Summary
Homologous recombination deficiency (HRD) is a predictive biomarker for poly(ADP-ribose) polymerase 1 inhibitor (PARPi) sensitivity. Routine HRD testing relies on identifying BRCA mutations, but additional HRD-positive patients can be identified by measuring genomic instability (GI), a consequence of HRD. However, the cost and complexity of available solutions hamper GI testing. We introduce a deep learning framework, GIInger, that identifies GI from HRD-induced scarring observed in low-pass whole-genome sequencing data. GIInger seamlessly integrates into standard BRCA testing workflows and yields reproducible results concordant with a reference method in a multisite study of 327 ovarian cancer samples. Applied to a BRCA wild-type enriched subgroup of 195 PAOLA-1 clinical trial patients, GIInger identified HRD-positive patients who experienced significantly extended progression-free survival when treated with PARPi. GIInger is, therefore, a cost-effective and easy-to-implement method for accurately stratifying patients with ovarian cancer for first-line PARPi treatment.
Keywords: homologous recombination deficiency, HRD, low-pass whole-genome sequencing, lpWGS, convolutional neural network, ovarian cancer, breast cancer, cancer, PARPi, biomarker
Graphical abstract

Highlights
-
•
Implementation hurdles restrict adoption of HRD testing, a PARPi response biomarker
-
•
GIInger leverages HRD-induced genome scarring from low-coverage sequencing data
-
•
GIInger accurately stratifies ovarian cancer patients for PARPi treatment
-
•
GIInger is cost effective and seamlessly integrates into BRCA status workflows
Pozzorini et al. describe GIInger, a deep learning method leveraging low-coverage sequencing data to identify HRD-induced scarring. Their multicenter study demonstrates reproducibly high concordance with the reference method. GIInger is a cost-effective method to identify patients with PARPi-responsive ovarian cancer.
Introduction
Dysregulation of the mechanisms that safeguard genome integrity, such as the homologous recombination repair (HRR) pathway, is a common hallmark of cancer.1 Such dysregulation can be exploited to induce cancer cell death,2 as illustrated by the response of HRR-deficient (HRD) tumors to poly(ADP-ribose) polymerase 1 inhibitors (PARPi).3 PARPis induce the formation of DNA double-strand breaks (DSBs) and cause synthetic lethality in HRD cells that are unable to repair this type of DNA damage.4
PARPi revolutionized the management of patients with BRCA-mutated or HRD-advanced ovarian cancer, significantly increasing life expectancy.5,6,7 However, the diversity of loss-of-function events in HRR genes (including BRCA1 and BRCA28,9,10) seen in PARPi responders11 renders patient stratification based only on genotyping information challenging. HRD detection can also rely on identifying the consequences of the loss of HRR function, including increased frequency of loss of heterozygosity (LOH),12 telomeric-allelic imbalance (TAI),13 and large-scale state transitions (LSTs),14 and on the identification of the specific mutational signatures15,16,17 associated with loss of this pathway function. Methods that integrate multiple classes of mutational and genomic scarring signatures, such as HRDetect,18 are among the most accurate in predicting HRD status in breast cancer. However, identification of mutational signatures with these methods requires high-coverage (>30×) whole-genome sequencing (WGS) data from tumor-normal pairs, which is costly, hard to implement in routine testing, and only validated on fresh frozen tissue, while remaining challenging with formalin-fixed paraffin-embedded (FFPE) tissues, limiting the current use of WGS for cancer patient management.19
The combined number of LOH, LST, and TAI events detected in the tumor genome reflects the level of HRD-caused genomic instability and can be combined into the genomic instability score (GIS).20 GIS can be used to determine the patient’s HRD status and identify PARPi responders with high confidence from tumor-only data.21 However, detecting LOH and TAI requires deep genomic profiling data. Alternative methods that rely on the detection of copy-number changes, including LST events, from WGS at low (∼1×) sequencing depth (low-pass WGS [lpWGS]) can also predict tumor HRD status22,23 and provide a cost-effective and easy-to-implement HRD detection alternative. However, the sensitivity of methods that solely rely on this type of genomic scar to identify HRD samples is limited, and their utility in a clinical context remains untested.23
Unlocking the full potential of lpWGS in HRD detection will require going beyond the enumeration of biomarker events by using alternative features that result from the cell’s inability to repair DNA damage. For example, specific genomic regions are more frequently affected by DSBs24 and other mutational signatures.25 We hypothesized that differences in the spatial distribution of HRD-related genomic scars could be exploited for patient stratification by analyzing the shallow sequencing coverage profiles derived from lpWGS data. Therefore, we developed “GIInger,” a novel convolutional neural network (CNN)-based method that leverages genomic-scar-induced differences between coverage profiles obtained from lpWGS data of HRD-positive and -negative samples. The model computes a genomic instability index (GII), which is predictive of a sample’s HRD status.
We validated GIInger using lpWGS data from fresh frozen breast cancer samples and demonstrate that it supports HRD prediction with accuracies similar to methods that rely on >30× deeper sequencing data. Furthermore, in a multicenter setting, we evaluated GIInger’s analytical performance on formalin-fixed parafin-embeded ovarian cancer samples. We show that it yields robust and reproducible results across laboratories and is highly concordant with the reference method. Retrospective analysis of a cohort of patients with ovarian cancer demonstrated GIInger’s ability to stratify patients according to their PARPi response, providing preliminary evidence for its clinical utility. In clinical routine, tests integrate information on BRCA status with HRD genomic scars since the enumeration of genomic scars alone can lead, for example, to false positive calls.8 In this study, we demonstrate how GIInger can generate accurate results and be easily integrated into an end-to-end workflow that, in addition to genomic scarring, also supports BRCA status analysis.
Results
Development of a CNN model for HRD detection from lpWGS coverage profiles
To obtain lpWGS (∼1×) data, we down-sampled high-coverage (∼30–40×) publicly available WGS data from 100 HRD-positive and 174 HRD-negative (as classified by HRDetect18) fresh frozen (FF) breast cancer samples to a uniform depth of 10 million mapped paired-end reads (i.e., fragments). We observed that the normalized coverage profiles of HRD-positive samples, including those with somatic or germline loss-of-function mutations in BRCA1 or BRCA2, were distinct from HRD-negative samples (Figure S1). Consistent with the heterogeneous consequences of HRD-induced GI across the genome, the frequent and sharp shifts in coverage observed in the genome of HRD-positive samples were heterogeneously distributed across the coverage profiles (Figure S1).
We reasoned that a machine learning algorithm would be suitable to leverage differences caused by genome instability on the coverage profiles obtained from lpWGS data to classify HRD-positive and -negative samples. The development of algorithms to detect clinically relevant genomic biomarkers is often limited by the relatively small number of well-characterized clinical samples available for training. Therefore, we focused on image classification algorithms, particularly CNN, that require fewer parameters than other machine learning methods, thus limiting the risk of overfitting.26 To further overcome the challenge posed by the relatively small number of samples available for training, we implemented a data augmentation strategy to increase the number and diversity of samples in our training dataset. Augmented training data were generated by randomly sampling chromosomes from samples sharing the same purity/ploidy within a subset (173 breast cancer samples; 61 HRD positive, of which 47 were BRCA mutated) of the original data18 (Figure S2; details in STAR Methods). The normalized coverage profiles from the data-augmented samples (Figure S3) display the characteristic chromosome- and HRD-status-specific coverage patterns obtained for the original samples (Figure S1).
We used 3,933 samples (173 patient derived and 3,760 data augmented) to train a CNN model using a supervised learning framework that inputs lpWGS information to classify samples according to their HRDetect18 HRD status. We attributed the same HRD status to the data-augmented samples as the one for the patient-derived samples at the origin of their assembly (refer to STAR Methods). As previously noted, genome instability caused by HRD is not observed homogenously across the genome. For example, LSTs occurring at chromosome centromeres correlate poorly with genome instability caused by HRD.14 We thus hypothesized that retaining information on where changes in coverage occur in the genome by spatially arranging data from autosomes would facilitate the identification of genome instability patterns specific to HRD by the CNN model. We created two-dimensional heatmaps of the normalized coverage data where each row corresponds to one autosome, ordered from 1 to 22, and each column represents a genome bin of ∼3 Mbp, aligned to its respective centromere (Figure 1B). Throughout the article, we refer to these heatmaps as smoothed coverage profiles. The CNN model “GIInger,” which inputs smoothed coverage profiles, was trained following a 5-fold cross-validation procedure. The CNN contains 3 convolution blocks that output a scalar, maximizing the differences between HRD-positive and -negative samples in the training set (Figure 1C; see STAR Methods).
Figure 1.
GIInger predicts HRD status using spatially organized coverage profiles from lpWGS data
(A) Study overview.
(B) Example of GIInger input. Heatmap of the smoothed normalized coverage across ∼3 Mbp bins (columns) for all autosomes (rows) aligned with respect to their centromere (vertical dashed line). Bins are colored (blue-white-red scale) based on their normalized coverage relative to the mean coverage of the sample (set to 1, white). Color scale is depicted on the bottom. NaN (“not a number”) refers to non-existing relative chromosome locations.
(C) GIInger architecture schematic. Input features are extracted through a series of convolution and pooling operations. The vector containing the 48 extracted features is provided to a set of fully connected layers trained to output the GIInger score, which predicts the sample’s genomic instability (GI) status based on the score threshold.
GIInger validation in FF breast cancer samples
To evaluate GIInger’s performance, we used WGS data for 101 FF breast cancer samples18 that were not used to train the algorithm. We randomly down-sampled sequencing libraries, originally at ∼30–40× coverage, to the equivalent of ∼1× coverage and used GIInger to predict the HRD status based on the corresponding lpWGS smoothed coverage profiles.
Comparing score distributions between samples revealed GIInger’s ability to distinguish breast cancer samples classified by HRDetect18 as HRD positive or negative (Figure 2A). When this analysis was extended to other HRD classification methods, we found that the GIInger scores classified as either HRD-positive or -negative were distinct and corresponded well to the relative comparator methods, supporting the relevance of HRD classifications based on GIInger (Figure S4).
Figure 2.
GIInger yields comparable results in predicting HRD status to tools that rely on high-coverage datasets
(A) 101 breast cancer samples (x axis) and GIInger scores (y axis), ordered by GIInger score and colored according to their HRD status according to HRDetect as positive (blue) and negative (gray) and patterned according to their BRCA status as mutated (cross-hatching) and wild type (no hatching).
(B) ROC curves for breast cancer HRD classification obtained using GIInger (red), LST (orange), GIS (green), LOH score (pink), and BRCA status (blue) using HRD status reported by HRDetect as reference. AUC values for each method are given within the insets.
To quantitatively evaluate GIInger’s performance in distinguishing positive and negative HRD status and to compare it with other HRD detection methods, we determined the area under the curve (AUC) of the model’s receiver operating characteristic (ROC), allowing us to assess classification performance of the method at all thresholds (see STAR Methods). Classification performance of HRD status in breast cancer was most similar (AUC = 0.996) to HRDetect, used here as ground truth (Figure 2B). GIInger’s concordance with HRDetect classification was higher than its concordance with GIS classification (AUC = 0.951), which is based on the enumeration of HRD biomarker events LST, TAI, and LOH. In addition, we found lower concordance with other methods that explore individual features previously associated with HRD. In particular, the concordance with the number of LST events, which can also be computed using lpWGS data,23 was lower than what was obtained with GIInger, indicating that the latter leverages HRD-relevant features from lpWGS data that are distinct from LST. For predicting BRCA1/2 status in breast cancer samples, GIInger yielded the highest AUC (AUC = 0.858) after the best-performing method HRDetect (AUC = 0.862) (Figure S5).
These analyses show that the GIInger score is generally predictive of HRD status. In order to allow classification of individual samples, we defined a GIInger score classification threshold. To aid interpretability, we shifted the score so that samples with a GIInger score higher than 0 are classified as HRD positive and the remaining samples as HRD negative (see STAR Methods section convolutional neural network for details on threshold definition).
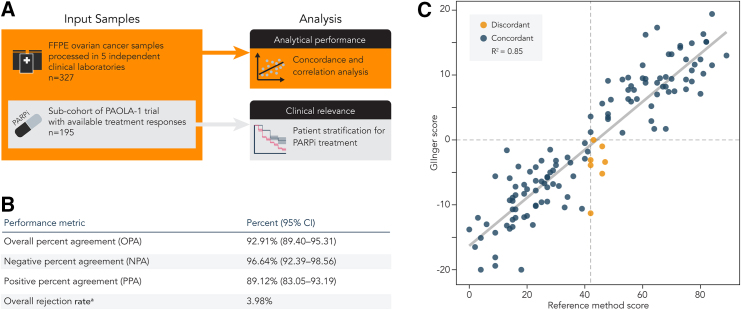
GIInger accurately detects genomic scars caused by HRD in clinical samples
Next, we aimed to establish the value of GIInger for HRD detection in the clinical setting. To evaluate its analytical performance, lpWGS data from 327 FFPE ovarian cancer clinical samples were generated in 5 independent clinical laboratories (Figure 3A; STAR Methods). Supporting the robustness of GIInger’s approach, we found that data for only 3.98% (13) samples did not pass GIInger’s quality assessment, resulting in an undetermined GIInger score (Figure 3B; STAR Methods). This rejection rate is 2 times lower (two-tailed Fisher’s exact test, p < 0.05) than what was observed for the 327 samples for which HRD status had also been tested using the reference method (7.95%, 26 samples).
Figure 3.
GIInger analytical performance analysis in clinical samples
(A) Representation of the multicenter study design.
(B) Concordance of GI status between GIInger and the reference method (n = 296 samples). For details on rejection criteria, see STAR Methods.
(C) GIInger scores (y axis) relative to the corresponding reference method GI score (x axis). Each point corresponds to a cancer patient sample (n = 125) and is labeled according to the GI status concordance between the two methods. Dashed lines indicate respective classification thresholds of GIInger and the reference method. Solid line indicates linear regression best fit; R2 = 0.85.
We found that the overall percent agreement (OPA) between classifications obtained using GIInger and the reference method was high at 92.91% (89.40–95.31, 95% confidence interval [CI]). Negative percent agreement (NPA) was 96.64% (92.39–98.56, 95% CI), and positive percent agreement (PPA) was 89.12% (83.05–93.19, 95% CI) (Figure 3B). In addition, and for the subset of samples in the cohort for which the GI score, obtained using the reference method, was known (38.4%, 125 samples), we found that GI and GIInger scores were highly correlated (R2 = 0.85; Figure 3C; STAR Methods). The high classification concordance of GIInger with a reference method that relies on the enumeration of HRD genomic biomarkers (including TAI and LOH events), which are not accessible from lpWGS data, demonstrates GIInger’s ability to leverage lpWGS data for accurate HRD detection in clinical samples. Most classification discordances are attributed to samples with scores located close to the respective classification thresholds (Figure 3C). When samples with a score close to the classification threshold, for example, ±4% or ±5%, the score dynamic range (the difference between the largest and smallest values a score can assume), were not considered, the OPA increased to 99.1% and 100%, respectively (Figures S6A and S6B). These discordant results likely reflect a general limitation of sample stratification methods that convert a continuous feature, i.e., the level of genome instability, into a binary classification, i.e., HRD-positive or -negative status, to support decision-making in the clinical setting.
Whereas more extensive reproducibility analysis is still required, no significant difference in OPA compared to the reference method was found across different clinical laboratories (Figure S6C), indicative of the method’s robustness.
GIInger can support patient stratification in the clinical setting
To gain initial insights regarding the value of GIInger in identifying patients who are likely to respond to PARPis, we generated and analyzed lpWGS data from a subcohort of ovarian cancer patient samples from the PAOLA-1 study.7 The PAOLA-1 (ClinicalTrials.gov: NCT02477644) was a randomized, double-blind, phase 3 trial investigating the benefit of the PARPi olaparib as first-line maintenance treatment (bevacizumab) in patients with advanced ovarian cancer. The PAOLA-1 study demonstrated that maintenance treatment with PARPi specifically benefited the progression-free survival (PFS) of patients with HRD-positive ovarian cancer.7
From the total PAOLA-1 cohort of 806 FFPE patient samples, we analyzed a subset of 195 samples. We chose the sample subset such that the median patient age, the ratio of olaparib/placebo, and the ratio of HRD-positive/-negative samples were identical between the subset analyzed here and the full PAOLA-1 cohort (Figure 4A; Table S2). Testing for the presence of germline and somatic BRCA mutations is well established, and BRCA-mutated patients are eligible for PARPi treatment.27,28,29 Patients with BRCA wild-type HRD-positive tumors have also been found to benefit from PARPi treatment. For this reason, we enriched our cohort in BRCA wild-type samples from patients with ovarian cancer who are more likely to benefit from HRD testing (Figure 4A).
Figure 4.
GIInger clinical relevance analysis
(A) Properties of samples included in the full PAOLA-1 cohort and the subset included in the present study. PFS curves stratified by treatment arms (orange: olaparib plus bevacizumab; gray: placebo plus bevacizumab) in the PAOLA-1 subcohort (n = 195) according to (B) GI-positive status assigned by GIInger (HR: 0.49, 95% CI: 0.28–0.85; p = 0.01) and (C) GI-negative status assigned by GIInger (HR: 0.82, 95% CI: 0.52–1.31; p = 0.41); y axis: probability of patients being free from disease progression and death; x axis: months since randomization; dashed lines indicate median survival times; and tables of absolute number of individuals at risk are given below each plot.
We generated lpWGS libraries from DNA extracted from patient FFPE samples. We used the Kaplan-Meier estimator to evaluate the PFS in olaparib+bevacizumab- and placebo+bevacizumab-treated GI-positive and negative patients. Patients classified by GIInger as GI positive had significantly (over two times) longer PFS when receiving olaparib+bevacizumab relative to those receiving placebo+bevacizumab (median 33.3 months, 95% CI: 21.9–NA, versus median 15.9 months, 95% CI: 13.4–38.6; hazard ratio (HR): 0.49, 95% CI: 0.28–0.85; p = 0.01) (Figure 4B).
In contrast, there was no significant difference in PFS between treatment arms for patients with negative GI status (olaparib group median PFS 16.8 months, 95% CI: 14.0–22.0, versus placebo group median PFS 14.6 months, 95% CI: 9.7–19.3; HR: 0.82, 95% CI: 0.52–1.31; p = 0.41) (Figure 4C). In addition, when the 8 samples that did not meet GIInger’s quality assessment are added to the set of samples classified by GIInger as negative, we see no significant difference in PFS between treatment arms in this group (non-positive, Figure S7A).
PFS analysis of the same PAOLA-1 sample subset stratified by the reference method returned highly similar results: longer PFS in the olaparib group than in the placebo group (olaparib group median PFS 29.8 months, 95% CI: 21.0–NA, versus placebo group median PFS 15.4 months, 95% CI: 13.0–38.6; HR: 0.58, 95% CI: 0.35–0.98, p = 0.04) for samples with positive GI + BRCA status (Figure S7B) and no detectable difference (olaparib group median PFS 17.6 months, 95% CI: 14.0–22.1, versus placebo group median PFS 15.1 months, 95% CI: 10.3–19.3; HR: 0.70, 95% CI: 0.42–1.19, p = 0.18) for samples with negative status (Figure S7C). In the analysis of the full PAOLA-1 cohort, HRD-positive BRCA-mutated patients displayed a significantly higher response to treatment.7 The difference in hazard ratio (HR) obtained in this study and the one reported for the full cohort7 is likely a consequence of the smaller fraction, relative to the full cohort, of samples from BRCA-mutated patients in the cohort subset analyzed here (Figure 4A).
Together, the PFS results suggest that GIInger-based classification was non-inferior to the reference method for stratifying 195 PAOLA-1 patients with ovarian cancer with regards to PARPi treatment response while relying solely on lpWGS, which is more practical to implement and less costly.
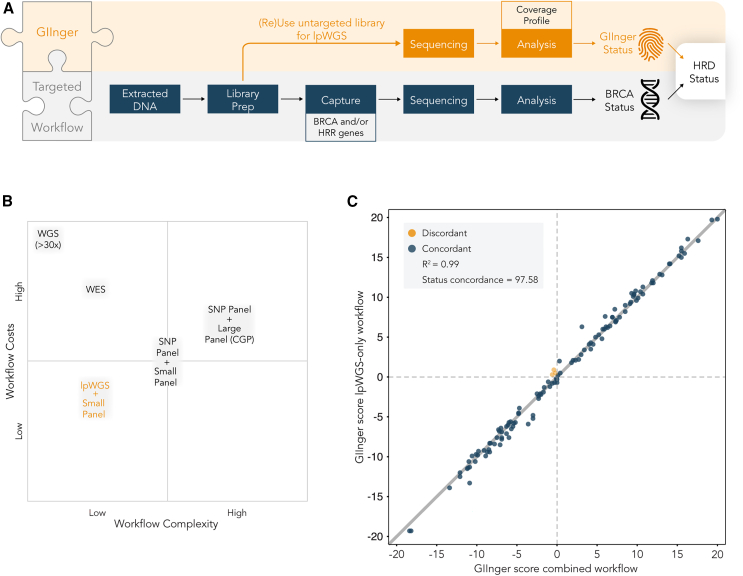
GIInger can be easily integrated in end-to-end HRD detection workflows
Integrating the identification of the genome instability scars caused by HRR deficiency with detection of pathogenic mutations in BRCA is important for the comprehensive identification of patients most likely to benefit from PARPi treatment.30 Determination of BRCA status requires variant calling and thus high sequencing depth at a multitude of positions, commonly achieved by targeted sequencing. The data used by GIInger cannot be used to support variant calling at these genes. However, because lpWGS data can be obtained by sequencing of the pre-enriched libraries used to determine BRCA status, GIInger can be easily integrated into existing workflows (Figure 5A). This feature provides further benefits relative to alternative GIS-based methods (Figure 5B; Table S3). If sequenced at sufficiently high depth, >30× for clonal high tumor content samples or even higher (∼500×) for most somatic applications, WGS data can simultaneously support the detection of genome instability and BRCA status.18 However, the costs of generating such high-coverage sequencing data outweigh the benefits of a single data generation workflow and limit the clinical implementation of WGS-based solutions. Recently, analysis of whole-exome sequencing (WES) data on a small cohort of patients was shown to allow simultaneous determination of GIS and BRCA status. However, in addition to the high cost of generating WES data, the accuracies demonstrated for such approaches relative to the reference method so far are low, further limiting their use in the clinical setting.31
Figure 5.
GIInger is a flexible and robust patient stratification method
(A) Workflow schematics illustrating flexible use of GIInger in combined workflow with existing targeted sequencing approaches. Pre-capture library aliquots can be sequenced at ∼1× depth and inform sample HRD status together with data on BRCA status.
(B) Qualitative workflow cost and complexity comparison of different methodologies for determining HRD-related biomarkers genome instability and BRCA status. CGP, comprehensive genomic profiling. Inferences are based on relative differences detailed in Table S3.
(C) Concordance of GIInger scores between lpWGS-only (y axis) and combined (i.e., targeted sequencing with HRDv1 panel + lpWGS) workflow (x axis). Each point corresponds to a PAOLA-1 cancer patient sample (n = 124) and is labeled according to the GI status concordance between the two workflows. Dashed lines represent GIInger classification threshold, and solid line indicates linear regression best fit; R2 = 0.99; overall concordance: 97.58% (95.31%, 99.55%).
Alternatively, data to determine GIS can be obtained by SNP genotyping methods, including SNP arrays32 or SNP capture.20 SNP capture workflows can be integrated with targeted sequencing methodologies, such as comprehensive genomic profiling (CGP) solutions or small panels that are commonly used to establish BRCA status. Relative to WGS/WES, these targeted approaches reduce data generation costs but elevate implementation complexity due to the need to ensure balanced sequencing depth between SNP positions and BRCA genes (Figure 5B). In contrast, GII can be obtained from low-coverage sequencing data of the pre-enrichment libraries used to determine BRCA status, removing the need for an additional capture experiment or establishing settings that support differentially enrichment of exonic and SNP regions and thereby reducing implementation complexity and data generation costs. Specifically, and due to the high coverage of sequencing required to support LOH determination, we estimate that 2 times more fragments are needed (STAR Methods) when GIS is used. Genotyping using SNP arrays, an alternative approach to determine LOH and GIS, is lengthy, requires specific detection equipment, introduces several biases that decrease repeatability, and is at least 50% more costly than lpWGS-based methods.33
We assessed the impact on result accuracy of GIInger score using an end-to-end workflow that combines genome instability detection and a capture panel used to determine BRCA status (Figure 5A) for a subset of 124 samples in the PAOLA-1 cohort. We compared the results obtained for the same samples using either an lpWGS-only (standalone) or an lpWGS and targeted capture (combined) workflow. Targeted capture was performed using a probe panel that covers a set of clinically relevant genes, including BRCA1 and BRCA2, to enrich these regions and generate data to support variant calling (STAR Methods). We found a high correlation (R2 = 0.99) between the GIInger scores obtained using the standalone and combined workflows (Figure 5C). As expected, OPA between classifications obtained using the standalone or the combined workflow was high at 97.58% (95% CI: 95.31–99.55). Among the 124 samples, only 3 (2.4%) samples, all with a GIInger score near the classification threshold, were differently classified (discordant) across the two workflows.
In addition, we also compared the BRCA status obtained using the combined workflow or target capture alone. We found a high OPA between the BRCA status obtained using the capture-only or combined workflow (99.19% [95% CI: 99.19–99.19]). In addition, the reported variant fraction for BRCA pathogenic variants, or variants of unknown significance (VUSs; Figure S8), found using data generated by the two workflows was also highly correlated (coefficient of determination, R2 = 1.0) between experiments.
These results illustrate the ease and robustness of GIInger integration with targeted capture solutions for the determination of BRCA status, providing a cost-effective and easier-to-implement alternative to other workflows.
Discussion
PARPi is an approved treatment for patients with cancer with HRD-positive tumors. Evidence that some HRD-negative patients also derive benefit, albeit to a lesser extent, from PARPi6 and that epistatic interactions34 between HR-related genes, tissue of origin,35,36 and cancer subtype37 can impact PARPi response warrant further research on biomarkers for PARPi sensitivity. However, whereas all determinants of PARPi response may not yet be known, consensus expert guidelines currently recommend HRD testing to identify patients who are likely to benefit from PARPi treatment. Most diagnostic laboratories have established standardized tests, often using targeted sequencing, to identify BRCA-mutated tumors. However, the diversity of genetic and epigenetic events underlying HRD3 limit the sensitivity of these approaches. Detection of GI increases the effectiveness of HRD testing and improves patient stratification. The most used approach to detect GI, GIS, relies on enumerating HRD biomarker events, such as LOH, TAI, and LST. However, the detection of some of these genomic scars, including LOH or TAI, requires deep genomic profiling to support variant calling. Whereas the value of HRD testing to identify which patients with ovarian cancer are most likely to benefit from PARPi is well recognized, the challenges of implementing available methods currently limit HRD testing in clinical settings.38 Therefore, we aimed to develop a cost-effective sample stratification method, named GIInger, that exploits the consequences of HRD on genome stability.
To ensure this method can be deployed in a clinical setting, GIInger leverages the impact on genomic lesions from lpWGS (∼1× coverage) coverage profiles. In addition to the reduction in data-generation costs afforded by the decrease in sequencing depth, lpWGS is also the easiest-to-implement genome profiling method, thus facilitating robust data collection. In addition, lpWGS can be obtained for pre-capture libraries used for targeted BRCA sequencing, which further simplifies its integration with established methods.
We used publicly available data to benchmark GIInger against well-established methods of HRD detection. In data from FF breast cancer samples, we found that GIInger results are most similar to those of HRDetect, which enumerates large-scale events and genomic signatures characteristic of HRD. Concordance was also high with GIS, which integrates the number of HRD biomarker events and is the most commonly used method for GI status determination in the clinical setting.20 Both these methods rely on the analysis of deep genome profiling data. The similar classification performance obtained by GIInger using datasets with lower sequencing depth indicates that this approach is a cost-effective alternative for HRD detection. Of the three biomarker genomic events commonly used in HRD testing,3 the number of LSTs can be quantified from lpWGS data and has been shown to allow sample classification according to their HRD status.23 The higher analytical performance of GIInger relative to LST we obtained in the present analysis indicates that the deep learning framework that supports GIInger goes beyond prior knowledge and expert design and supports the identification of features that distinguish HRD-positive and -negative samples using low-coverage data.
To gain initial insights into the value of the deep learning approach in the clinical setting, we set up a multicenter study to assess GIInger’s analytical performance in a cohort of 327 FFPE ovarian cancer samples. Despite having been trained using data from FF breast cancer patient samples, GIInger results were highly concordant with those of a reference method in FFPE ovarian cancer samples, suggesting that the features used by GIInger are not specific to one cancer type. The similarly high overall concordance obtained between independent centers supports the method’s robustness. As a testimony to the test’s ease of implementation, we observe an increase in the number of patients for which HRD status could be established relative to the alternative method. In addition, when the present results are compared to the results of recently developed alternative HRD detection solutions validated using a subset of the original PAOLA-1, we found that GIInger displays a higher accuracy, a lower rejection rate than the alternative tests, or both39,40,41,42,43,44 (Table S4).
GIInger shares with other methods that rely on detecting the genomic impact of HRD the limitation that these genome instability scars accumulate over time, which can result in false negatives calls due to insufficient time since the loss of HRR function or false positives due to the evolution of PARPi resistance in HRD tumors. Like for other methods, integrating the GIInger score with the results of variant-calling analysis in HRR genes should limit, to some extent, the impact of this constraint. We demonstrate how GIInger can be integrated in an end-to-end workflow that also allows determination of BRCA status from targeted capture with negligible impact on result accuracy. GIInger can be obtained from the analysis of various types of WGS data. This means that it is, in principle, possible to integrate GIInger data generation with any targeted capture method routinely used in the clinics to identify variants in HRR genes, including BRCA1 and BRCA2, and establish BRCA status. This ease of integration with targeted sequencing workflows already routinely used adds a further advantage to the decrease in implementation and data generation costs afforded by the sufficiency of using lpWGS data to establish genome instability using GIInger.
Providing initial support of GIInger’s value in the clinical setting, we found in a cohort of 195 samples that responses to PARPi treatment of patients differed depending on the GI status as classified by GIInger. PFS of patients classified by GIInger as HRD positive and receiving PARPi olaparib (+bevacizumab) was extended relative to their placebo (+bevacizumab) counterparts. This contrasts with patients classified by GIInger as HRD negative who presented similar PFS as the placebo control group. These preliminary results support the use of GIInger for patient stratification. It is important to note that in this study, GIInger has been tested on retrospective data and found to be effective in determining whether a patient will benefit from PARPi. Future randomized trials on larger cohorts are required to fully establish its clinical utility.
In summary, GIInger is a deep learning method that allows accurate stratification of HRD samples, including FFPE, based on lpWGS data. The accuracy of this method is comparable to that obtained using well-established methods that typically require genomic data that are either more expensive or harder to obtain, making this a promising solution for standardized HRD testing in the clinical setting.
Limitations of the study
Similar to other methods that rely on the evaluation of genomic scarring caused by HRD, GIInger alone does not provide real-time information regarding HR function in patients. For example, patients where reversal mutations lead to PARPi resistance would result in false positive calls for clinical response if genome scarring alone was used to predict response to treatment. This limitation can be accounted for by integrating information on HRR gene status, particularly BRCA1 and BRCA2, or by using methods that measure HRR function.
In addition, and like other methods that use genomic scarring caused by loss of HRR function to stratify patients, analysis of samples with low tumor content may lead to false negatives due to the reduced signal coming from tumor cells. Approaches that allow flagging of results from samples with likely low tumor content minimize the impact of this limitation.
Further work is now required to assess the value of GIInger in other cancer types, such as prostate cancer,45 where PARPi has already been shown to be valuable as the first line of treatment for HRD-positive patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Maxwell® CSC DNA FFPE Kit | Promega | Cat#AS1350 |
| Maxwell® 16 FFPE Plus LEV DNA Purification Kit | Promega | Cat#AS1135 |
| GeneRead™ DNA FFPE Kit | Qiagen | Cat#180134 |
| AllPrep DNA/RNA FFPE Kit | Qiagen | Cat#80234 |
| Qubit™ dsDNA HS Quantification Assay Kit | Thermo Fisher Scientific | Cat#Q32851 |
| SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution | SOPHiA GENETICS | Cat#BS0121ILLCSMY08-32 |
| Myriad myChoice® CDx HRD Companion Diagnostics test | Myriad Genetics | N/A |
| Deposited data | ||
| Breast cancer WGS FF sample data and associated patient information | European Genome-phenome Archive (EGA) | EGA: EGAS00001001178 |
| HRD scores for breast and ovarian cancer samples | Davies et al.18 | N/A |
| PAOLA-1 ovarian cancer clinical trial data with available treatment responses | Ray-Coquard et al.7 | N/A |
| Software and algorithms | ||
| biopython 1.78 | Python | https://biopython.org/ |
| cython 0.29.2 | Python | http://cython.org/ |
| h5py 2.10.0 | Python | http://www.h5py.org |
| keras 2.4.3 | Python | https://github.com/keras-team/keras |
| lifelines | Python | https://github.com/CamDavidsonPilon/lifelines |
| matplotlib-base 3.3.4 | Python | https://matplotlib.org/ |
| more-itertools 8.0.5 | Python | https://github.com/more-itertools/more-itertools |
| numpy 1.18.5 | Python | https://www.numpy.org |
| pandas 1.2.0 | Python | https://pandas.pydata.org/ |
| pillow 8.1.0 | Python | https://python-pillow.org |
| pybedtools 0.8.1 | Python | https://github.com/daler/pybedtools |
| pylatex 1.4.1 | Python | https://github.com/JelteF/PyLaTeX |
| pysam 0.15.3 | Python | https://github.com/pysam-developers/pysam |
| pytest 6.1.1 | Python | https://docs.pytest.org/en/latest/ |
| python 3.7.6 | Python | https://www.python.org/ |
| scikit-learn 0.24.0 | Python | http://scikit-learn.org |
| scipy 1.4.1 | Python | https://www.scipy.org |
| seaborn 0.11.1 | Python | https://seaborn.pydata.org |
| statsmodels 0.12.1 | Python | https://www.statsmodels.org/stable/index.html |
| tensorboard 2.4.1 | Python | https://github.com/tensorflow/tensorboard |
| tensorflow 2.3.0 | Python | https://www.tensorflow.org/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Zhenyu Xu (zxu@sophiagenetics.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
WGS data from FF breast cancer samples and associated patient information data were downloaded from the European Genome-phenome Archive (EGA), and the accession number is listed in the key resources table. They are available upon request if access is granted. To request access, contact EGA and apply for access via the ICGC Data Access webpage.
-
•
The data generated for FFPE ovarian cancer clinical samples reported in this study cannot be deposited in a public repository because they were used under license for the current study and therefore are not publicly available. To request access, contact the authors of the PAOLA-1 study7 and/or the relevant contacts (for details, see list of authors and affiliations) of the following institutions: Vienna General Hospital, Center Léon Bérard, European Institute of Oncology, Istituto Nazionale Tumori Pascale, and Institut de Cancérologie de Lorraine.
-
•
GIInger is available as part of the SOPHiA GENETICS SOPHiA DDM platform.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Helsinki Declaration. All patients involved in the study gave their informed consent, and the study was approved by:
-
•
The institutional ethics board of the Institut de Cancérologie de Lorraine.
-
•
The Medical University Vienna (2295/2020 positive vote),
-
•
The European Institute Review Board (UID 2386),
-
•
The Ethical Committee of the Pascale Institute,
-
•
The Comité de Protection des Personnes (EudraCT number: 2014-004027-52; reference: GINECO-OV125b/ENGOT Ov25).
In the original FF breast cancer sample set,18 4 out of 560 samples were derived from male patients, reflecting the sex-related incidence rate in the general population. The subset used in the present study included a single male sample (PD18730) which was used in the test dataset. While both GIInger and HRDetect reported the same GI status for this sample, the absence of male samples in the training dataset possibly limits GIInger’s explanatory power for this sample group. Due to its sex-specific incidence, ovarian cancer FFPE samples were exclusively derived from female patients.
Method details
DNA extraction
The sample tumor content was obtained by estimating the percentage of tumor cells on hematoxylin and eosin-stained slides. Methods for extracting DNA from FFPE tissue sections used according to the manufacturer’s instruction in the multicenter study include: Maxwell CSC DNA FFPE Kit (Promega; Cat#AS1350), GeneRead DNA FFPE Kit (Qiagen; Cat#180134), AllPrep DNA/RNA FFPE Kit (Qiagen; Cat#80234), and Maxwell 16 FFPE Plus LEV DNA Purification Kit (Promega; Cat#AS1135).
DNA was quantified using the Qubit dsDNA HS Quantification Assay Kit (Thermo Fisher Scientific; Cat|Q32851).
DNA quality was assessed by analyzing the fragment size distribution of the sample using the Fragment Analyzer (Agilent). The DNA quality number (DQN) for each sample was determined as the fraction of DNA fragments larger than 300 bp using the DQN function of the Fragment Analyzer software, with the DQN fragment length threshold set to 300 bp.
At least 50 ng DNA extracted from representative FFPE tumor tissue blocks (except one sample where 44 ng were reported) were used for library preparation. The clinical samples’ properties are listed in Table S1.
Library preparation and sequencing
Whole-genome and targeted libraries were prepared using SOPHiA GENETICS library preparation kit LPIII. First, 50 or 100 ng of FFPE DNA input was fragmented, end-repaired, and A-tailed, followed by ligation to Illumina-compatible adapters. Ligation products were purified using AMPure beads (Beckman Coulter) and amplified by PCR for 8 or 10 cycles. Amplified libraries were cleaned up using AMPure beads. The obtained libraries were used either at this step as WGS libraries or further enriched for targeted sequencing as follows: Libraries were pooled and mixed with human Cot-1 DNA (Life Technologies) and xGen Universal Blockers-TS Mix oligos (Integrated DNA Technologies) and lyophilized. Pellets were resuspended in a hybridization mixture, denatured for 10 min at 95°C, and incubated for 16 h at 65°C in the presence of biotinylated probes (xGEN Lockdown IDT). The probe panel spanned 157 kb and covers a set of clinically relevant genes, including genes implicated in HRR-related genes. Probe-hybridized library fragments were captured with Dynabeads M270 Streptavidin (Invitrogen) and subsequently washed. The captured libraries were amplified by PCR for 15 cycles and cleaned up using AMPure beads (Beckman Coulter).
Whole-genome sequencing libraries were sequenced to approximately 10 M fragments per sample. When whole-genome and targeted libraries were sequenced for the same sample, libraries were loaded to a flow cell in a 67–33% ratio and sequenced to approximately 16 M fragments per sample corresponding to median ∼1–2x whole-genome coverage and ∼5500x coverage in the targeted regions using Illumina instruments (NovaSeq 6000, NextSeq 550, or NextSeq 550 Dx) with 150 bp paired-end reads. Results for the analysis of the target-enriched libraries are not included in the present study. With the exception of the 124 samples for which whole-genome sequencing and targeted capture were also generated separately and for the purpose of comparison of the results between combined and standalone data, the results presented in this study were obtained using the SOPHiA DDM Dx Homologous Recombination Deficiency Solution (SOPHiA GENETICS; Cat#BS0121ILLCSMY08-32).
Clinical sample sequencing data processing and interpretation
Read alignment to the reference human genome (hg19), read filtering, and variant calling was performed using the SOPHiA GENETICS analysis workflow. Briefly, after read mapping adaptors were trimmed, mis-priming events were removed and read softclipped regions were realigned. Read fragments shorter than 21 bp were excluded. The resulting alignment was used for variant calling using the SOPHiA GENETICS pipeline and to prepare coverage profiles.
Processing of publicly available WGS data for training GIInger
BAM files for WGS data for FF breast and ovarian cancer with BRCA-deficiency status were downloaded from the Wellcome Trust Sanger Institute and the International Cancer Genome Consortium ICGC available at the European Genome-phenome Archive EGA (https://www.ebi.ac.uk/ega/studies/EGAS00001001178). This cohort was divided into training (173 breast cancer samples) and test sets (101 breast cancer samples). We obtained HRD scores for these samples from the HRDetect publication.18
Coverage profile preparation
The human reference genome (hg19) was divided into contiguous non-overlapping intervals of 100 kb (hereafter referred to as “bins”). Bins containing blocklisted regions, including regions known to introduce bias in coverage, were excluded. These regions included: i) centromere and telomere positions as obtained from UCSC for hg19, ii) genome assembly gaps in the reference genome (N’s), iii) regions with extreme GC content and high mappability, and iv) sex chromosomes. In the case of NGS data generated in a workflow combining both lpWGS and targeted sequencing (an optional step in SOPHiA DDM Dx HRD solution), bins overlapping with enriched regions or regions highly homologous to enriched regions were also discarded. The raw coverage count for each bin was obtained by counting the number of mapped pair-end reads (fragments) to the respective bin. Fragments mapping across two bins were assigned to the bin with the largest overlap. Only unambiguously mapped fragments were considered in the coverage calculation.
For TCGA datasets and to mimic lpWGS, datasets were down-sampled to 10 million pair-end reads (∼1x coverage). The probability of observing k mapped fragments within a given genomic bin according to the multinomial distribution was considered:
where n denotes the number of fragments after down-sampling and p is the prior probability that a fragment drawn will fall within the considered bin of the coverage profile defined as p = Cbin/Ctot, where Cbin and Ctot is the observed coverage for the bin considered and the sum of the raw coverage across all bins, respectively.
Coverage across 100kb bins was normalized by dividing the raw coverage count for the bin by the sample’s mean coverage. We fitted a lowess regression function to the raw coverage versus GC content of the bins and obtained corrected normalized coverage by dividing the normalized coverage by the result of the lowess fit.
To generate the input matrix of the CNN, we further smoothed the 100kb corrected normalized coverage, per chromosome arm, by considering the median corrected normalized coverage across non-overlapping smoothing windows (in 5′ to 3′ direction). The smoothing window width (between 2.5 and 3.5Mbp) used for each chromosome arm was chosen to minimize the size of the last incomplete smoothing bin. The normalized coverage information in this last incomplete smoothing bin was not included in the CNN input matrix.
We arranged the smoothed normalized coverage bins into a 2D array with a width of 87 and a height of 22 bins. The width of the array corresponds to the sum of the maximum number of smoothed bins seen in chromosome p- and q-arms. The height corresponds to the number of autosomes. Smoothed normalized coverage bins for the 22 autosomes were aligned with respect to their centromeric bins. For each chromosome, we attributed undefined values (e.g., “Not A Number” float values) to the bins (undefined bins) that would extend the chromosome size beyond the end of the chromosome’s telomere position. We calculated the mean and the standard deviation across all defined smoothed normalized coverage bins. Next, we used backward and forward filling to assign values to undefined bins in each chromosome’s p- and q-arm, respectively. Finally, we applied a Z score transformation to the entire spatially arranged smoothed normalized coverage matrix using the mean and standard deviation calculated using defined bins.
Data augmentation
173 breast cancer samples were considered in the training set (35% HRD-positive, 27% BRCA1/2 mutated) to generate the data augmented (DA) dataset. To generate one DA sample, 22 autosomes were randomly sampled from N original tumor samples with the same HRD status (Figure S2A). N is a number between 2 and 22 drawn from an exponential distribution with an associated probability density function:
where denotes the mean and setting it to 3 expands the tail of the probability density function to higher range which ultimately allows us to include more samples in the mix: .
We accounted for the impact of sample-specific differences in purity and ploidy by normalizing the purity/ploidy ratio (m) of all samples (N) used to generate one data augmented sample. Among N, a sample with minimal m was determined, and the other samples were in silico diluted (decreased the sample purity) until their purity/ploidy ratio was equal to m (Figures S2B and S2C). Purity and ploidy information for each sample was provided in the HRDetect manuscript.18
DA samples (Figure S2D) were obtained by combining chromosomes from samples with normalized purity/ploidy ratio, ensuring the amplitude of coverage differences observed for a given ploidy was constant across chromosomes. This approach introduced biases in the purity/ploidy ratios of DA samples that tended to be lower than what was observed in the original samples (Figure S2E). To account for this bias, we applied a Metropolis-Hastings and Gibbs sampling method and ensured that the purity/ploidy distribution of the 3,760 retained DA training samples matched that of the original samples (Figure S2F). The rejection sampling algorithm stopped when no new samples could be drawn after 100,000 iterations. This approach ensured that after data augmentation, the properties of the training data reflected those of the real data.
Convolutional neuronal network
173 patient-derived and 3,760 data augmented samples were used in training the CNN model (GII) to predict a sample’s HRD status. The input to GII were smoothed coverage profiles. A 2-dimensional heatmap of the smoothed normalized coverage depth was obtained for the sample (see Figure 1A). Each row in the heatmap corresponds to one autosome, ordered from 1 to 22, and each column is a genome bin of 3Mbp (ordered from 5′ to 3′). Chromosomes were aligned with respect to their respective centromeres.
The GIInger architecture included 3 convolution blocks (convolution, batch normalization, and max-pooling) whose output was aggregated by an average pooling layer. The 48 features extracted by these convolution steps were flattened into a 1D feature vector and passed to a fully connected layer with a single output node (whose output is a scalar) that was passed through a corresponding sigmoid activation function. Please note that throughout this study, we are referring to the CNN output’s pre-sigmoid score, which has the same predictive power as the post-sigmoid score but allows easier sample discrimination, as its dynamic range is not restricted to [0, 1].
Five CNN models with the described architecture and initiated using a different random seed were trained following a 5-fold cross-validation procedure. Each model was trained using a different set of data used for training (80%) and validation (20%).
Each model was trained for a maximum of 1000 epochs with a batch size of 64 on eight Intel Xeon(R) Platinum 8168 CPU @ 2.70GHz CPUs. During training, categorical cross-entropy loss between the target and predicted outputs was minimized using Adam optimizer.46 Class weights, defined according to Equations 1 and 2, were used in the loss function to account for the difference in the number of HRD positive and negative samples available for training.
| (Equation 1) |
| (Equation 2) |
Wneg is the weight associated to HRD negative samples.
Wpos is the weight associated to HRD positive samples.
Nneg is the number of HRD negative samples available.
Npos is the number of HRD positive samples available.
The learning rate of the Adam optimizer was first set to 0.005 and reduced by a factor of 5 (minimal accepted value = 0.00001) if the decrease of loss function value was <0.0005 for 10 epochs. In addition, early stopping was triggered if the decrease in loss function value was <0.0001 over 100 epochs. Using this approach, on average, early stopping for the 5 models was triggered after 244 epochs.
When used for testing, GIInger considers each input using the 5 trained models. The GIInger output is the average of outputs of the 5 trained models used as the predicted output. Demonstrating the absence of overfitting to the training data of the GIInger CNN model, the binary cross-entropy functions obtained using the training and the validation datasets for the models are comparable (Figure S9).
To establish the GIInger classification cut-off threshold, we considered 138 clinical ovarian cancer FFPE samples with an established HRD status. The analysis results for these clinical samples that were processed as described here are not included in the present manuscript. We defined the GIInger threshold as the median cut-off value that maximized the overall percent agreement between GIInger classification and the HRD status assigned to the samples. To aid the interpretation of the GIInger scores, we subsequently shifted the threshold to 0.0, so positive GIInger scores correspond to HRD-positive status and vice versa. The score shift does not affect the analytical performance.
Code availability
The deep learning frameworks used here (TensorFlow resp. Keras) are available at https://www.tensorflow.org/resp. https://keras.io/. The Python libraries used for computation and plotting of the performance metrics (Pandas, Numpy, SciPy, Scikit-Learn, Lifelines, MatPlotLib and Seaborn) are available under https://pandas.pydata.org/, https://numpy.org/, https://www.scipy.org/, https://scikit-learn.org/stable/, https://github.com/CamDavidsonPilon/lifelines/, https://matplotlib.org/and https://seaborn.pydata.org/, respectively.
Quantification and statistical analysis
Analytical performance evaluation
Algorithm performance was assessed through the Area Under the Curve (AUC) in Receiver Operating Characteristic (ROC) profiles with either BRCA mutational status or HRD classification (according to HRDetect) as reference method using the scikit-learn python library (v.0.24.0) ROC and AUC functions (default parameters). LOH, TAI,13 and LST14 for breast FF samples used in the analysis were obtained from Supplementary Table 17 of Nik-Zainal et al.47 HRD12 scores for these samples were obtained through HRDetect manuscript.18
To establish the percent agreement between GIInger and the reference method, we classified patient samples as follows: patients classified by GIInger and reference method as GI positive and negative were classified as True Positive and Negative (TP and TN respectively); GIInger GI positive samples classified using reference method as negative were considered to be False Positives (FP); and GIInger GI negative samples classified by reference method as positive were considered to be False Negatives (FN). We determined overall percent agreement (OPA) as (TP + TN)/(TP + TN + FP + FN); Positive percent agreement (PPA) as TP/(TP + FN) and Negative percent agreement (NPA) as TN/(TN + FP).
The 95% confidence interval for the OPA was computed with respect to CLSI guideline EP12A2E:
The overall rejection rate for analytical performance evaluation was calculated as N_rejected/N_tot with N_rejected being the number of samples reported as "Inconclusive" or "Rejected" and N_tot being the total number of samples considered. The sample inclusion criteria used to select the 327 samples were the following (samples were still included if the following information was not available): sample type: FFPE; cancer type: Ovarian; DQN ≥3; tumor content ≥30%; NGS library yield ≥250 ng. Replicate samples were removed such that only the replicate with the highest coverage available for GIInger analysis was retained.
Association with clinical outcome
Progression-free survival, defined as the duration between randomization and investigator-assessed disease progression or death, was the primary endpoint of the trial PAOLA-1.7 Progression-free survival was assessed in the intention-to-treat population, defined as all patients who were randomized, regardless of the treatment received.
In each subgroup of patients (HRD-positive and HRD-non-positive), progression-free survival was compared between the olaparib group and the placebo group using the stratified log rank test. The Kaplan-Meier method was used to estimate PFS curves and median PFS times with associated 95% CIs. Hazard ratios were estimated from Cox proportional hazards models adjusted on treatment group, and associated 95% CIs and p values were provided.
To assess the association between HRD status and PFS outcome, the statistical analyses were done with R version 4.2.0 (main packages: survival v.3.3–1, survminer v.0.4.9, ggplot2 v.3.4.0).
Comparison between the number of fragments
The number of 2x150 bp fragments needed for 1x whole genome sequencing coverage is ∼10 million. Due to the requirement for determining LOH events genome-wide of GIS, this method relies on targeted capture of ∼26,500 SNPs at an average depth of >400x.48 Assuming an on-target rate between 50 and 80%, which is typical for this type of experiment, between 21.2 and 13.2 million fragments need to be sequenced to ensure appropriate coverage on these positions.
Comparison between GIInger and BRCA status between standalone and combined workflow
To establish percentage agreement between GIInger score and BRCA status obtained using data generated using lpWGS or targeted capture alone or as part of the lpWGS+capture workflow, we classified patient samples as follows: patients classified in the same way using standalone or combined workflow were classified as True Positive and Negative (TP and TN respectively); Samples classified as BRCA or GI positive using the standalone method and as negative using the combined workflow were considered to be False Negatives (FN); Samples classified as BRCA or GI negative using the standalone method and as positive using the combined workflow were considered False Positives (FP).
Twenty (20) variants of unknown significance (VUS) or pathogenic variants in BRCA were reported by SOPHiA DDM (VAF> 5%) in the 124 samples analyzed using data collected from either standalone or combined workflows. Nineteen (19, 95%) were found using data collected using both workflows. One variant of unknown significance was detected in the combined workflow (VAF = 5%), but not in the capture-only workflow was below 5%. This variant was excluded from the analysis of variant fraction concordance between BRCA variants found using the 2 workflows.
Acknowledgments
We thank Jean-Baptiste Mignardot for his help with figure design and Niamh O’Connor for supporting the assembly of the manuscript. We thank Life Science Editors for editorial assistance. The authors received no specific funding for this work.
Author contributions
G.A., T.C., C.P., and Z.X. conceived and planned the study. G.A, T.C., and C.P. developed GIInger. G.A., T.C., J.B., and L.F. performed statistical analysis. C.P., G.A., T.C., J.B., and L.F. had unrestricted access to data. A.B., P.S., A.H., D.V., M.B., P.G., C.R., N.N., C.G., L.M., P.H., S.P., A.G.-M., U.D., K.F., I.V., N.C., E.P.-L., P.-A.J., and I.R.-C. conducted the selection, collection, and characterization of ovarian cancer samples. A.B. obtained ethical approval for the use of the clinical samples for this study and provided clinical expertise. A.B., A.S., E.S., F.C.B., L.R., C.R., and R.K. generated next-generation sequencing (NGS) data. G.A., T.C., J.B., A.A., and R.M. analyzed NGS data. L.F. analyzed the association between GIInger status and clinical outcome. M.F. and A.C.M. wrote the first draft of the manuscript and reviewed and edited it. C.P., A.W., and Z.X. coordinated the study. All authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility for its content, including the accuracy of the data.
Declaration of interests
G.A., T.C., C.P., J.B., R.K., L.F., A.S., F.C.B., L.R., A.A., R.M., M.F., A.C.M., E.S., A.W., and Z.X. are SOPHiA GENETICS employees. A.H. received consultant fees from SOPHiA GENETICS and honoraria from AstraZeneca, Janssen, and GSK. P.G. received honoraria from AstraZeneca. M.B. has received honoraria for consulting, advisory role, speakers’ bureau, travel, accommodation, and expenses from MSD Oncology, Roche/Genetech, AstraZeneca, Thermo Fisher Scientific, and Illumina. S.P. has received honoraria and research funding from AstraZeneca, MSD, GSK, and Roche and honoraria from Clovis. E.P.-L. served on a data safety monitoring board for Agenus Incyte, as a consultant for Roche, and as an advisory board member for AstraZeneca and GSK. I.V. as a consulting or advisory role at AstraZeneca. P.-A.J. has conducted teaching events for GSK, Roche, and EISAI. N.C. has reported fees for advisory board membership for AstraZeneca, Clovis Oncology, Eisai, GSK, Immunogen, Mersana, MSD/Merck, Nuvation Bio, Onxerna, Pfizer, Pieris, and Roche; fees as an invited speaker for AstraZeneca, Novartis, Clovis Oncology, GSK, and MSD/Merck; and institutional research grants from AstraZeneca and Roche. She has also reported non-remunerated activities as a member of the ESMO Guidelines Steering Committee and chair of the Scientific Committee of ACTO (Alleanza contro il tumore ovarico). A.G.-M. has received fees for different educational or advisory-related activities from Alkermes, AstraZeneca, Clovis, Genmab, GSK, HederaDx, Immunogen, Illumina, Mersana, MSD, Novartis, Novocure, Oncoinvent, PharmaMar, Roche, SOTIO, SUTRO, Seagen, and Takeda. P.H. has received honoraria from AstraZeneca, Roche, Sotio, Tesaro, Stryker, ASCO, Zai Lab, and MSD and has acted in advisory/consultancy for AstraZeneca, Roche, Tesaro, Lilly, Clovis, Immunogen, and MSD/Merck.
Published: December 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101344.
Supplemental information
∗Treatment response available for 195 samples from PAOLA-1 sub-cohort.
References
- 1.Hanahan D., Weinberg R.A. Holland-Frei Cancer Medicine. John Wiley & Sons, Ltd; 2017. Biological Hallmarks of Cancer; pp. 1–10. [Google Scholar]
- 2.O’Kane G.M., Connor A.A., Gallinger S. Characterization, Detection, and Treatment Approaches for Homologous Recombination Deficiency in Cancer. Trends Mol. Med. 2017;23:1121–1137. doi: 10.1016/j.molmed.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Lord C.J., Ashworth A. BRCAness revisited. Nat. Rev. Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 4.Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., D’Andrea A.D. Homologous recombination deficiency: Exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore K., Colombo N., Scambia G., Kim B.-G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 6.González-Martín A., Pothuri B., Vergote I., DePont Christensen R., Graybill W., Mirza M.R., McCormick C., Lorusso D., Hoskins P., Freyer G., et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 7.Ray-Coquard I., Pautier P., Pignata S., Pérol D., González-Martín A., Berger R., Fujiwara K., Vergote I., Colombo N., Mäenpää J., et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 8.Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lord C.J., Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abkevich V., Timms K.M., Hennessy B.T., Potter J., Carey M.S., Meyer L.A., Smith-McCune K., Broaddus R., Lu K.H., Chen J., et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birkbak N.J., Wang Z.C., Kim J.-Y., Eklund A.C., Li Q., Tian R., Bowman-Colin C., Li Y., Greene-Colozzi A., Iglehart J.D., et al. Telomeric Allelic Imbalance Indicates Defective DNA Repair and Sensitivity to DNA-Damaging Agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popova T., Manié E., Rieunier G., Caux-Moncoutier V., Tirapo C., Dubois T., Delattre O., Sigal-Zafrani B., Bollet M., Longy M., et al. Ploidy and Large-Scale Genomic Instability Consistently Identify Basal-like Breast Carcinomas with BRCA1/2 Inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 15.Tutt A., Bertwistle D., Valentine J., Gabriel A., Swift S., Ross G., Griffin C., Thacker J., Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia F., Taghian D.G., DeFrank J.S., Zeng Z.C., Willers H., Iliakis G., Powell S.N. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. USA. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moynahan M.E., Pierce A.J., Jasin M. BRCA2 Is Required for Homology-Directed Repair of Chromosomal Breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 18.Davies H., Glodzik D., Morganella S., Yates L.R., Staaf J., Zou X., Ramakrishna M., Martin S., Boyault S., Sieuwerts A.M., et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobain E.F., Wu Y.-M., Vats P., Chugh R., Worden F., Smith D.C., Schuetze S.M., Zalupski M.M., Sahai V., Alva A., et al. Assessment of Clinical Benefit of Integrative Genomic Profiling in Advanced Solid Tumors. JAMA Oncol. 2021;7:525–533. doi: 10.1001/jamaoncol.2020.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timms K.M., Abkevich V., Hughes E., Neff C., Reid J., Morris B., Kalva S., Potter J., Tran T.V., Chen J., et al. Association of BRCA1/2defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014;16:475. doi: 10.1186/s13058-014-0475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tutt A., Tovey H., Cheang M.C.U., Kernaghan S., Kilburn L., Gazinska P., Owen J., Abraham J., Barrett S., Barrett-Lee P., et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat. Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macintyre G., Goranova T.E., De Silva D., Ennis D., Piskorz A.M., Eldridge M., Sie D., Lewsley L.-A., Hanif A., Wilson C., et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018;50:1262–1270. doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eeckhoutte A., Houy A., Manié E., Reverdy M., Bièche I., Marangoni E., Goundiam O., Vincent-Salomon A., Stoppa-Lyonnet D., Bidard F.-C., et al. ShallowHRD: detection of homologous recombination deficiency from shallow whole genome sequencing. Bioinformatics. 2020;36:3888–3889. doi: 10.1093/bioinformatics/btaa261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratto F., Brick K., Khil P., Smagulova F., Petukhova G.V., Camerini-Otero R.D. Recombination initiation maps of individual human genomes. Science. 2014;346 doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86:2278–2324. [Google Scholar]
- 27.Tew W.P., Lacchetti C., Kohn E.C. Poly(ADP-Ribose) Polymerase Inhibitors in the Management of Ovarian Cancer: ASCO Guideline Rapid Recommendation Update. J. Clin. Oncol. 2022;40:3878–3881. doi: 10.1200/JCO.22.01934. [DOI] [PubMed] [Google Scholar]
- 28.Vergote I., González-Martín A., Ray-Coquard I., Harter P., Colombo N., Pujol P., Lorusso D., Mirza M.R., Brasiuniene B., Madry R., et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann. Oncol. 2022;33:276–287. doi: 10.1016/j.annonc.2021.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Colombo N., Sessa C., du Bois A., Ledermann J., McCluggage W.G., McNeish I., Morice P., Pignata S., Ray-Coquard I., Vergote I., et al. ESMO-ESGO consensus conference recommendations on ovarian cancer: pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease. Ann. Oncol. 2019;30:672–705. doi: 10.1093/annonc/mdz062. [DOI] [PubMed] [Google Scholar]
- 30.Miller R.E., Leary A., Scott C.L., Serra V., Lord C.J., Bowtell D., Chang D.K., Garsed D.W., Jonkers J., Ledermann J.A., et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020;31:1606–1622. doi: 10.1016/j.annonc.2020.08.2102. [DOI] [PubMed] [Google Scholar]
- 31.Lin P.-H., Kuo K.-T., Hwu W.-L., Huang H.-N., Lin T.-Y., Chen C.-M., Cheng W.-F., Chiang Y.-C. Developing a whole exome sequencing-based homologous recombination deficiency test of epithelial ovarian cancer. Research Square. 2023 Preprint at. [Google Scholar]
- 32.Cristescu R., Liu X.Q., Arreaza G., Chen C., Albright A., Qiu P., Marton M.J. Concordance between single-nucleotide polymorphism–based genomic instability assays and a next-generation sequencing–based homologous recombination deficiency test. BMC Cancer. 2022;22:1310. doi: 10.1186/s12885-022-10197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaubey A., Shenoy S., Mathur A., Ma Z., Valencia C.A., Reddy Nallamilli B.R., Szekeres E., Stansberry L., Liu R., Hegde M.R. Low-Pass Genome Sequencing: Validation and Diagnostic Utility from 409 Clinical Cases of Low-Pass Genome Sequencing for the Detection of Copy Number Variants to Replace Constitutional Microarray. J. Mol. Diagn. 2020;22:823–840. doi: 10.1016/j.jmoldx.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Gruber J.J., Afghahi A., Timms K., DeWees A., Gross W., Aushev V.N., Wu H.-T., Balcioglu M., Sethi H., Scott D., et al. A phase II study of talazoparib monotherapy in patients with wild-type BRCA1 and BRCA2 with a mutation in other homologous recombination genes. Nat. Cancer. 2022;3:1181–1191. doi: 10.1038/s43018-022-00439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coussy F., Bidard F.-C. Expanding biomarkers for PARP inhibitors. Nat. Cancer. 2022;3:1141–1143. doi: 10.1038/s43018-022-00440-8. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Villatoro F., Oikkonen J., Casado J., Chernenko A., Gulhan D.C., Tumiati M., Li Y., Lavikka K., Hietanen S., Hynninen J., et al. Optimized detection of homologous recombination deficiency improves the prediction of clinical outcomes in cancer. npj Precis. Oncol. 2022;6 doi: 10.1038/s41698-022-00339-8. 96–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takaya H., Nakai H., Takamatsu S., Mandai M., Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci. Rep. 2020;10:2757. doi: 10.1038/s41598-020-59671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngoi N.Y.L., Tan D.S.P. The role of homologous recombination deficiency testing in ovarian cancer and its clinical implications: do we need it? ESMO Open. 2021;6 doi: 10.1016/j.esmoop.2021.100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buisson A., Saintigny P., Constantoulakis P., Oikonomaki K., Samara S., Harter P., Pignata S., Gonzalez Martin A., Schauer C., Fujiwara K., et al. Blinded-assessment of a solution to evaluate olaparib maintenance treatment efficacy in patients with ovarian cancer from the GINECO/ENGOT PAOLA-1 trial. J. Clin. Oncol. 2023;41:5588. [Google Scholar]
- 40.Christinat Y., Ho L., Clément S., Genestie C., Sehouli J., Cinieri S., Gonzalez Martin A., Denison U., Fujiwara K., Vergote I., et al. Normalized LST Is an Efficient Biomarker for Homologous Recombination Deficiency and Olaparib Response in Ovarian Carcinoma. JCO Precis. Oncol. 2023;7 doi: 10.1200/PO.22.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loverix L., Vergote I., Busschaert P., Vanderstichele A., Venken T., Boeckx B., Harter P., Brems H., Van Nieuwenhuysen E., Pignata S., et al. PARP inhibitor predictive value of the Leuven HRD test compared with Myriad MyChoice CDx PLUS HRD on 468 ovarian cancer patients from the PAOLA-1/ENGOT-ov25 trial. Eur. J. Cancer. 2023;188:131–139. doi: 10.1016/j.ejca.2023.04.020. [DOI] [PubMed] [Google Scholar]
- 42.Willing E.-M., Vollbrecht C., Vössing C., Weist P., Schallenberg S., Herbst J.M., Schatz S., Jóri B., Bataillon G., Harter P., et al. Development of the NOGGO GIS v1 Assay, a Comprehensive Hybrid-Capture-Based NGS Assay for Therapeutic Stratification of Homologous Repair Deficiency Driven Tumors and Clinical Validation. Cancers. 2023;15 doi: 10.3390/cancers15133445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boidot R., Blum M.G.B., Wissler M.-P., Gottin C., Ruzicka J., Duforet-Frebourg N., Jeanniard A., Just P.-A., Harter P., Pignata S., et al. 39MO Clinical evaluation of a low-coverage whole-genome test for homologous recombination deficiency detection in ovarian cancer. ESMO Open. 2023;8 [Google Scholar]
- 44.Callens C., Rodrigues M.J., Briaux A., Browaeys E., Eeckhoutte A., Pujade-Lauraine E., Renault V., Stoppa-Lyonnet D., Bieche I., Bataillon G., et al. 38MO Validation study of the ShallowHRDv2 assay for homologous recombination deficiency (HRD) detection in high-grade ovarian carcinomas (HGOC) in the first-line setting, from the phase III PAOLA-1/ENGOT-ov25 trial. ESMO Open. 2023;8 [Google Scholar]
- 45.Antonarakis E.S., Gomella L.G., Petrylak D.P. When and How to Use PARP Inhibitors in Prostate Cancer: A Systematic Review of the Literature with an Update on On-Going Trials. Eur. Urol. Oncol. 2020;3:594–611. doi: 10.1016/j.euo.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Kingma D.P., Ba J. Adam: A Method for Stochastic Optimization. arXiv. 2017 Preprint at. [Google Scholar]
- 47.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denkert C., Romey M., Swedlund B., Hattesohl A., Teply-Szymanski J., Kommoss S., Kaiser K., Staebler A., du Bois A., Grass A., et al. Homologous Recombination Deficiency as an Ovarian Cancer Biomarker in a Real-World Cohort: Validation of Decentralized Genomic Profiling. J. Mol. Diagn. 2022;24:1254–1263. doi: 10.1016/j.jmoldx.2022.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
∗Treatment response available for 195 samples from PAOLA-1 sub-cohort.
Data Availability Statement
-
•
WGS data from FF breast cancer samples and associated patient information data were downloaded from the European Genome-phenome Archive (EGA), and the accession number is listed in the key resources table. They are available upon request if access is granted. To request access, contact EGA and apply for access via the ICGC Data Access webpage.
-
•
The data generated for FFPE ovarian cancer clinical samples reported in this study cannot be deposited in a public repository because they were used under license for the current study and therefore are not publicly available. To request access, contact the authors of the PAOLA-1 study7 and/or the relevant contacts (for details, see list of authors and affiliations) of the following institutions: Vienna General Hospital, Center Léon Bérard, European Institute of Oncology, Istituto Nazionale Tumori Pascale, and Institut de Cancérologie de Lorraine.
-
•
GIInger is available as part of the SOPHiA GENETICS SOPHiA DDM platform.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.