Summary:
In this issue of Blood Cancer Discovery, Neri, Barwick, and colleagues and Welsh, Barwick, and colleagues performed RNA sequencing, chromatin immunoprecipitation sequencing, assay for transposase-accessible chromatin using sequencing, and genetic studies to characterize the underlying mechanisms of immunomodulatory drug (IMiD) resistance in multiple myeloma. They demonstrated that IMiD resistance is driven by sustained expression of MYC and IRF4 via transcriptional plasticity that involves induction of ETV4 and BATF proteins, the binding of these proteins to their super-enhancers, and the recruitment of BRD4 and p300. Finally, these studies suggest IMiD and p300 inhibitor combination as a promising therapeutic strategy in multiple myeloma.
Multiple myeloma is a recalcitrant neoplastic B-cell disorder that is characterized by abnormal proliferation of malignant plasma cells in the bone marrow, leading to marked increases of monoclonal immunoglobulin (paraprotein or M-spike) proteins in the blood and urine, cytopenia, bone fracture, and organ dysfunction (1). The development and progression of multiple myeloma remains rather poorly understood, where patients can present with monoclonal gammopathy of unknown significance (MGUS) that is characterized by elevated paraprotein, or with smoldering multiple myeloma, which is an asymptomatic precursor to frank disease. Complex genetic aberrations are a hallmark of multiple myeloma and include a large cast of recurrent chromosomal rearrangements and somatic mutations that promote the proliferation of monoclonal plasma cells and their transformation into frank multiple myeloma (1). Among known genetic abnormalities that drive multiple myeloma, MYC, and IRF4 are the most commonly dysregulated and overexpressed oncogenes and B-lineage factors in this disease, where their high levels of expression are sustained by immunoglobulin light and heavy chain enhancers (2) or plasma cell specific super-enhancers (SE; e.g., FAM46C, PRDM1, and DUSP22, ref. 3), that are enriched for BRD4 and p300 (4, 5), nonredundant transcriptional coactivators that bind to chromatin via bromodomains and augment transcription by binding to the transcription elongation factor P-TFEb, activating P-TFEb kinase activity and phosphorylation of the C-terminal tail of RNA polymerase-II (BRD4), or by acetylating the tails of histones, thereby promoting open chromatin, and by binding to components of the transcriptional machinery (p300).
Although nearly all patients with multiple myeloma ultimately relapse with refractory disease, the development of the immunomodulatory drugs (IMiD), including lenalidomide, thalidomide, and pomalidomide, was a breakthrough that significantly improved clinical outcomes in treatment-naïve and relapsed myeloma (1). Mechanistically, IMiDs compromise multiple myeloma cell survival by binding to an E3 ubiquitin ligase substrate adapter coined Cereblon (CRBN), which provokes the ubiquitination and proteasomal destruction of the essential B-cell master transcriptional regulatory proteins IKZF1 and IKZF3 by the CRBN/DDB1/CUL4/ROC1 E3 ubiquitin ligase (6). Notably, IKZF1 and IKZF3 are essential for multiple myeloma cell survival; thus, multiple myeloma is addicted to IKZF1/IKZF3 that are disabled by IMiDs. The tragic therapeutic dilemma is that, despite robust clinical responses, acquired resistance to IMiDs occurs in most patients with multiple myeloma and relapsed disease is often difficult to treat. Furthermore, although acquired mutations or splice variants in CRBN confer IMiD resistance in a small group of multiple myeloma cases, the mechanisms driving resistance remain largely unknown in the majority of multiple myeloma cases (7).
A hallmark of IMiD resistance in multiple myeloma is elevated expression of MYC and IRF4, which connote poor prognosis, and which are essential for the maintenance of IMiD-resistant cells (3, 7, 8). Furthermore, MYC and IRF4 form an autoregulatory circuit in multiple myeloma, where they induce each other's transcription to sustain multiple myeloma cell growth and survival (3). Heretofore, it was not clear whether there were links between IKZF1/IKZF3, MYC, and IRF4 that explained their coessentiality in multiple myeloma. Importantly, in this issue of Blood Cancer Discovery, two back-to-back studies by Neri and colleagues (9) and Welsh and colleagues (10) have shown that IMiD resistance in multiple myeloma requires an IKZF1/IKZF3-to-MYC/IRF4 circuit, where IKZF1/IKZF3 promotes MYC and IRF4 expression by binding to SEs and recruiting the coactivators BRD4 and p300 that drive their transcription. Furthermore, the comprehensive analyses of this circuit revealed that IMiDs downregulate IKZF1/IKZF3, MYC, and IRF4 in IMiD-sensitive multiple myeloma cells, but only IKZF1/IKZF3 in IMiD-resistant multiple myeloma cells, and that the IKZF1/IKZF3 dependence of MYC and IRF4 transcription in IMiD-resistant multiple myeloma is circumvented via transcriptional plasticity that involves the induction and binding of select ETS (i.e., ETV4) or AP-1 (i.e., BATF) family transcription factors to their SEs and the recruitment of BRD4 and p300. Finally, the authors show that these alternative regulators of MYC and IRF4 are overexpressed in patients with IMiD-resistant multiple myeloma, connote poor prognosis, and represent exciting new vulnerabilities to disable IMiD-resistant disease (9, 10).
The exciting new insights provided by the authors came from a comprehensive battery of ex vivo experiments [RNA sequencing, chromatin immunoprecipitation sequencing (ChIP-seq), assay for transposase-accessible chromatin using sequencing (ATAC-seq), and genetic studies], in vivo efficacy studies, and deep analyses of multiple myeloma patient samples. First, Neri and colleagues (9) showed that IKZF1 binds to canonical MYC enhancers, immunoglobulin enhancers, and other SEs in multiple myeloma cell lines, and that IMiD treatment reduces not only the levels of IKZF1/IKZF3 and their binding to these enhancers and SEs, but also to the eviction of BRD4 and p300, leading to rapid downregulation of MYC and death of IMiDs-sensitive multiple myeloma cells (Fig. 1). Surprisingly, IMiD treatment of IMiD-resistant multiple myeloma cells still provoked downregulation of IKZF1/IKZF3 and their binding to these enhancers and SEs but did not affect the binding of p300 and BRD4 to these elements (9). Second, further inspection of the binding motif enriched in IKZF1-bound regions revealed that the ETS family member ETV4 shares a common “AGGAA” binding motif with IKZF1, and ChIP-seq studies showed that ETV4 indeed binds to elements bound by IKZF1 in multiple myeloma cells (with an overlap of nearly 80%; Fig. 1). Importantly, ETV4 binding to these enhancer elements was refractory to IMiD treatment of IMiD-resistant multiple myeloma cells, and CRISPR-mediated knockout of ETV4 led to downregulation of MYC and to rapid death of IMiD-resistant cells following IMiD treatment (9). Finally, underscoring the clinical relevance of ETV4 in multiple myeloma and IMiD resistance: (i) levels of ETV4 are significantly elevated in patients with relapsed and refractory multiple myeloma versus patients with treatment-naïve multiple myeloma; (ii) ETV4 expression increases as disease progresses in paired analysis of serial multiple myeloma patient samples; and, accordingly, (iii) elevated ETV4 levels connote inferior survival outcomes (9).
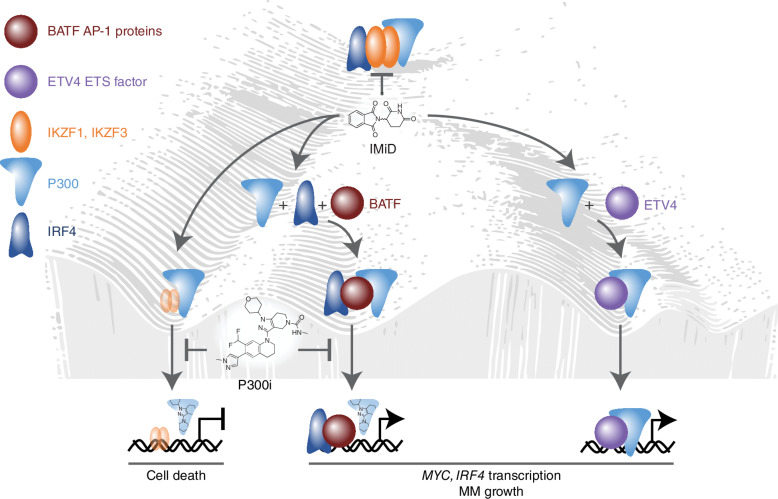
Figure 1.
Transcriptional plasticity sustains SE activities that drive MYC-IRF4-dependent IMiD resistance in multiple myeloma (MM). IMiDs induce multiple myeloma cell death by promoting CRBN-mediated downregulation of IKZF1/IKZF3 and subsequent downregulation of MYC and IRF4 transcription. In IMiD-resistant multiple myeloma cells, IKZF1/IKZF3 dependence of MYC and IRF4 transcription is circumvented via transcriptional plasticity that involves the induction of ETV4 and BATF transcription factors, which bind and recruit p300 to MYC and IRF4 super-enhancers. Accordingly, p300i potentiates the efficacy of IMiDs by downregulating IRF4 and MYC transcription in IMiD-resistant multiple myeloma cells. Figure concept and design by Ben Barwick.
In parallel studies, Welsh and colleagues (10) first showed that IMiDs display significant synergy with inhibitors of p300 (p300i; i.e., GNE-781, CCS1477) for the majority of multiple myeloma cell lines, including IMiD-resistant cells, and that this was associated with downregulation of MYC and IRF4, profound growth arrest and cell death. Importantly, this combination also showed potent efficacy versus multiple myeloma in vivo, and with minimal toxicity (10). Interestingly, ATAC-seq analyses showed that the IMiD/p300i combination provoked a significant loss of chromatin accessibility, especially in regions enriched for IKZF1/IKZF3, p300, and BATF proteins (the AP-1 family members BATF, BATF2, BATF3), including an enrichment at enhancers and SEs that drive MYC and IRF4 transcription (Fig. 1). Supporting critical roles of BATF in IMiD resistance in multiple myeloma: (i) BATF family members are highly expressed in IMiD-resistant versus IMiD-sensitive multiple myeloma cells; (ii) BATF levels are significantly higher in advanced versus early stage multiple myeloma; and (iii) high BATF levels connote significantly shorter progression-free and overall survival (10). Moreover, in genetic validation studies, short hairpin RNA-mediated knockdown of BATF in IMiD-resistant multiple myeloma cells was shown to be sufficient to resensitize them to IMiDs, whereas overexpression of BATF in IMiD-sensitive cells prevented IMiD-induced cell death and the downregulation of MYC expression. These findings suggest essential roles of BATF transcription factors in IMiD resistance in multiple myeloma. Finally, in a series of additional definitive studies the authors showed that: (i) overexpression of either BATF or IRF4 was sufficient to prevent IMiD-induced multiple myeloma cell death; (ii) overexpression of a mutant form of BATF that cannot heterodimerize with IRF4 (BATFH55Q) fails to prevent IMiD-induced cell death; (iii) enforced BATF or IRF4 expression prevents IMiD/p300i-induced downregulation of IRF4 and MYC, or of MYC, respectively; and (iv) BATF2 and IRF4 colocalize at IgH and the DUSP22-IRF4 SEs (10). These findings support a model whereby p300i potentiates the efficacy of IMiDs by downregulating IRF4 and MYC transcription, and that increased expression of BATF and its heterodimerization with IRF4 in IMiD-resistant multiple myeloma can compensate to overcome the antimyeloma activity of IMiDs and/or p300 inhibitors (Fig. 1).
Collectively, these studies indicate that transcriptional plasticity manifest in multiple myeloma facilitates the rapid evolutionary selection for cells that overexpress functionally redundant transcription factors (ETV4, BATF) that can bind to and sustain the activity of key oncogenic enhancers and SEs that normally require the binding and activity of IKZF1/IKZF3 for co-occupancy of coactivators such as BRD4 and p300 (Fig. 1). While these findings are viewed as a highly significant advance that suggest exciting therapeutic strategies (IMiD/p300i) and new vulnerabilities (ETV4, BATF family members) several key issues remain. First, mechanistically it is not clear how ETV4 and BATF family members are overexpressed in multiple myeloma and in IMiD-resistant disease. Specifically, as noted by the authors, although ETV4 copy-number gain occurs in 9.3% of newly diagnosed multiple myeloma cases, this is not associated with increased ETV4 mRNA levels. Furthermore, although elevated BATF2 expression might be driven by IgH translocation and SEs in multiple myeloma, translocations or other chromosomal aberrations near the BATF or BATF3 genes are not evident in multiple myeloma. Second, as noted by Neri and colleagues (9), ETV4 overexpression in IMiD-sensitive multiple myeloma cells is not sufficient to confer IMiD resistance, suggesting that not only ETV4 levels, but also the dependency on ETV4-binding enhancers, contributes to IMiD resistance. Third, the mechanisms of how BATF confers IMiD resistance (through transcriptional activation of IRF4, enhancing IRF4 binding to its motifs, etc.) needs to be resolved. Fourth, the roles of ETV4 and BATF family members in promoting the natural course of disease (i.e., MGUS to smoldering multiple myeloma to multiple myeloma) deserve investigation, as this could provide insights regarding the roles of transcriptional plasticity in disease development and transformation. Finally, and importantly, the authors findings suggest that these new mechanisms of resistance are highly selective to IMiDs, underscoring the nefarious means by which this highly plastic malignancy evades agents that target transcriptional or signaling circuits, the proteasome, and immune surveillance. As such it seems that successful treatment of drug-resistant multiple myeloma should include strategies that will restrict evolutionary trajectories (e.g., drugs that will fix the epigenetic or metabolic state) to ensure that promising combination treatments such as IMiDs plus p300i show the most benefit.
Authors’ Disclosures
J.L. Cleveland reports grants from NCI outside the submitted work. No disclosures were reported by the other author.
References
- 1. Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, et al. Diagnosis and management of multiple myeloma: a review. JAMA 2022;327:464–77. [DOI] [PubMed] [Google Scholar]
- 2. Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 2014;28:1725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature 2008;454:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 2015;521:366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014;343:305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bird S, Pawlyn C. IMiD resistance in multiple myeloma: current understanding of the underpinning biology and clinical impact. Blood 2023;142:131–40. [DOI] [PubMed] [Google Scholar]
- 8. Holien T, Våtsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood 2012;120:2450–3. [DOI] [PubMed] [Google Scholar]
- 9. Neri P, Barwick BG, Jung D, Patton JC, Maity R, Tagoug I, et al. ETV4-dependent transcriptional plasticity maintains MYC expression and results in IMiD resistance in multiple myeloma. Blood Cancer Discov 2024;5:56–73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Welsh SJ, Barwick BG, Meermeier EW, Riggs DL, Shi C-X, Zhu YX, et al. Transcriptional heterogeneity overcomes super-enhancer disrupting drug combinations in multiple myeloma. Blood Cancer Discov 2024;5:34–55 . [DOI] [PMC free article] [PubMed] [Google Scholar]