1128/jb.169.7.2913-2916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seki T, Yoshikawa H, Takahashi H, Saito H. Nucleotide sequence of the Bacillus subtilis phoR gene. J Bacteriol. 1988;170:5935–5938. doi: 10.1128/jb.170.12.5935-5938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorokin A, Zumstein E, Azevedo V, Ehrlich S D, Serror P. The organization of the Bacillus subtilis 168 chromosome region between the spoVA and serA genetic loci, based on sequence data. Mol Microbiol. 1993;10:385–395. doi: 10.1111/j.1365-2958.1993.tb02670.x. [DOI] [PubMed] [Google Scholar]
- 27.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive response in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 29.Suvorov A N, Ferretti J J. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J Bacteriol. 1996;178:5546–5549. doi: 10.1128/jb.178.18.5546-5549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vagner, V., and S. D. Ehrlich. Unpublished data.