Abstract
NPM1-mutated acute myeloid leukemia (AML) represents the largest molecular subgroup of adult AML. NPM1-mutated AML is recognizable by molecular techniques and immunohistochemistry, which, when combined, can solve difficult diagnostic problems (including identification of myeloid sarcoma and NPM1 mutations outside exon 12). According to updated 2022 European LeukemiaNet (ELN) guidelines, determining the mutational status of NPM1 (and FLT3) is a mandatory step for the genetic-based risk stratification of AML. Monitoring of measurable residual disease (MRD) by qRT-PCR, combined with ELN risk stratification, can guide therapeutic decisions at the post-remission stage. Here, we review the criteria for appropriate diagnosis and molecular monitoring of NPM1-mutated AML.
Significance:
NPM1-mutated AML represents a distinct entity in the 2022 International Consensus Classification and 5th edition of World Health Organization classifications of myeloid neoplasms. The correct diagnosis of NPM1-mutated AML and its distinction from other AML entities is extremely important because it has clinical implications for the management of AML patients, such as genetic-based risk stratification according to 2022 ELN. Monitoring of MRD by qRT-PCR, combined with ELN risk stratification, can guide therapeutic decisions at the post-remission stage, e.g., whether or not to perform allogeneic hematopoietic stem cell transplantation.
INTRODUCTION
NPM1-mutated acute myeloid leukemia (AML) represents the largest molecular subgroup of AML in adults, accounting for 30% to 35% of cases (1). NPM1 mutations are driver genetic events that are AML-specific (2) and promote leukemia acting in concert with mutations of other genes, usually associated with clonal hematopoiesis, such as DNMT3A, IDH1/2, and TET2 (2). NPM1 mutations are characterized by the aberrant cytoplasmic localization of the NPM1 mutant (NPM1c+; refs. 1, 3) and a unique gene-expression profile (2). Because of its unique features (Table 1), NPM1-mutated AML is recognized as a distinct leukemia entity in both the 2022 International Consensus Classification (ICC; ref. 4) and World Health Organization (WHO) 5th edition (5) classifications of myeloid neoplasms.
Table 1.
Features of NPM1-mutated AML.
| Definition and epidemiology |
|
| Pathology and immunophenotype |
|
| Gene-expression profile |
|
| Response to therapy and prognosis |
|
Abbreviations: FAB, French–American–British; GEP, gene-expression profile; WBC, white blood cell count.
The aberrant accumulation of NPM1c in the cytoplasm of AML cells (1, 3) plays a key role in leukemogenesis (2). This is supported by the observation that all NPM1 mutants, regardless of the affected exons (3, 6), lead to the cytoplasmic localization of nucleophosmin. In addition, all NPM1 mutations are “born to be exported” since the nuclear export activity of NPM1c is strictly regulated by the strength of the C-terminal nuclear export signal (NES; ref. 3). Moreover, NPM1c is critical for HOX gene expression and leukemic state maintenance (7). Additionally, a gain-of-function in the cytoplasm, leading to the inhibition of caspase-6 and -8 with deregulation of cell death and myeloid differentiation, has been reported (8), and NPM1c was found to hamper the formation of promyelocytic leukemia nuclear bodies, which are regulators of mitochondrial fitness (9). However, the function of NPM1c in the cytoplasm still remains elusive.
NPM1c is recruited to chromatin through Exportin-1 (XPO1) (10) and controls HOX/MEIS expression (11, 12). HOX/MEIS overexpression can be blocked using either menin or XPO1 inhibitors. Menin inhibitors disrupt the menin–MLL1 network (11) and show strong antileukemic activity in KMT2A-rearranged and NPM1-mutated AML, both in vitro and in vivo (13). The menin inhibitor revumenib led to 21% complete remissions in NPM1-mutated AML patients, often with measurable residual disease (MRD) negativity, downregulation of MEIS and HOXA9 genes and increased expression of CD14 (14). XPO1 inhibitors cause the release of NPM1c from its targets with a consequent decrease in the expression of HOX/MEIS. NPM1c is particularly enriched at active chromatin sites, where MLL1 and RNA Polymerase II (Pol II) are also located (11). Specific degradation of NPM1c leads to reduced HOX/MEIS transcription within 15 minutes, due to the loss of Pol II from these loci (11, 12). Notably, the NPM1 acidic domain (∼aa 120–150) also plays an important role in recruiting NPM1c to chromatin (11).
Based on the above findings, we have hypothesized that NPM1c can promote leukemogenesis by acting at both the nuclear and cytoplasmic level, i.e., “killing two birds with one stone” (15). Haploinsufficiency for wild-type NPM1 at the nucleolar level (because of heterozygosity of NPM1 mutation and aberrant localization of the NPM1 native protein in the cytoplasm through the formation of heterodimers with the NPM1 mutant) may also play a role in the mechanism of leukemogenesis.
Frameshift indel mutations (such as NPM1 mutation A) are also responsible for the unique immunologic features of NPM1-mutated AML cells. In fact, these somatic mutations can generate tumor-specific neoepitopes, which, after proteasomal degradation, processing in the endoplasmic reticulum, and loading as neoantigens onto the cell's major histocompatibility complex, are recognized by a patient's autologous CD4+ and CD8+ T cells, giving rise to an immune response. The ectopic cytoplasmic location of mutated NPM1 protein may enhance its processing by the human leukocyte antigen (HLA) class 1 pathway, leading to efficient antigen presentation. We proposed for the first time that C-terminus peptides of the NPM1 mutants can bind HLA class I molecules (16). Other investigators have subsequently confirmed this original report (17).
A number of observations suggest that neoantigens generated from mutated NPM1 protein are potential targets for immunotherapy. First, NPM1 mutations are very frequent and specific for AML, are driver genetic events that are not associated with clonal hematopoiesis, and are stable at relapse (2). Second, NPM1 neoepitopes are not subject to central immune tolerance and are not expressed in normal tissues. Third, despite the large number of mutation sequences reported, two of these (types A and B) account for >80% of patients. Fourth, the C-terminal sequence of type A mutated NPM1 protein generates a strong immune response, including specific antibodies, in animal models. Finally, T cells, which can generate a response to peptides from mutated NPM1, can be observed in patients treated for NPM1-mutated AML (18, 19), including those in molecular complete remission (20), and it has been speculated that this might contribute to the favorable outcomes of this AML subtype (21). Therefore, cellular therapies (22, 23) and cancer vaccines that target mutated NPM1 epitopes are of great interest.
The distinction of NPM1-mutated AML from other AML genotypes can sometimes be difficult (24), and the percentage of blasts required for its diagnosis remains controversial (25). Immunohistochemistry techniques for detecting cytoplasmic NPM1 (26) can provide complementary information to molecular techniques. Determining the mutation status of NPM1 is essential for risk stratification in the European LeukaemiaNet (ELN) guidelines (27); moreover, this identifies patients for monitoring of measurable residual disease (MRD) by qRT-PCR (28, 29), which can further refine risk stratification and guide therapeutic decisions after remission. Here, we review the criteria for the appropriate diagnosis, risk stratification, and molecular monitoring of NPM1-mutated AML.
DETECTION TECHNIQUES OF NPM1-MUTATED AML
NPM1 mutations can be identified by molecular assays or by surrogate techniques, including IHC. These methods are complementary and allow a flexible approach to the diagnosis of NPM1-mutated AML which is critical for implementing the use of the ICC and WHO classifications worldwide.
Qualitative Detection of NPM1 Mutations by Molecular Techniques
NPM1 mutations occur in about one-third of adult AML patients (1). Conversely, they are uncommon in childhood (about 8% of cases; ref. 30), where they are usually non-type A (31). NPM1 mutation A (a duplication of TCTG at position 860–863 of the reference sequence) occurs in about 75%–80% of adult cases (1). Mutations B and D account for approximately 10% and 5% of cases, while other mutations are rare. More than 100 different types of NPM1 mutations are now recognized. One distinguishing feature of NPM1 mutations is that they do not drive clonal hematopoiesis. Thus, their presence at remission indicates active disease that can cause relapse and is associated with inferior outcomes. In this respect, NPM1 mutations clearly differ from those involving the DNMT3A, TET2, and ASXL1 genes that are associated with clonal hematopoiesis. These mutations can persist at remission, but they do not have prognostic value, simply reflecting the reestablishment of a preleukemic state following therapy for AML that does not require further treatment.
Qualitative assays for NPM1 mutations are most commonly based on genomic DNA as a substrate and use PCR followed by fragment length analysis to detect the insertion, although assays based on melting curve analysis and qRT-PCR are also available (32–35). In general, PCR fragment analysis methods are preferred because they can detect all insertions within the PCR amplicon regardless of the mutation sequence, and they are simple and rapid, affording a sensitivity of ∼5%, which is adequate in almost all cases (32). The exception to this is myeloid sarcoma, where the establishment of submicroscopic bone marrow (BM) involvement requires a more sensitive technique such as qRT-PCR (32).
Qualitative assays are best applied to fresh BM or peripheral blood (PB) leukemic cells (34) but plasma (36) is also suitable. Molecular detection of NPM1 mutations in paraffin-embedded trephines is unreliable due to the denaturing effect of decalcifying agents on nucleic acids. However, DNA extracted from paraffin-embedded tissue biopsies in cases of myeloid sarcoma is usually adequate for PCR fragment analysis (33).
Following the identification of an NPM1 mutation, for patients where MRD monitoring is planned, it is essential to establish the mutation sequence and to store both DNA and RNA to allow the determination of baseline transcript levels to permit comparison with post-remission samples (discussed further below; ref. 37). Next-generation sequencing (NGS) is increasingly used in the diagnosis of AML (27) and has the advantages of providing the insertion sequence, the ability to detect mutations outside exon 12 if an appropriate panel is used, and to identify comutations that may also have a prognostic impact. A recent study investigated the interlaboratory concordance in identifying driver mutations in AML (DNMT3A, FLT3, IDH1, IDH2, NPM1, TET2, TP53, and WT1) using different NGS platforms and found concordance >95%, with perfect agreement for NPM1 mutations (38). Given the increasing need to rapidly molecularly stratify patients prior to treatment initiation, it is likely that rapid PCR-based methods for NPM1 and other mutations that could influence first-line treatment choice will play a role in the foreseeable future.
IHC Detection of Cytoplasmic NPM1
The aberrant export of NPM1 (1) to the cytoplasm of leukemic cells (also referred to as NPM1c+) is the result of the mutation-induced changes at the C-terminus of NPM1, i.e., loss of one or two of the tryptophans at positions 288 and 290 and addition of a de novo NES motif (3, 39) that enhances the interaction with the nuclear exporter XPO1. New C-terminal NES motifs of different strength are inserted to further tune the nuclear-cytoplasmic shuttling of the mutant (3).
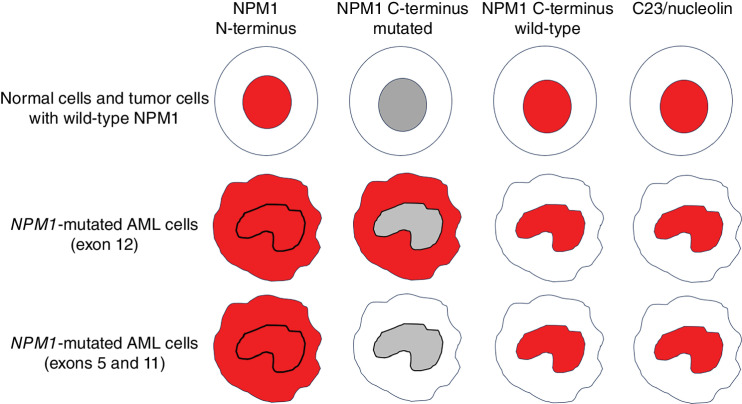
IHC detection of NPM1c+ is a simple, low-cost, very sensitive, and specific alternative assay to diagnose NPM1-mutated AML (26) that can serve as a surrogate to molecular assays. Interestingly, IHC also allows the study of the genetic lesion at the protein level in tissue sections and may provide information on the topographical distribution of the NPM1-mutated leukemic cells (e.g., in the paratrabecular area or close to the vessels). The pattern of reactivity of monoclonal antibodies directed against different domains of NPM1 and C23/nucleolin is shown in Fig. 1.
Figure 1.
Representative examples of subcellular (nuclear and/or cytoplasmic) expression of NPM1 and nucleolin (red staining) in normal tissues, NPM1 wild-type tumors, and NPM1-mutated AML.
IHC is critical for defining multilineage dysplasia, diagnosis of myeloid sarcoma (40, 41), recognition of cases with aplastic or necrotic BM, resulting in dry tap and identification of patients carrying NPM1 mutations outside exon 12 that may be missed by standard molecular assays (see below; refs. 6, 42). This technique can also serve as a surrogate for the detection of NPM1 mutations when molecular assays are not available, for example in some developing countries (43).
NPM1-mutated AML can be further characterized by molecular IHC using a monoclonal antibody specifically directed against the IDH1-R132H mutant protein (44), as NPM1 mutations associate more frequently with IDH1-R132H than with other amino acid changes (i.e., IDH1-R132C, R132G, R132S; ref. 44).
Detection of NPM1-Mutated Proteins by Flow Cytometry
Diagnosis of NPM1-mutated AML by flow cytometry is based either on the detection of cytoplasmic NPM1 (45) or a particular phenotype. A recent study highlighted unique immunophenotypic patterns associated with NPM1-mutated AML, including the presence of (i) immature CD34lo/HLA-DRlo/CD15+/CD7+ AML cells and/or, (ii) neutrophil lineage AML cells displaying low CD34, CD71, and CD64, while upregulating CD105 and/or, (iii) monocytic leukemia cells with CD34lo and asynchronous (CD300e+CD14−; CD35+CD14−) phenotypes (46). Moreover, the NPM1-mutated/FLT3-ITD genotype was closely associated with a CD7hi CD38lo phenotype on immature leukemia cells and/or CD117het and CD123hi expression on neutrophil lineage-committed AML cells (46).
DIAGNOSTIC PITFALLS IN NPM1-MUTATED AML
NPM1-mutated AML can be diagnosed when NPM1 mutation and/or aberrant cytoplasmic expression of NPM1 are detected in a patient meeting other criteria for diagnosis of AML. Other features that may help to predict NPM1-mutated AML pending molecular confirmation include occurrence in a middle-age or older patient, relatively preserved number of platelets despite high white blood cell (WBC) count, blasts with myelomonocytic/monocytic (M4–M5) differentiation, multilineage involvement, cup-like morphology, CD34 and HLA-DR negativity, normal cytogenetics, and skin involvement (24, 47). Patients comutated for NPM1 and FLT3-ITD or RAS frequently present with hyperleukocytosis (40) and may show a starry sky pattern (48). Conversely, NPM1-mutated AML without FLT3 or RAS mutations usually presents with low–normal WBC count (40). The BM in NPM1-mutated AML is characteristically hypercellular but reticulin fibers are usually not increased. Pitfalls in the diagnosis of NPM1-mutated AML are discussed below.
AML with Multilineage Dysplasia
About 23% of NPM1-mutated AML display multilineage dysplasia (ref. 49; i.e., dysplasia ≥50% of cells, in at least two BM cell lineages). These cases may be misdiagnosed as myelodysplasia (MDS) or AML/MDS. Demonstration of NPM1 mutation establishes the diagnosis because the genetic lesion supersedes morphology in importance (4, 40). IHC confirms the multilineage involvement by demonstrating aberrant cytoplasmic expression of nucleophosmin in precursors of erythroid and myeloid lineages and even in mature megakaryocytes.
Myeloid Sarcoma
Extramedullary involvement is not infrequent in NPM1-mutated AML, with the skin being one of the most commonly involved anatomic sites (24, 47, 50), especially in cases with monocytic or myelomonocytic features. As compared with NPM1-mutated AML, NPM1-mutated myeloid sarcoma appears to show differences in genomic landscape, including a higher frequency of cytogenetic abnormalities and mutations affecting epigenetic modifiers such as ASXL1, and a lower frequency of mutations affecting PTPN11, DNMT3A, and IDH1 (51). NPM1-mutated myeloid sarcoma also has poorer overall survival than NPM1-mutated AML (51).
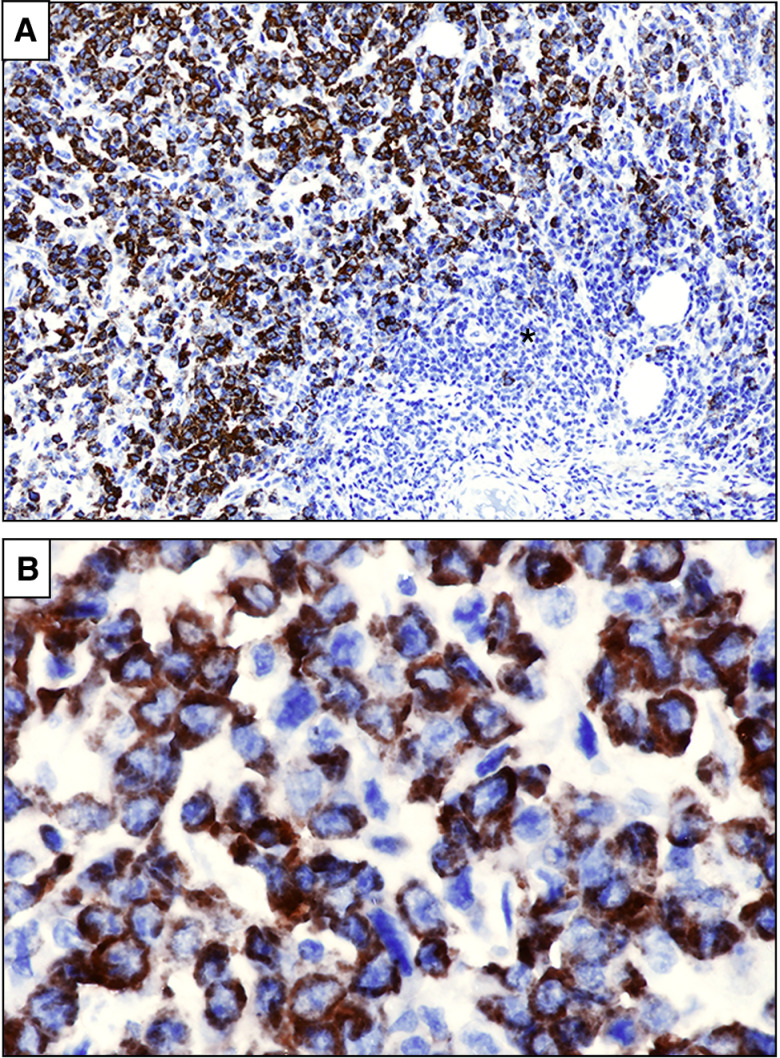
IHC detection of cytoplasmic NPM1 is more reliable than molecular assays in establishing the diagnosis of NPM1-mutated myeloid sarcoma (refs. 24, 41; Fig. 2A and B), particularly when only a small biopsy sample is available for analysis (e.g., punch biopsy). Whether extramedullary involvement represents a poor prognostic factor in NPM1-mutated AML remains controversial. In a large study on >3,000 patients with AML, extramedullary disease did not emerge as an independent prognostic factor (52) but mutational status was not considered. Molecular discordance between myeloid sarcomas and concurrent BM (defined as the occurrence of different mutations in either sample) was associated with worse overall survival, probably due to increased clonal heterogeneity and resistance to therapy (53).
Figure 2.
NPM1-mutated myeloid sarcoma (lymph node). A, Partial infiltration of the lymph node (asterisk) by leukemic cells showing aberrant cytoplasmic expression of nucleophosmin (brown; immunoperoxidase; hematoxylin counterstaining; × 100. B, The same field showing NPM1 cytoplasmic positive tumor cells at higher magnification (immunoperoxidase; hematoxylin counterstaining; × 400).
Identification of NPM1 Mutation Outside Exon 12
NPM1 mutations almost exclusively affect exon 12 (1). Mutations involving exons 9 (3), 11 (3), and 5 (6, 42) may rarely occur. Independently of the exon involved, all NPM1 mutations lead to similar changes at the C-terminus that result in the increased export of NPM1 mutant protein and its accumulation in the cytoplasm of leukemic cells (3). For this reason, IHC is an excellent method for detecting NPM1 mutations occurring outside exon 12 (26). If IHC and molecular assays are used in combination, a discrepancy between the two techniques (i.e., detection of cytoplasmic NPM1 in the absence of mutation at exon 12) should prompt analysis of the entire NPM1 coding sequence, to identify mutations in other exons (Fig. 3). Aberrant cytoplasmic localization of exon 11 and exon 5 NPM1 mutants is identified by antibodies directed against the N-terminus of NPM1 but not by antibodies specific for the NPM1 mutant (Fig. 1). In fact, exon 11 and exon 5 mutations result in the translation of either truncated proteins or longer mutants retaining the same C-terminus sequence as the NPM1 wild-type (6). Although NGS can identify mutations occurring in any exon, many commercially available NGS panels only target exon 12; therefore, custom panels may be required for the detection of other mutations (Fig. 3).
Figure 3.
IHC and molecular procedure for recognizing NPM1 mutations occurring outside exon 12 and NPM1-containing fusion proteins.
Inability to recognize exon 11 and 5 mutations may lead to the incorrect assignment of these cases to the ELN intermediate-risk (NPM1 wild-type without FLT3-ITD) rather than to the favorable-risk group (NPM1-mutated without FLT3-ITD). Because of the low number of cases analyzed, it remains unclear whether these patients have the same outcome as the typical patients with exon 12 NPM1 mutations. So far there are little data regarding MRD monitoring for NPM1 mutations outside exon 12, which requires patient-specific qRT-PCR assays.
AML with Concomitant NPM1 Mutations and BCR::ABL
The association of NPM1 mutations with BCR::ABL1 has been rarely reported in AML (40, 54, 55). These cases should be classified as NPM1-mutated AML, annotating the presence of BCR::ABL1. This is also supported by their CD34 negativity, which is unusual in BCR::ABL1 AML. NPM1-mutated AML without FLT3-ITD has a relatively good outcome (27), whereas AML with BCR::ABL1 is a high-risk leukemia. ELN does not provide prognostic information on cases carrying both NPM1 mutations and BCR::ABL1. These cases immunophenotypically and clinically behave more like an NPM1-mutated AML than AML with BCR::ABL1, but further studies are warranted to clarify this issue.
Therapy-Related NPM1-Mutated AML
NPM1 mutations are characteristically detected in AML of de novo origin (1). About, 15% of cases of therapy-related AML that occur after previous cytotoxic chemotherapy and/or radiotherapy harbor NPM1 mutations (56, 57). These cases have many overlapping biological and clinical features with de novo NPM1-mutated AML. In fact, they consistently show normal cytogenetics (56), DNMT3A (58–60), and TET2 (60) mutations, cytoplasmic NPM1, and a gene-expression profile characterized by upregulation of HOX genes and downregulation of CD34 (60). Moreover, the rate of TP53 (61) and PPM1D mutations (62) that are responsible for chemoradiotherapy-driven selection is much lower in therapy-related NPM1-mutated AML (3% and 4%, respectively; ref. 60) than in therapy-related AML with wild-type NPM1 (25% and up to 20%, respectively; refs. 61, 63). Finally, the survival of therapy-related and de novo NPM1-mutated AML was similar but differed significantly from that of therapy-related AML with wild-type NPM1 (60, 64). Collectively, these findings clearly indicate that the leukemic mechanism underlying “therapy-related” NPM1-mutated AML differs from that of other therapy-related AMLs and most likely represents a de novo leukemia with a coincidental history of prior therapy (60). Based on these findings, therapy-related NPM1-mutated AML is now regarded in both the 2022 ICC (4) and WHO-5 (5) classifications as NPM1-mutated AML (with the addition of “therapy-related” or “post-cytotoxic therapy” as qualifier). Therapy-related NPM1-mutated AML without FLT3-ITD should be assigned to the ELN favorable group and transplant decisions guided by MRD assessment, as in patients with typical de novo NPM1-mutated AML (27).
Percentage of Blasts Defining NPM1-Mutated AML
According to the 5th edition of WHO (5), the diagnosis of NPM1-mutated AML can be made irrespective of the percentage of blasts while the 2022 ICC (4) still requires ≥10% blasts. Thus, the question of how an NPM1-mutated myeloid neoplasm with <10% blasts should be diagnosed still remains open. In the past, such cases were usually classified as NPM1-mutated myelodysplastic syndrome (MDS) or chronic myelomonocytic leukemia (CMML; refs. 65, 66) with mutated NPM1. However, they generally resemble more closely NPM1-mutated AML than MDS or CMML with wild-type NPM1. In fact, they usually show a normal karyotype and lack the typical mutations (ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2; refs. 65, 66) that define MDS or AML with myelodysplasia-related mutations (4). Moreover, they show CD34 negativity, a tendency to evolve rapidly to AML (67) especially when the NPM1 mutation allelic burden is high (65, 66), and good response to chemotherapy but suboptimal response to hypomethylating agents (65, 66, 68). Collectively, these findings strongly suggest that NPM1-mutated myeloid neoplasms with <10% blasts may represent NPM1-mutated AML diagnosed at an early stage and that the NPM1 mutation defines AML irrespective of blast count (25). This concept is also supported by the IHC study of BM biopsies that show aggregates of NPM1 cytoplasmic blasts, usually outnumbering blast cells detectable by morphologic criteria alone (25). Thus, we suggest that these patients should be treated as typical NPM1-mutated AML.
PROGNOSTIC IMPACT OF NPM1 MUTATIONS
Genetic-based risk stratification is part of routine work-up for the management of patients with AML. The median number of pathogenetic variants in newly diagnosed AML is four to five (69), with a frequent scenario being the stepwise accumulation of mutations beginning with those associated with clonal hematopoiesis (e.g., DNMT3A and TET2), followed by an AML-defining mutation (such as NPM1) with the acquisition of mutations in signaling pathway components (e.g., FLT3, NRAS, KRAS) as the final events in leukemogenesis (2). However, a large variety of mutational combinations, sometimes associated with secondary chromosomal abnormalities, may contribute to the genotype of NPM1-mutated AML (69).
Once the diagnosis of NPM1-mutated AML has been established, patients should be risk stratified according to the ELN 2022 guidelines (27). Patients with NPM1 mutation in the absence of FLT3-ITD are assigned to the favorable-risk group. Those with FLT3-ITD (regardless of the allelic ratio) are assigned to the intermediate group because of the difficulty in reproducing measurement of the allelic ratio between laboratories and the recognition that MRD status plays an increasingly important role in risk stratification. Those with adverse karyotype are assigned to the high-risk group on the basis of a large meta-analysis (70). Accumulating evidence indicates that the outcome of NPM1-mutated AML may vary according to accompanying mutations other than FLT3-ITD. Very large studies of uniformly treated patients are required to confidently assign risk associations to mutational subgroups, and current data should be regarded as provisional. In a study including 435 patients with NPM1 mutation enrolled in sequential intensive treatment protocols (69), patients with both FLT3-ITD and DNMT3A comutations (n = 93) showed a particularly poor outcome, while those with either NRAS codon 12 (n = 69) or RAD21 (n = 33) mutations appeared to show improved overall survival. A similar study involving 297 NPM1-mutated patients identified a poor prognosis in those with WT1 comutation (71). An analysis using combined data from multiple cooperative group studies (72), including 1,093 patients with NPM1 mutation, demonstrated complex interactions between comutations but again identified a very poor outcome in those with both FLT3-ITD and DNMT3A comutations. NRAS, KRAS, PTPN11, and RAD21 mutations were associated with favorable prognosis in the absence of FLT3-ITD and IDH1.
Both the ICC and WHO classifications of AML prioritize NPM1 mutation status above myelodysplasia-related (MR; “secondary type”) mutations (SRSF2, SF3B1, U2AF1, ZRSR2, ASXL1, EZH2, BCOR, and STAG2) in cases where these co-occur (73, 74). The suggestion of the ELN panel (27) that myelodysplasia-related mutations should not overrule the favorable impact of a cooccurring NPM1 mutation is further supported by a recent study including 936 patients with NPM1-mutated AML in which myelodysplasia-related mutations did not affect outcomes in this cohort (75). NPM1-mutated AML cases with cooccurring myelodysplasia-related mutations were significantly older and showed lower levels of WBC and platelets than cases without cooccurring mutations (75); however, there were no differences in rates of complete response (CR), relapse-free survival (RFS), or overall survival (OS; ref. 75). Therefore, these patients should still be considered ELN favorable risk and treated accordingly (75).
To date, the only genotype consistently associated with poor outcome (76) not fully captured in the ELN scheme is the combination of NPM1, FLT3-ITD, and DNMT3A mutations. However, it remains unclear whether or how this finding should influence treatment.
Decisions regarding postremission therapy are usually guided by the ELN scheme. However, it is proposed that assessment of MRD should be used to reclassify patients; thus favorable-risk patients with a poor MRD response could be considered candidates for allogeneic hematopoietic stem cell transplantation (allo-HSCT), whereas intermediate-risk patients with a good MRD response could potentially avoid this procedure. Although the limited data published to date support this approach, the selection of patients for allo-HSCT remains somewhat controversial. There is currently no direct evidence that patients with favorable-risk NPM1 mutated AML testing MRD-positive benefit from allo-HSCT in CR1, although results from trial protocols where these patients were directed to transplant have been very encouraging (77). For intermediate-risk patients, a post-hoc analysis from the ALFA 0702 study (78) suggested that only patients with an unfavorable MRD response benefited from allo-HSCT. More recently, this has been further supported by data from the HOVON (79) and GIMEMA (80) groups where patients in the intermediate-risk group (regardless of baseline NPM1 status) were allocated to either autograft or allo-HSCT on the basis of MRD results after the second cycle of intensive chemotherapy, resulting in identical outcomes between the MRD-positive and negative groups. In the context of patients with NPM1 and FLT3-ITD mutations, this remains controversial because of earlier studies showing a benefit of transplant, especially in patients with a high allelic ratio (81). These studies did not incorporate MRD assessment, and therefore the benefit of allo-HSCT for patients with NPM1 and FLT3-ITD mutations who achieve MRD negativity remains uncertain, although protocols wherein only MRD-positive patients were directed to allo-HSCT have shown excellent results in this group (77). Similarly, for patients with other high-risk genotypes including “triple-hit” there is a lack of data regarding the benefit of transplant in patients achieving MRD negativity; however, in one study these patients had a relatively favorable outcome (3-year OS 70%; ref. 29).
In summary, although further data are clearly needed, the current ELN risk stratification scheme, with appropriate reclassification between the favorable and intermediate-risk groups based on MRD status, should remain the basis for decisions regarding post-remission therapy.
MOLECULAR MRD ASSESSMENT IN NPM1-MUTATED AML
NPM1 mutations are an ideal target for monitoring subclinical levels of leukemia (i.e., MRD), because they are common, AML-specific, not expressed in normal tissues and absent in the preleukemic state (e.g., clonal hematopoiesis; Table 2). Moreover, they correlate with therapeutic response, recur at disease relapse, and rising levels of MRD (called MRD relapse) reliably predict clinical relapse within a period of weeks/months, allowing time for preemptive intervention (Table 2). MRD evaluation, including that of NPM1 mutant transcripts, is now routinely recommended by the European LeukemiaNet (27) to evaluate the molecular response to treatment in AML, while associated preleukemic mutations “should be excluded from MRD analysis.” This approach may also be used to guide MRD-directed therapy with novel, less toxic drugs that have been approved in AML. Finally, the approval of NPM1 MRD as a regulatory endpoint is expected to markedly change the clinical trial landscape, such as biomarker-driven adaptive design.
Table 2.
Features of MRD marker mutations in acute myeloid leukemia.
| Feature | DTA a | NPM1 | FLT3-ITD |
|---|---|---|---|
| Clonal hierarchy | Preleukemic | Leukemia-initiating | Usually subclone |
| High frequency | Yes | Yes | Yes |
| AML specificity | No | Yes | No |
| Distinct GEP | No | Yes | No |
| Clonal hematopoiesis | Yes | No | No |
| Clearance post-morphologic CR | No | Yes | Yes |
| Prediction of relapse | No | Yes | No |
| Stability at relapse | Yes | Yes | No |
| Sample source | gDNA | Usually cDNA | gDNA |
Abbreviations: GEP, gene-expression profile; CR, complete remission; gDNA, genomic DNA.
a DTA: DNMT3A, TET2, ASLX1 mutations.
NPM1 mutant transcripts are almost always insertions in a small hotspot, making them ideal targets for qRT-PCR detection using RNA as a substrate, and this is the currently recommended method (37). This method provides very high sensitivity, particularly when BM samples are used (sensitivity is approximately one-log lower when using PB). In a recent study, a significant proportion of false-positive results in the NPM1 wild-type sample was reported among 29 laboratories (82). False-positive results may result in erroneous clinical decisions, e.g., planning unnecessary additional chemotherapy and/or allotransplant with the consequent risk of morbidity and mortality, underlying the need for extensive validation, rigorous negative controls, and external quality assurance when these assays are used to inform clinical decision-making.
There is increasing interest in the use of digital droplet PCR (83–85) and NGS-based ultra-deep sequencing (86, 87). Advantages of digital droplet PCR include high sensitivity (when using RNA as input) and the ability to quantify rare mutations (i.e., non-type A, B, or D) without the need for a standard curve (88). NGS-based strategies can detect all types of NPM1 mutants, but they currently lack standardization and currently have lower sensitivity, particularly those using genomic DNA as a substrate; therefore, they are currently recommended only in the context of clinical trials (37).
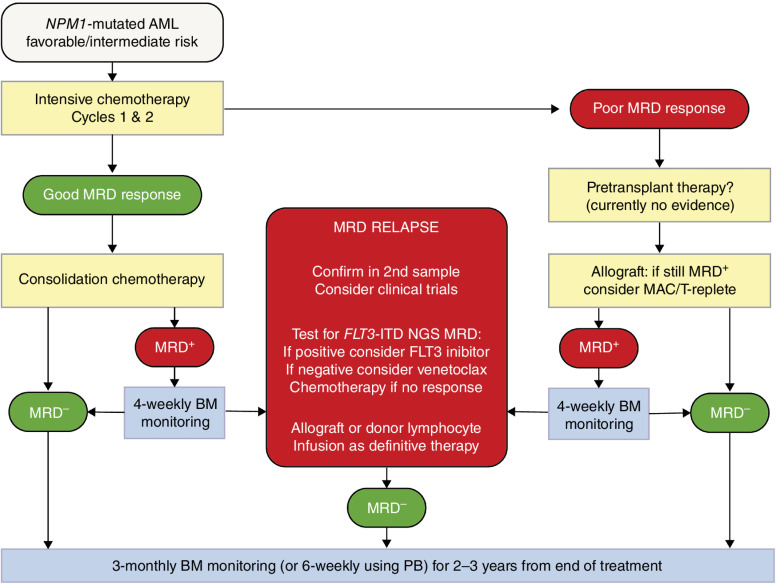
Molecular assessment of MRD in NPM1-mutated AML has a number of different clinical applications: (i) MRD status at post-induction time points is strongly predictive of relapse and OS (29, 80, 86, 89) and therefore can be used to refine risk assessment within the current ELN scheme, allowing more rational selection of patients for allo-HSCT; (ii) pretransplant MRD assessment is highly predictive of outcome, and may inform peritransplant management; (iii) sequential MRD assessment allows the detection of MRD relapse, which predicts impending clinical relapse (90, 91) but provides a window period allowing for preemptive intervention (92–94); (iv) it can be used to define a new response category of CRMRD− for use in both routine practice and clinical studies; and (v) it can serve as surrogate endpoint to accelerate drug testing and approval (27). A possible approach to the incorporation of MRD into treatment algorithms for patients with NPM1-mutated AML is shown in Fig. 4.
Figure 4.
Possible approach to the incorporation of MRD into treatment algorithms for patients with NPM1-mutated AML.
Multiparameter flow cytometry (MFC) has a limited role in monitoring MRD in NPM1-mutated AML because leukemia-associated immunophenotypes (LAIP) are not entirely specific. Moreover, persistent clonal hematopoiesis following eradication of the NPM1 mutation may lead to spurious detection of MRD by MFC.
NPM1 MRD Assessment at Postinduction Time Points
Several large prospective clinical trials have demonstrated the prognostic effect of MRD measured by qRT-PCR at early time points, although precise cutoffs defining high-risk groups differ between studies. In the German AMLSG 0704 study (90), patients achieving MRD negativity in the BM after cycle 2 had a 4-year cumulative incidence of relapse (CIR) 6% versus 53% and 4-year OS 90% versus 51%. The subsequent AMLSG 0909 study (95) reported 4-year CIR for BM MRD negativity after cycle 2 of 25% versus 38% (n = 370) and also showed the prognostic impact of PB MRD negativity (4-year CIR 18% vs. 53%, n = 341). This study also identified a 3-log reduction in both PB and BM as prognostically important. The French ALFA 0702 study (78) identified a >4-log reduction in PB MRD after cycle 1 as most predictive of outcome (3 year CIR 21% vs. 66%, 3y OS 92% vs. 41%). Finally, the UK NCRI AML17 study (29) reported that the PB MRD status after cycle 2 had the strongest prognostic impact (3-year CIR 28% vs. 83%, 3-year OS 77% vs. 25%). Thus, although clearly of prognostic value, further work is needed to identify a unified threshold most predictive of poor outcomes.
Evaluation of NPM1 MRD has a strong prognostic value even in patients treated with venetoclax-based nonintensive therapies. In particular, patients achieving BM MRD negativity by the end of cycle 4 had a 2-year OS of 84% compared with 46% if MRD-positive. On multivariable analyses, MRD negativity was the strongest prognostic factor (96).
NPM1-mutated AML shows high expression of CD33 and a good response to gemtuzumab ozogamicin (GO). In the ALFA-0701 study, patients with NPM1 mutation treated with GO showed deeper MRD responses at multiple time points compared with those not receiving GO (97). The same findings were observed in the AMLSG 09-09 trial, which also showed a lower 4-year CIR and higher RFS in patients who received GO (95).
NPM1 MRD Assessment At End of Treatment
Although MRD positivity at the end of treatment is associated with increased relapse risk, a proportion of these patients will remain in long-term remission, some even converting to CRMRD− without further therapy. The molecular detection of persistent NPM1 mutation is notably agnostic to the cell type(s) carrying the mutant allele. Thus, at least in some patients with persistently low expression of NPM1 mutant transcripts, the residual NPM1-mutated cells may include at least a subset that are incapable of driving relapse. This is consistent with the observation that IHC of posttreatment MRD-positive samples shows terminally differentiated monocytic elements and mature megakaryocytes with cytoplasmic expression of NPM1. These differentiated cell types contribute to the measurement of mutant NPM1 transcripts but may be irrelevant to the risk of relapse. Therefore, MRD positivity at the end of treatment should not automatically trigger further therapy.
In a UK–Australian study (98), 42% of patients with detectable NPM1-mutant transcripts at the end of treatment remained progression free at one year, and of these, 30% spontaneously achieved molecular negativity. Risk factors for progression included baseline FLT3-ITD and <4.4-log MRD reduction from diagnosis; 93% of patients with both of these factors relapsed and died within one year. The AMLSG 0704 study identified a threshold of 200 copies/104ABL with 100% specificity for relapse; however, this threshold appeared less specific in the AMLSG 0909 study (4-year CIR 67%; refs. 90, 95). Again, efforts to define a unified threshold for intervention are now required. Here, a major concern is the danger of overtreating patients who are not destined to relapse. However, MRD positivity should certainly prompt closer monitoring (e.g., every 4–6 weeks) allowing intervention in the case of MRD relapse.
NPM1 MRD Assessment Before Transplant
Although many studies show that pretransplant MRD is strongly associated with poor outcome (99), few studies have investigated this specifically in NPM1-mutated AML. Here, the relationship between MRD and outcome appears more complex, being affected by both the level of MRD and FLT3 mutation status. In the NCRI AML17 study (n = 107; ref. 100), a threshold of 200 copies/105ABL in the PB or 1,000 copies in the BM defined a group with poor outcome (3-year OS 13%). The same BM threshold was identified in a German study (n = 67, 5-year OS 40% vs. 89%; ref. 101). In AML17, patients who were MRD-positive below these levels and who were FLT3-ITD-negative at baseline had the same outcome as those testing MRD-negative (2-year OS 82%), whereas those who had FLT3-ITD at baseline had poor outcomes (2-year OS 17%). A subsequent study using a sensitive NGS-based FLT3-ITD MRD assay (102) could further stratify patient outcome: 2-year OS for patients testing MRD−, MRD+ for NPM1 only and MRD+ for NPM1 and FLT3-ITD in the pretransplant sample was 82%, 68%, and 25%. The US “Pre-Measure” study (103) also showed poor outcomes in patients with detectable pretransplant NPM1 MRD (3-year CIR 63% vs. 22%, 3-year OS 35% vs. 66%). This study used a genomic DNA-based assay with less sensitive PB samples, supporting the concept of a threshold effect, and confirmed the additional prognostic impact of FLT3 MRD status (3-year CIR 64% vs. 75% and 3-year OS 40% vs. 25% for patients testing MRD+ for NPM1 only and MRD+ for NPM1 and FLT3).
Whether myeloablative conditioning can reduce relapse risk in NPM1 MRD-positive patients remains controversial (100, 104, 105). In one study, myeloablative HLA-haploidentical transplantation with regulatory and conventional T cell–adoptive therapy (106) was shown to dramatically reduce the CIR. Studies evaluating the use of hypomethylating agents plus venetoclax during salvage therapy are also of interest (107), especially because NPM1-mutated AML is particularly sensitive to this combination therapy.
Sequential Monitoring for MRD Relapse
Regardless of first-line treatment approach and MRD status, all patients remain at a nontrivial risk of relapse for the first 2–3 years after therapy. Sequential MRD monitoring can be used to reliably identify patients destined to relapse (29) for preemptive intervention. However, overtreatment of patients who will not relapse remains a primary concern; therefore, stringent criteria for diagnosing MRD relapse have been proposed (37). These require two consecutive samples confirming conversion from MRD− to MRD+ for patients who have previously tested MRD− in >1 technically adequate sample. Otherwise, a 1-log increase, confirmed in a second sample, is required.
The optimal treatment for patients with MRD relapse remains undefined. Although standard salvage chemotherapy appears effective, with MRD negativity achieved in ∼60% (100), targeted therapies may provide a less toxic alternative without requiring hospital admission. One of the first examples of MRD-directed therapy (excluding acute promyelocytic leukemia) was the use of 5-azacytidine to treat patients with NPM1-mutated AML in molecular relapse (108) or experiencing a molecular relapse following allo-HSCT (phase II RELAZA2 study; ref. 92). In the latter study, 60% of 53 patients were NPM1-mutated, and 31 (58%) patients had an MRD response, including 19 (36%) who achieved MRD negativity (92). Notably, 58% of patients remained relapse free at 6 months from therapy initiation, and the 2-year RFS was 46% (92).
Venetoclax-based regimens have also been applied as MRD-directed therapy, especially in NPM1-mutated AML, which is recognized as a predictive biomarker of response to venetoclax (109). Venetoclax with azacytidine or low-dose cytarabine produced rates of molecular negativity of 80% to 90% in small retrospective studies (93, 94). Based on these findings, the off-label combination of venetoclax plus azacitidine as a bridge-to-transplant strategy has been retrospectively evaluated in NPM1-mutated MRD-positive fit AML patients (94). After a median number of two cycles (range, 1–4), 9 of 11 patients (81.8%) achieved MRD-negative CR. All 11 patients proceeded to allo-HSCT; 10 of 11 patients are alive, with 9 of 10 being in MRD-negative status.
For patients with baseline FLT3 mutation, a retrospective study (110) showed a molecular response to FLT3 inhibitors (>1-log reduction in MRD) in 60% and MRD negativity in 45%. FLT3-ITD NGS MRD could identify patients more likely to respond. For patients who have MRD relapse after allo-HSCT, donor lymphocyte infusion provides an additional option for eradicating MRD, with evidence of immune response against epitopes derived from mutated NPM1 protein (20).
MRD relapse may provide an extremely useful setting for early-phase evaluation of novel targeted agents. Historically, early-phase studies have usually been performed in patients with frank relapse. A major potential advantage of the MRD relapse setting includes usually normal baseline hematopoietic function (removing the difficulty of deconvoluting disease versus treatment-related hematologic toxicity and infections) and lower baseline disease burden (reducing the chance for clonal evolution or other adaptive therapy resistance). Additionally, sequential MRD monitoring provides a rapid, objective, and highly sensitive method for efficacy evaluation. We anticipate that studies of new targeted agents and immunotherapies will be developed to exploit these advantages in the coming years.
Acknowledgments
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC IG 2019 number 23604) and the European Research Council (Advanced Grant 2016 number 740230).
Authors’ Disclosures
B. Falini reports personal fees from European patent EP1944316B1 during the conduct of the study; in addition, B. Falini has a Eurepean patent, EP1944316B1, with royalties paid. R. Dillon reports grants and personal fees from AbbVie, grants from Amgen, Pfizer, and Jazz, and personal fees from Astellas outside the submitted work.
References
- 1. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 2005;352:254–66. [DOI] [PubMed] [Google Scholar]
- 2. Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood 2020;136:1707–21. [DOI] [PubMed] [Google Scholar]
- 3. Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia 2009;23:1731–43. [DOI] [PubMed] [Google Scholar]
- 4. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood 2022;140:1200–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia 2022;36:1703–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martelli MP, Rossi R, Venanzi A, Meggendorfer M, Perriello VM, Martino G, et al. Novel NPM1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in AML. Blood 2021;138:2696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 2018;34:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leong SM, Tan BX, Bte Ahmad B, Yan T, Chee LY, Ang ST, et al. Mutant nucleophosmin deregulates cell death and myeloid differentiation through excessive caspase-6 and -8 inhibition. Blood 2010;116:3286–96. [DOI] [PubMed] [Google Scholar]
- 9. Wu HC, Rerolle D, Berthier C, Hleihel R, Sakamoto T, Quentin S, et al. Actinomycin D targets NPM1c-primed mitochondria to restore PML-driven senescence in AML therapy. Cancer Discov 2021;11:3198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oka M, Mura S, Otani M, Miyamoto Y, Nogami J, Maehara K, et al. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife 2019;8:e46667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uckelmann HJ, Haarer EL, Takeda R, Wong EM, Hatton C, Marinaccio C, et al. Mutant NPM1 directly regulates oncogenic transcription in acute myeloid leukemia. Cancer Discov 2023;13:746–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang XQD, Fan D, Han Q, Liu Y, Miao H, Wang X, et al. Mutant NPM1 hijacks transcriptional hubs to maintain pathogenic gene programs in acute myeloid leukemia. Cancer Discov 2023;13:724–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Issa GC, Ravandi F, DiNardo CD, Jabbour E, Kantarjian HM, Andreeff M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia 2021;35:2482–95. [DOI] [PubMed] [Google Scholar]
- 14. Issa GC, Aldoss I, DiPersio J, Cuglievan B, Stone R, Arellano M, et al. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature 2023;615:920–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Falini B, Martelli MP, Brunetti L. Mutant NPM1: nuclear export and the mechanism of leukemogenesis. Am J Hematol 2023;98:550–2. [DOI] [PubMed] [Google Scholar]
- 16. Liso A, Colau D, Benmaamar R, De Groot A, Martin W, Benedetti R, et al. Nucleophosmin leukaemic mutants contain C-terminus peptides that bind HLA class I molecules. Leukemia 2008;22:424–6. [DOI] [PubMed] [Google Scholar]
- 17. Greiner J, Ono Y, Hofmann S, Schmitt A, Mehring E, Gotz M, et al. Mutated regions of nucleophosmin 1 elicit both CD4(+) and CD8(+) T-cell responses in patients with acute myeloid leukemia. Blood 2012;120:1282–9. [DOI] [PubMed] [Google Scholar]
- 18. Narayan R, Olsson N, Wagar LE, Medeiros BC, Meyer E, Czerwinski D, et al. Acute myeloid leukemia immunopeptidome reveals HLA presentation of mutated nucleophosmin. PLoS One 2019;14:e0219547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Lee DI, Reijmers RM, Honders MW, Hagedoorn RS, de Jong RC, Kester MG, et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J Clin Invest 2019;129:774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann S, Gotz M, Schneider V, Guillaume P, Bunjes D, Dohner H, et al. Donor lymphocyte infusion induces polyspecific CD8(+) T-cell responses with concurrent molecular remission in acute myeloid leukemia with NPM1 mutation. J Clin Oncol 2013;31:e44–7. [DOI] [PubMed] [Google Scholar]
- 21. Greiner J, Schneider V, Schmitt M, Gotz M, Dohner K, Wiesneth M, et al. Immune responses against the mutated region of cytoplasmatic NPM1 might contribute to the favorable clinical outcome of AML patients with NPM1 mutations (NPM1mut). Blood 2013;122:1087–8. [DOI] [PubMed] [Google Scholar]
- 22. Armistead PM. Cellular therapy against public neoantigens. J Clin Invest 2019;129:506–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie G, Ivica NA, Jia B, Li Y, Dong H, Liang Y, et al. CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat Biomed Eng 2021;5:399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falini B, Brunetti L, Martelli MP. How I diagnose and treat NPM1-mutated AML. Blood 2021;137:589–99. [DOI] [PubMed] [Google Scholar]
- 25. Falini B, Martelli MP, Brunetti L, Gjertsen BT, Andresen V. The NPM1 mutant defines AML irrespective of blast count. Am J Hematol 2023;98:E187–E9. [DOI] [PubMed] [Google Scholar]
- 26. Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood 2006;108:1999–2005. [DOI] [PubMed] [Google Scholar]
- 27. Dohner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022;140:1345–77. [DOI] [PubMed] [Google Scholar]
- 28. Gorello P, Cazzaniga G, Alberti F, Dell'Oro MG, Gottardi E, Specchia G, et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carryng nuclephosmin 1 (NPM1) gene mutations. Leukemia 2006;20:1103–8. [DOI] [PubMed] [Google Scholar]
- 29. Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med 2016;374:422–33. [DOI] [PubMed] [Google Scholar]
- 30. Cazzaniga G, Dell'Oro MG, Mecucci C, Giarin E, Masetti R, Rossi V, et al. Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood 2005;106:1419–22. [DOI] [PubMed] [Google Scholar]
- 31. Thiede C, Creutzig E, Reinhardt D, Ehninger G, Creutzig U. Different types of NPM1 mutations in children and adults: evidence for an effect of patient age on the prevalence of the TCTG-tandem duplication in NPM1-exon 12. Leukemia 2007;21:366–7. [DOI] [PubMed] [Google Scholar]
- 32. Ottone T, Ammatuna E, Lavorgna S, Noguera NI, Buccisano F, Venditti A, et al. An allele-specific rt-PCR assay to detect type A mutation of the nucleophosmin-1 gene in acute myeloid leukemia. J Mol Diagn 2008;10:212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szankasi P, Jama M, Bahler DW. A new DNA-based test for detection of nucleophosmin exon 12 mutations by capillary electrophoresis. J Mol Diagn 2008;10:236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wertheim G, Bagg A. Nucleophosmin (NPM1) mutations in acute myeloid leukemia: an ongoing (cytoplasmic) tale of dueling mutations and duality of molecular genetic testing methodologies. J Mol Diagn 2008;10:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Q, Chen W, Gaal KK, Slovak ML, Stein A, Weiss LM. A rapid, one step assay for simultaneous detection of FLT3/ITD and NPM1 mutations in AML with normal cytogenetics. Br J Haematol 2008;142:489–92. [DOI] [PubMed] [Google Scholar]
- 36. Ma W, Kantarjian H, Zhang X, Jilani I, Sheikholeslami MR, Donahue AC, et al. Detection of nucleophosmin gene mutations in plasma from patients with acute myeloid leukemia: clinical significance and implications. Cancer Biomark 2009;5:51–8. [DOI] [PubMed] [Google Scholar]
- 37. Heuser M, Freeman SD, Ossenkoppele GJ, Buccisano F, Hourigan CS, Ngai LL, et al. 2021 Update on MRD in acute myeloid leukemia: a consensus document from the European LeukemiaNet MRD Working Party. Blood 2021;138:2753–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Borate U, Yang F, Press RD, Ruppert AS, Jones D, Caruthers S, et al. AML Samples show high concordance in detection of mutations by NGS at local institutions versus central laboratories. Blood Adv 2023;7:6048–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood 2006;107:4514–23. [DOI] [PubMed] [Google Scholar]
- 40. Falini B, Sciabolacci S, Falini L, Brunetti L, Martelli MP. Diagnostic and therapeutic pitfalls in NPM1-mutated AML: notes from the field. Leukemia 2021;35:3113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Falini B, Lenze D, Hasserjian R, Coupland S, Jaehne D, Soupir C, et al. Cytoplasmic mutated nucleophosmin (NPM) defines the molecular status of a significant fraction of myeloid sarcomas. Leukemia 2007;21:1566–70. [DOI] [PubMed] [Google Scholar]
- 42. Wang P, Segal J, Drazer MW, Venkataraman G, Arber DA, Gurbuxani S. NPM1 exon 5 mutations in acute myeloid leukemia: Implications in diagnosis and minimal residual monitoring. EJHaem 2022;3:962–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Falini B, Martelli MP, Pileri SA, Mecucci C. Molecular and alternative methods for diagnosis of acute myeloid leukemia with mutated NPM1: flexibility may help. Haematologica 2010;95:529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Falini B, Spinelli O, Meggendorfer M, Martelli MP, Bigerna B, Ascani S, et al. IDH1-R132 changes vary according to NPM1 and other mutations status in AML. Leukemia 2019;33:1043–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gruszka AM, Lavorgna S, Consalvo MI, Ottone T, Martinelli C, Cinquanta M, et al. A monoclonal antibody against mutated nucleophosmin 1 for the molecular diagnosis of acute myeloid leukemias. Blood 2010;116:2096–102. [DOI] [PubMed] [Google Scholar]
- 46. Matarraz S, Leoz P, Yeguas-Bermejo A, van der Velden V, Bras AE, Sanchez Gallego JI, et al. Baseline immunophenotypic profile of bone marrow leukemia cells in acute myeloid leukemia with nucleophosmin-1 gene mutation: a EuroFlow study. Blood Cancer J 2023;13:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luskin MR, Huen AO, Brooks SA, Stewart C, Watt CD, Morrissette JJ, et al. NPM1 mutation is associated with leukemia cutis in acute myeloid leukemia with monocytic features. Haematologica 2015;100:e412–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Falini B, Cardinali V, Brunetti L, Martelli MP. NPM1-mutated AML with starry sky pattern. Br J Haematol 2023;202:8. [DOI] [PubMed] [Google Scholar]
- 49. Falini B, Macijewski K, Weiss T, Bacher U, Schnittger S, Kern W, et al. Multilineage dysplasia has no impact on biologic, clinicopathologic, and prognostic features of AML with mutated nucleophosmin (NPM1). Blood 2010;115:3776–86. [DOI] [PubMed] [Google Scholar]
- 50. Niscola P, Mazzone C, Fratoni S, Ardu NR, Cesini L, Giovannini M, et al. Acute myeloid leukemia with NPM1 mutation and disseminated leukaemia cutis: achievement of molecular complete remission by venetoclax/azacytidine combination in a very old patient. Acta Haematol 2023:146:408–12. [DOI] [PubMed] [Google Scholar]
- 51. Ramia de Cap M, Wu LP, Hirt C, Pihan GA, Patel SS, Tam W, et al. Myeloid sarcoma with NPM1 mutation may be clinically and genetically distinct from AML with NPM1 mutation: a study from the Bone Marrow Pathology Group. Leuk Lymphoma 2023;64:972–80. [DOI] [PubMed] [Google Scholar]
- 52. Ganzel C, Manola J, Douer D, Rowe JM, Fernandez HF, Paietta EM, et al. Extramedullary disease in adult acute myeloid leukemia is common but lacks independent significance: analysis of patients in ECOG-ACRIN Cancer Research Group trials, 1980–2008. J Clin Oncol 2016;34:3544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Werstein B, Dunlap J, Cascio MJ, Ohgami RS, Fan G, Press R, et al. Molecular discordance between myeloid sarcomas and concurrent bone marrows occurs in actionable genes and is associated with worse overall survival. J Mol Diagn 2020;22:338–45. [DOI] [PubMed] [Google Scholar]
- 54. Konoplev S, Yin CC, Kornblau SM, Kantarjian HM, Konopleva M, Andreeff M, et al. Molecular characterization of de novo Philadelphia chromosome-positive acute myeloid leukemia. Leuk Lymphoma 2013;54:138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim MJ, Ahn S, Jeong SH, Jang JH, Han JH, Choi JR, et al. Minor BCR-ABL1-positive acute myeloid leukemia associated with the NPM1 mutation and FLT3 internal tandem duplication. Ann Lab Med 2016;36:263–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Andersen MT, Andersen MK, Christiansen DH, Pedersen-Bjergaard J. NPM1 mutations in therapy-related acute myeloid leukemia with uncharacteristic features. Leukemia 2008;22:951–5. [DOI] [PubMed] [Google Scholar]
- 57. Kayser S, Dohner K, Krauter J, Kohne CH, Horst HA, Held G, et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood 2011;117:2137–45. [DOI] [PubMed] [Google Scholar]
- 58. Pedersen-Bjergaard J, Andersen MK, Andersen MT, Christiansen DH. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia 2008;22:240–8. [DOI] [PubMed] [Google Scholar]
- 59. Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015;125:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Othman J, Meggendorfer M, Tiacci E, Thiede C, Schlenk R, Dillon R, et al. Overlapping features of therapy-related and de novo NPM1-mutated AML. Blood 2023;141:1846–57. [DOI] [PubMed] [Google Scholar]
- 61. Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kahn JD, Miller PG, Silver AJ, Sellar RS, Bhatt S, Gibson C, et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 2018;132:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hsu JI, Dayaram T, Tovy A, De Braekeleer E, Jeong M, Wang F, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell 2018;23:700–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nilsson C, Linde F, Hulegardh E, Garelius H, Lazarevic V, Antunovic P, et al. Characterization of therapy-related acute myeloid leukemia: increasing incidence and prognostic implications. Haematologica 2023;108:1015–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Montalban-Bravo G, Kanagal-Shamanna R, Sasaki K, Patel K, Ganan-Gomez I, Jabbour E, et al. NPM1 mutations define a specific subgroup of MDS and MDS/MPN patients with favorable outcomes with intensive chemotherapy. Blood Adv 2019;3:922–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Patel SS, Ho C, Ptashkin RN, Sadigh S, Bagg A, Geyer JT, et al. Clinicopathologic and genetic characterization of nonacute NPM1-mutated myeloid neoplasms. Blood Adv 2019;3:1540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vallapureddy R, Lasho TL, Hoversten K, Finke CM, Ketterling R, Hanson C, et al. Nucleophosmin 1 (NPM1) mutations in chronic myelomonocytic leukemia and their prognostic relevance. Am J Hematol 2017;92:E614–E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prata PH, Bally C, Prebet T, Recher C, Venton G, Thomas X, et al. NPM1 mutation is not associated with prolonged complete remission in acute myeloid leukemia patients treated with hypomethylating agents. Haematologica 2018;103:e455–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Papaemmanuil E, Dohner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med 2016;375:900–1. [DOI] [PubMed] [Google Scholar]
- 70. Angenendt L, Rollig C, Montesinos P, Ravandi F, Juliusson G, Recher C, et al. Revisiting coexisting chromosomal abnormalities in NPM1-mutated AML in light of the revised ELN 2022 classification. Blood 2023;141:433–5. [DOI] [PubMed] [Google Scholar]
- 71. Eisfeld AK, Kohlschmidt J, Mims A, Nicolet D, Walker CJ, Blachly JS, et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia 2020;34:3215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sánchez AH, Ramiro AV, Sträng E, Gastone C, Heckman CA, Versluis J, et al. Machine learning allows the identification of new co-mutational pattern with prognostic implication in NPM1 mutated AML- Results of the European Harmony Alliance. Blood 2022;140:739–42. [Google Scholar]
- 73. Wright MF, Pozdnyakova O, Hasserjian RP, Aggarwal N, Shaver AC, Weinberg OK, et al. Secondary-type mutations do not impact prognosis in acute myelogenous leukemia AML with mutated NPM1. Am J Hematol 2022;97:E462–E5. [DOI] [PubMed] [Google Scholar]
- 74. Zhou Q, Zhao D, Zarif M, Yeung YWT, Richard-Carpentier G, Chang H. Impact of secondary-type mutations in NPM1 mutated AML. Eur J Haematol 2023;111:165–8. [DOI] [PubMed] [Google Scholar]
- 75. Eckardt JN, Bill M, Rausch C, Metzeler K, Spiekermann K, Stasik S, et al. Secondary-type mutations do not impact outcome in NPM1-mutated acute myeloid leukemia - implications for the European leukemia net risk classification. Leukemia 2023.. doi 10.1038/s41375-023-02016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bezerra MF, Lima AS, Pique-Borras MR, Silveira DR, Coelho-Silva JL, Pereira-Martins DA, et al. Co-occurrence of DNMT3A, NPM1, FLT3 mutations identifies a subset of acute myeloid leukemia with adverse prognosis. Blood 2020;135:870–5. [DOI] [PubMed] [Google Scholar]
- 77. Russell N, Wilhelm-Benartzi C, Knapper S, Batten LM, Canham J, Hinson EL, et al. FLAG-Ida combined with gemtuzumab ozogamicin (GO) improves event-free survival in younger patients with newly diagnosed acute myeloid leukaemia (AML) and shows an overall survival benefit in NPM1 and FLT3 mutated subgroups. Results from the UK NCRI AML19 Trial. Blood 2022;140:526–8.35951345 [Google Scholar]
- 78. Balsat M, Renneville A, Thomas X, de Botton S, Caillot D, Marceau A, et al. Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with NPM1 mutation: a study by the Acute Leukemia French Association Group. J Clin Oncol 2017;35:185–93. [DOI] [PubMed] [Google Scholar]
- 79. Tettero JM, Ngai LL, Bachas C, Breems DA, Fischer T, Gjertsen BT, et al. Measurable residual disease-guided therapy in intermediate-risk acute myeloid leukemia patients is a valuable strategy in reducing allogeneic transplantation without negatively affecting survival. Haematologica 2023;108:2794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Venditti A, Piciocchi A, Candoni A, Melillo L, Calafiore V, Cairoli R, et al. GIMEMA AML1310 trial of risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia. Blood 2019;134:935–45. [DOI] [PubMed] [Google Scholar]
- 81. Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, et al. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood 2014;124:3441–9. [DOI] [PubMed] [Google Scholar]
- 82. Scott S, Dillon R, Thiede C, Sadiq S, Cartwright A, Clouston HJ, et al. Assessment of acute myeloid leukemia molecular measurable residual disease testing in an interlaboratory study. Blood Adv 2023;7:3686–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bacher U, Dicker F, Haferlach C, Alpermann T, Rose D, Kern W, et al. Quantification of rare NPM1 mutation subtypes by digital PCR. Br J Haematol 2014;167:710–4. [DOI] [PubMed] [Google Scholar]
- 84. Mencia-Trinchant N, Hu Y, Alas MA, Ali F, Wouters BJ, Lee S, et al. Minimal residual disease monitoring of acute myeloid leukemia by massively multiplex digital PCR in patients with NPM1 mutations. J Mol Diagn 2017;19:537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wertheim GBW, Bagg A. NPM1 for MRD? Droplet like it's hot! J Mol Diagn 2017;19:498–501. [DOI] [PubMed] [Google Scholar]
- 86. Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med 2018;378:1189–99. [DOI] [PubMed] [Google Scholar]
- 87. Ritterhouse LL, Parilla M, Zhen CJ, Wurst MN, Puranik R, Henderson CM, et al. Clinical validation and implementation of a measurable residual disease assay for NPM1 in acute myeloid leukemia by error-corrected next-generation sequencing. Mol Diagn Ther 2019;23:791–802. [DOI] [PubMed] [Google Scholar]
- 88. Lesieur A, Thomas X, Nibourel O, Boissel N, Fenwarth L, De Botton S, et al. Minimal residual disease monitoring in acute myeloid leukemia with non-A/B/D-NPM1 mutations by digital polymerase chain reaction: feasibility and clinical use. Haematologica 2021;106:1767–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pratz KW, Jonas BA, Pullarkat V, Recher C, Schuh AC, Thirman MJ, et al. Measurable residual disease response and prognosis in treatment-naive acute myeloid leukemia with venetoclax and azacitidine. J Clin Oncol 2022;40:855–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kronke J, Schlenk RF, Jensen KO, Tschurtz F, Corbacioglu A, Gaidzik VI, et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol 2011;29:2709–16. [DOI] [PubMed] [Google Scholar]
- 91. Schuurhuis GJ, Heuser M, Freeman S, Bene MC, Buccisano F, Cloos J, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood 2018;131:1275–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Platzbecker U, Middeke JM, Sockel K, Herbst R, Wolf D, Baldus CD, et al. Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): an open-label, multicentre, phase 2 trial. Lancet Oncol 2018;19:1668–79. [DOI] [PubMed] [Google Scholar]
- 93. Tiong IS, Dillon R, Ivey A, Teh TC, Nguyen P, Cummings N, et al. Venetoclax induces rapid elimination of NPM1 mutant measurable residual disease in combination with low-intensity chemotherapy in acute myeloid leukaemia. Br J Haematol 2021;192:1026–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sartor C, Brunetti L, Audisio E, Cignetti A, Zannoni L, Cristiano G, et al. A venetoclax and azacitidine bridge-to-transplant strategy for NPM1-mutated acute myeloid leukaemia in molecular failure. Br J Haematol 2023;202:599–607. [DOI] [PubMed] [Google Scholar]
- 95. Kapp-Schwoerer S, Weber D, Corbacioglu A, Gaidzik VI, Paschka P, Kronke J, et al. Impact of gemtuzumab ozogamicin on MRD and relapse risk in patients with NPM1-mutated AML: results from the AMLSG 09-09 trial. Blood 2020;136:3041–50. [DOI] [PubMed] [Google Scholar]
- 96. Othman J, Tiong IS, O'Nions J, Dennis M, Mokretar K, Ivey A, et al. Molecular MRD is strongly prognostic in patients with NPM1-mutated AML receiving venetoclax-based non-intensive therapy. Blood 2023: blood.2023021579. [DOI] [PubMed] [Google Scholar]
- 97. Lambert J, Lambert J, Nibourel O, Pautas C, Hayette S, Cayuela JM, et al. MRD assessed by WT1 and NPM1 transcript levels identifies distinct outcomes in AML patients and is influenced by gemtuzumab ozogamicin. Oncotarget 2014;5:6280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tiong IS, Dillon R, Ivey A, Kuzich JA, Thiagarajah N, Sharplin KM, et al. Clinical impact of NPM1-mutant molecular persistence after chemotherapy for acute myeloid leukemia. Blood Adv 2021;5:5107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Short NJ, Zhou S, Fu C, Berry DA, Walter RB, Freeman SD, et al. Association of measurable residual disease with survival outcomes in patients with acute myeloid leukemia: a systematic review and meta-analysis. JAMA Oncol 2020;6:1890–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dillon R, Hills R, Freeman S, Potter N, Jovanovic J, Ivey A, et al. Molecular MRD status and outcome after transplantation in NPM1-mutated AML. Blood 2020;135:680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kayser S, Benner A, Thiede C, Martens U, Huber J, Stadtherr P, et al. Pretransplant NPM1 MRD levels predict outcome after allogeneic hematopoietic stem cell transplantation in patients with acute myeloid leukemia. Blood Cancer J 2016;6:e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Loo S, Dillon R, Ivey A, Anstee NS, Othman J, Tiong IS, et al. Pretransplant FLT3-ITD MRD assessed by high-sensitivity PCR-NGS determines posttransplant clinical outcome. Blood 2022;140:2407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dillon LW, Gui G, Page KM, Ravindra N, Wong ZC, Andrew G, et al. DNA sequencing to detect residual disease in adults with acute myeloid leukemia prior to hematopoietic cell transplant. JAMA 2023;329:745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Walter RB, Gyurkocza B, Storer BE, Godwin CD, Pagel JM, Buckley SA, et al. Comparison of minimal residual disease as outcome predictor for AML patients in first complete remission undergoing myeloablative or nonmyeloablative allogeneic hematopoietic cell transplantation. Leukemia 2015;29:137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jager P, Rautenberg C, Kaivers J, Kasprzak A, Geyh S, Baermann BN, et al. Allogeneic hematopoietic stem cell transplantation and pre-transplant strategies in patients with NPM1-mutated acute myeloid leukemia: a single center experience. Sci Rep 2023;13:10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Martelli MF, Di Ianni M, Ruggeri L, Falzetti F, Carotti A, Terenzi A, et al. HLA-haploidentical transplantation with regulatory and conventional T-cell adoptive immunotherapy prevents acute leukemia relapse. Blood 2014;124:638–44. [DOI] [PubMed] [Google Scholar]
- 107. Garcia JS, Kim HT, Murdock HM, Cutler CS, Brock J, Gooptu M, et al. Adding venetoclax to fludarabine/busulfan RIC transplant for high-risk MDS and AML is feasible, safe, and active. Blood Adv 2021;5:5536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sockel K, Wermke M, Radke J, Kiani A, Schaich M, Bornhauser M, et al. Minimal residual disease-directed preemptive treatment with azacitidine in patients with NPM1-mutant acute myeloid leukemia and molecular relapse. Haematologica 2011;96:1568–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020;135:791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Othman J, Potter N, Mokretar K, Taussig D, Khan A, Krishnamurthy P, et al. FLT3 inhibitors as MRD-guided salvage treatment for molecular failure in FLT3 mutated AML. Leukemia 2023;37:2066–72. [DOI] [PMC free article] [PubMed] [Google Scholar]