Abstract
A nonswarming Serratia liquefaciens mutant deficient in serrawettin W2 production was constructed by transposon mutagenesis. Sequence homology indicated that insertion had occurred in gene swrA, which encodes a putative peptide synthetase. Expression of swrA is controlled by quorum sensing.
Several bacterial species are able to form expanding colonies on the surface of semisolid growth media by means of swarming motility (1, 7, 8, 24). Serratia liquefaciens MG1 swarms on the surface of AB minimal medium (3) solidified with 0.5 to 1.0% agar supplemented with 0.5% Casamino Acids and 0.5% glucose (6, 7). Development of a swarming colony requires the sensing and integration of a variety of environmental, as well as intracellular, signals involving surface contact and local high population density. According to our current working hypothesis for the development of a swarming colony (11, 12), exposure of the cells to surfaces with a certain viscosity is recognized by an unknown sensor, and signal transduction then progresses via the flhDC master operon. Stimulation of this operon initiates swarm cell differentiation that involves development of characteristic traits such as cell elongation, multinucleation, and hyperflagellation (6). The population density is recognized by a homoserine lactone-dependent quorum-sensing system constituted by the swrI and swrR genes (7, 11, 12). On defined growth medium, expansion of the swarm colony is strictly dependent on a functional swrI gene, a member of the luxI family of autoinducer synthetase genes (7). The swrI gene product catalyzes the formation of N-butanoyl-l-homoserine lactone (BHL) and N-hexanoyl-l-homoserine lactone in a molar ratio of 10:1 (7). The flagellar master and the quorum-sensing system are global regulators which control two separated regulons (12). We are now in the process of identifying target genes of the quorum-sensing system, as well as macromolecules and compounds that are synthesized to facilitate swarming motility.
MG1 produces an extracellular biosurfactant.
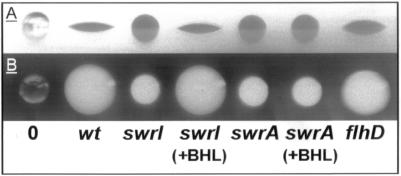
The ability to synthesize compounds with surface tension-reducing properties is widespread among bacteria, and the significance of surface-active compounds or surfactants in interaction with interfaces has been reviewed (27). The application of a drop-collapsing test has been used to identify bacteria that produce surfactants (14). A 10-μl drop of an outgrown bacterial culture or of a suspended bacterial colony is placed on a hydrophobic surface such as the lid of a petri dish. The drop either remains globular or collapses rapidly while increasing the wet area on the surface. The latter situation indicates the presence of a surfactant capable of lowering the surface tension of the water (Fig. 1). This simple test indicates that S. liquefaciens MG1 produces a biosurfactant. The swrI mutant MG44, which is defective in BHL and N-hexanoyl-l-homoserine lactone synthesis (7), is impaired in surfactant production (Fig. 1). However, addition of exogenous BHL to the growth medium restores surfactant production (Fig. 1). This strongly suggests that the quorum-sensing mechanism controls the production of a biosurfactant. The presence of an inactivating mutation at the flhD locus does not affect surfactant production (Fig. 1). After removal of the cells from the cell suspensions depicted in Fig. 1, the drop-collapsing test result was reproduced with the cell-free supernatants (data not shown). This indicated that the surface tension-reducing activity is caused by the presence of an extracellular compound(s).
FIG. 1.
Drop-collapsing test. Ten-microliter volumes of the following bacterial cultures were placed on the lid of a petri dish. (A) Side view. (B) Top view. 0, 0.9% NaCl; wt, S. liquefaciens MG1; swrI, S. liquefaciens MG44; swrA, S. liquefaciens PL10; flhD, S. liquefaciens MG3; +BHL, strain grown in the presence of 200 nM BHL.
Quorum sensing regulates expression of a gene necessary for surfactant production.
The swrI mutant MG44 was randomly mutagenized with a Tn5-derived transposon carrying a promoterless luxAB reporter as described by Kristensen et al. (18). The resulting double mutants were screened for BHL-induced bioluminescence on agar plates supplemented with 0.2 μM BHL. This gave rise to the isolation of 19 mutants in which expression of the luxAB reporter was BHL dependent. One mutant, designated PL10, was unable to swarm in the presence of externally added BHL. The drop-collapsing test indicated that the PL10 mutant is completely defective in surfactant production, even in the presence of BHL (Fig. 1). From this we concluded that the luxAB transposon had most likely been integrated into a gene, designated swrA, which is essential for surfactant production. A Southern blot analysis targeting luxAB in PL10 and the parent strain MG44 was performed. Chromosomal DNA was digested with five different restriction enzymes that had no recognition sites in the luxAB sequence. The Southern blot confirmed the presence of a single transposon insert in PL10 and no insertion in MG44 (data not shown).
PL10 cells were grown as colonies, washed off, and suspended in fresh growth medium. When expression of bioluminescence per unit of optical density at 450 nm was determined, cells that previously had been grown in the presence of 0.2, 2.0, and 20 μM BHL showed 50-fold higher light emission than cells from a colony grown in the absence of BHL (data not shown). We also monitored a liquid culture of PL10 and determined the expression of bioluminescence per unit of optical density at 450 nm throughout the growth cycle in the absence or presence of 0.2, 2.0, and 20 μM BHL. The presence of BHL in the medium caused a maximum fivefold stimulation of bioluminescence (data not shown).
The product of the gene swrA is a putative peptide synthetase.
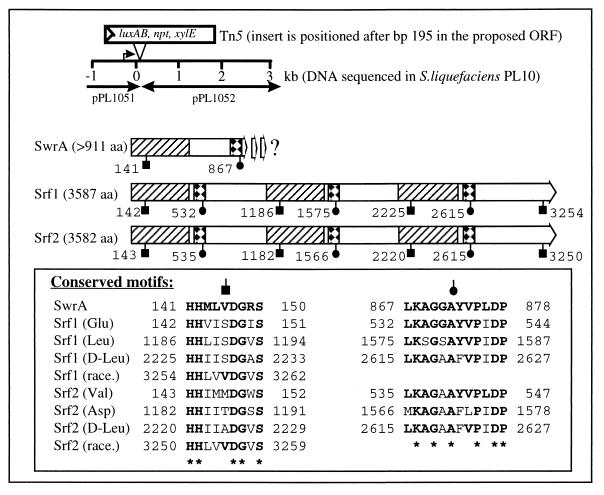
Chromosomal DNA of PL10 was prepared as described by Givskov et al. (10), restricted with either PmlI or BamHI, and ligated into the SmaI or BamHI site of pUC18Not (32), respectively. Ligation mixtures were electroporated into Escherichia coli MT102 [araD139 (ara-leu)7697 Δlac thi hsdR]). Two hybrid plasmids, pPL1051, carrying 3.5 kbp of PL10 DNA upstream of the transposon insert, and pPL1052, carrying 3 kbp downstream of the transposon insert, were isolated (Fig. 2). From plasmid 1051, approximately 1.5 kb was sequenced in one direction upward from the transposon. This DNA sequence exhibited no homology to known genes. The insert of pPL1052 was completely sequenced in both directions. A BLASTX homology search for the DNA sequence was done via the worldwide web at the BCM Nucleic Acid Sequence Searches of the Human Genome Center, Baylor College of Medicine (http://dot.imgen.bcm.tmc:9331/seq-search/nucleic_acid-search.html). Results of the BLASTX homology search (Fig. 2) indicate that swrA encodes a peptide synthetase with high homology to, e.g., the surfactin synthetase SrfA of Bacillus subtilis (5) and the tyrocidine synthetase TycABC of B. brevis (25), both of which are members of a superfamily of peptide synthetases all encoded by large operons (31). Several characteristic conserved amino acid motifs or core sequences of this superfamily have been defined (13) within the larger conserved modular domains. Two such motifs with homology to the consensus sequences LKAGGA and HHILDGV are found in the predicted SwrA open reading frame (ORF) (Fig. 2). The stretch of weak homology in the putative SwrA (Fig. 2, open region) separating the areas of high homology (Fig. 2, hatched and squared regions) seems to be larger than in most petide synthetases. This region might house a modification module, e.g., an epimerase, but this is purely speculative. The biosynthesis of lipodepsipeptides is often accomplished by nonribosomal, multifunctional peptide synthetases (16, 17, 19). The surfactin biosynthesis in B. subtilis is one of the best characterized of these systems. Surfactin is a small, cyclic peptide consisting of seven amino acids and a 3-hydroxy-13-methyltetradecanoic fatty acid side chain (2) which is synthesized by the Srf enzyme complex. The Srf complex contains seven highly homologous amino acid binding domains present in four subunits, Srf1-4, encoded by four ORFs. These domains determine the seven specific amino acids and their order in the final surfactin molecule; thus, the synthetase acts as a protein template during peptide formation. Also, the complex contains two thioesterase active site-like sequences, and finally, the third and sixth amino acid binding domains are followed immediately by modifying domains which epimerize the bound amino acid to the d form present in surfactin (5).
FIG. 2.
DNA region of S. liquefaciens PL10 (including size markers) containing the transposon insert. The extent of cloned and sequenced DNA on plasmids pPL1051 and pPL1052 is indicated by arrows. The predicted SwrA ORF of 911 amino acids (aa) is shown as a thick arrow. Homology between the putative SwrA and the SrfA surfactin synthetase subunits Srf1 and Srf2 of B. subtilis (7) is as follows: hatched regions of 450 aa are 28% identical and 42% similar and squared regions of 75 aa are 54% identical and 70% similar. The positions of motifs characteristic of peptide synthetases are indicated by black symbols and given by amino acid number. The motif sequences, as well as the specific amino acids (Glu, Leu, etc.) that the corresponding domains bind are shown in the box. Race denotes a domain which catalyzes the l- to d-form epimerization of the adjacent bound Leu.
Isolation and identification of the biosurfactant serrawettin W2.
Growth conditions were found to affect surfactant production. When cultured in liquid, MG1 gave rise to only small amounts compared to the large quantities produced when the strain was cultured on an agar surface (data not shown), an observation consistent with the observed light emission pattern of PL10. The combined contents of 15 petri dishes that had been completely colonized by swarming colonies of S. liquefaciens MG1 over 48 h were extracted three times with equal amounts of 1% formic acid in ethyl acetate for 24 h. The combined extracts were evaporated to dryness, suspended in water, lyophilized, extracted with methanol, and then filtered, and the filtrate was evaporated to dryness leaving a total of 358 mg of crude extract. A 300-mg sample of the extract was subjected to high-performance liquid chromatography (HPLC) separation on an RP18C column (Merck) isocratically with acetonitrile-water (80:20) containing 0.05% trifluoroacetic acid as the eluent and monitored at 215 nm. The chromatogram showed five fractions, the major one (56 mg) being pure serrawettin W2. Serrawettin W2 is a colorless solid. Fast atom bombardment (FAB)-mass spectrometry (MS): m/z 732.5 (M+H)+, 754.5 (M+Na)+, 1,463.9 (2M+H)+, 467.2 (M+H-C10H17COLeu)+, 266.3 (C10H17COLeu+H)+. In glycerol matrix: m/z 732.3 (M+H)+. Electron impact MS with direct inlet at 202°C: m/z (% relative intensity) 732 (M+, 8), 714 (9), 696 (100), 688 (25); compatible with the composition C28H61N5O9. FAB-MS data were determined in an m-nitrobenzylic alcohol–glycerol–thioglycerol 1:1:2 matrix containing 1% trifluoroacetic acid. The FAB-MS exhibited a cluster ion at m/z 1,463.9 representing the (2M+H)+ ion. Fragmentation of an acylated linear peptide formed by ring opening between the protonated C-terminal l-Ile and the β-hydroxydecanoyl moiety form the protonated linear N-decenoyl peptide ion which, in turn, loses the N-decenoylleucine moiety, forming m/z 467. The N-decenoylleucine pseudomolecular ion appears at m/z 266. This finding is analogous to the recorded fragmentation of massetolides (9).
Optical rotation and UV spectra were determined as [α]D20 −10.3° (c 0.0975, ethanol [EtOH]). UV (EtOH) λmax nm (log ɛ): 251 (2.51), 258 (2.51), 264 (2.44), 268sh (2.37). The circular dichroism (CD) spectra measured were CD (1.33 × 10−3 mol/liter, EtOH) λ (Δɛ): 270 (0.15), 263 (0.10), 255 (0.10), 246 (0.15), 223 (−3.45).
Nuclear magnetic resonance (NMR) spectra were recorded at 400.0 and 100.6 MHz for 1H- and 13C-NMR, respectively, on samples in dimethyl sulfoxide-d6. The 1H- and 13C-NMR data are compatible with this structure (Table 1). The partial assignments presented in Table 1 are supported by pulse gradient, multiquantum-filtered, phase-sensitive correlation spectroscopy and two-dimensional total correlation spectroscopy experiments, establishing the scalar couplings between neighboring protons. The carbon resonance assignments resulted from pulse gradient, reverse-detected heteronuclear multiple quantum coherence (HMQC) experiments optimized for a JC,H of 140 Hz and the connectivities from pulse gradient, reverse-detected heteronuclear multiple bond coherence (HMBC) experiments optimized for a JC,H of 7 Hz.
TABLE 1.
NMR data for serrawettin W2 in dimethyl sulfoxide-d6
| Residue and position(s) | δcf | δH(JH,H, Hz) |
|---|---|---|
| d-Leu (1) | ||
| NH | 7.82 (8.8) | |
| Cα | —a | |
| Cβ | 24.8 | —d |
| Cγ | 23.4 | 1.64 m |
| Cδ | —c | |
| Cδ | —c | |
| l-Ser (2) | ||
| NH | 8.08 (8.2) | |
| Cα | —a | |
| Cβ | 61.3 | 3.63 (11.0, 6.4), 3.56 (11.0, 6.8) |
| l-Thr (3) | ||
| NH | 8.03 (8.2) | |
| Cα | 58.5 | 4.11 (8.4, 2.9) |
| Cβ | 65.4 | 4.24 m |
| Cγ | 20.2 | 0.97 (6.4) |
| d-Phe (4)e | ||
| NH | 7.46 (7.0) | |
| Cα | —a | |
| Cβ | 37.2 | 3.14 (13.7, 4.5), 2.89 (13.7, 7.2) |
| l-Ile (5) | ||
| NH | 8.49 (6.4) | |
| Cα | 57.6 | 3.76 (8.2, 6.8) |
| Cβ | 34.7 | 1.77 m |
| Cγ | 40.4 | —d |
| Cδ | —c | |
| Cδ | —c | |
| 3-Hydroxy-decanoyl | ||
| C-2 | 39.0 | 2.64 (14.5, 3.0), 2.36 (14.5, 5.9) |
| C-3 | 71.8 | 4.92b m |
| C-4 | 31.1 | 1.53 br m |
| C-5–C-9 | 1.23 br | |
| C-10 | —c |
—, the α protons of Phe, Ser, and Leu appear at δ 4.48 to 4.32 as an unresolved multiplet (m).
Partly obscured by the water resonance at δ 4.83 br.
—, five unresolved methyl resonances appear at δ 0.86 to 0.78.
—, the Leuβ and Ileγ protons appear as overlapping multiplets at δ 1.45 to 1.34, and the Leuβ′ and Ileγ′ protons appear as overlapping multiplets at δ 1.18 to 1.09.
The aromatic protons appear as an unresolved multiplet at δ 7.16 to 7.05, and the aromatic carbons appear at δ C2′, 6′, 3′ and 5′ 128.1, 129.1; C1′ 126.5; and C4′ 136.8
The six carbonyl carbon signals appear at δ 172.1, 171.3, 170.6, 170.6, 169.5, and 168.8.
The sequence of the amino acid units and the points of attachment of the hydroxyacyl group were revealed by a rotating frame nuclear Overhauser and exchange spectroscopy (ROESY) experiment using a mixing time of 300 ms. Cross peaks were present between Leu-NH to the α protons of the decanoyl moiety and to Ser-NH, thus placing the Leu moiety at the acylated N-terminal position followed by Ser. The Ser β protons are close to Thr-NH which, in turn, magnetically relaxes with Phe-NH. The C terminus is occupied by Ile, since Ile-NH interacts with the Phe α proton. The same experiment served to verify the assignments of the proton resonances of the various constituting units.
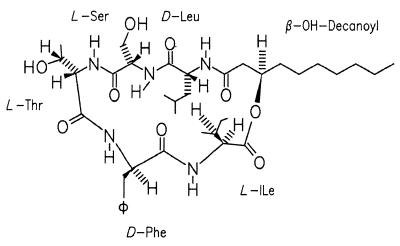
We conclude that the surface-active compound is identical to serrawettin W2, a compound that was originally isolated from S. marcescens NS 25 (21) with the chemical structure proposed in Fig. 3. Based on automated amino acid analysis, MS, and NMR (1H and nuclear Overhauser enhancement spectroscopy) data on a sample containing at least 20% impurity, the structure of W2 was proposed by Matsuyama and coworkers (21). The sterochemistry of the amino acid units was determined, as well as the absolute configuration of the 3-D-hydroxydecanoic moiety (23). The spectroscopic investigations described here confirm the proposed structure.
FIG. 3.
Molecular structure of serrawettin W2.
An inherent complication of the identification of this type of structure is the lack of a single physical parameter allowing its identity to be established unambiguously. A realistic method must present a unique fingerprint in order to be generally reliable. The 13C-NMR spectrum, recorded under carefully controlled conditions, seems to be ideally suited for this purpose, since it displays, in this case, 28 distinct signals distributed on a scale covering around 160 ppm (Table 1). CD data have not been recorded much for this type of compound either. In order to establish a reference for future discussions concerning the stereochemistry and conformation of this type of cyclodepsipeptides, these data have been included in this report.
Surfactants promote swarming motility of S. liquefaciens.
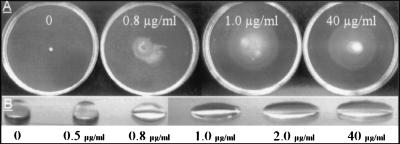
The following complementation experiments completely restored the swarming motility of the PL10 and MG44 mutants. The strains were inoculated on 0.6% agar plates supplemented with three purified biosurfactants: either 1-μg/ml surfactin (Sigma) produced by B. subtilis (2) (data not shown), 10-μg/ml serrawettin W1 produced by S. marcescens (20) (data not shown), or HPLC-purified serrawettin W2 (Fig. 4A). The presence of W2 above the critical concentration of 1 μg/ml restores the swarming motility of PL10 to the level of the wild-type strain (Fig. 4A). When W2 is dissolved in water at 1 μg/ml and above, rapid drop collapse follows, as demonstrated in Fig. 4B. This critical concentration of W2 could reflect a physical property of the compound itself, e.g., a critical micelle-forming concentration.
FIG. 4.
(A) Swarming motility of S. liquefaciens PL10 on medium supplemented with HPLC-purified serrawettin W2 at the concentrations indicated. (B) Drop-collapsing test of water supplemented with HPLC-purified serrawettin W2 at the concentrations indicated.
Conclusion.
The present study is in agreement with previously published data demonstrating that biosurfactant production is crucial for the swarming motility of Serratia spp. (22). B. subtilis also exhibits swarming motility on surfaces, and mutants unable to produce the biosurfactant are defective in swarming motility (24). Swarming motility appears to be an intrinsically linked surface and cell density phenomenon. It is therefore not surprising to find that S. liquefaciens employs a quorum-sensing regulatory circuit in the process of surface conditioning. Since the swarming motility of the swrI and swrI swrA mutants could be restored simply by supplementing the medium with pure surfactant, the only quorum-sensing-controlled gene involved in swarming motility may be swrA. The fact that external addition of BHL leads to 10-fold higher induction of the swrA-luxAB fusion on the surface of agar plates compared to liquid is in accordance with the finding that only small surfactant amounts are produced in a liquid culture. These observations are intriguing; we speculate that either the surface itself, i.e., a form of surface recognition, or dense growth within a colony, i.e., intimate cell-cell contact, accounts for the observed differences. In this context, it is interesting that the recently identified quorum-sensing system in Rhodobacter sphaeroides controls the switch from aggregated to dispersed growth (29). A screening of our Serratia strain collection revealed a correlation between swarming motility and surfactant production. Many produced detectable amounts of acylated homoserine lactone (our unpublished observations), and this indicates that surfactant production in Serratia in general is responsive to cell density by quorum-sensing mechanisms that may control the expression of biosurfactant synthetases. Recently it was shown that B. subtilis regulates surfactin production by a cell density-responsive mechanism not based on homoserine lactone but utilizing a peptide pheromone, ComX (4), as is usually the case in gram-positive bacteria (15). From an evolutionary point of view, the ubiquitous peptide synthetases are highly interesting, being present in both gram-negative and gram-positive bacteria, as well as in certain filamentous fungi (30). The different products of the peptide synthetases display a range of powerful biological properties, such as antibiotic, hemolytic, antitumor, and surface-conditioning activities (16, 26, 28).
Nucleotide sequence accession number.
The nucleotide sequence of swrA has been assigned GenBank accession no. AF039572.
Acknowledgments
We are indebted to S. E. Harnung for determination of the CD data.
The CD data were measured on a modified JASCO 710 instrument financed by the Danish Natural Science Research Council (grant 0373-1). We thank Linda Stabell for excellent technical assistance and Tohey Matsuyama for kindly providing a purified sample of serrawettin W1.
REFERENCES
- 1.Allison C, Hughes C. Closely linked genetic loci required for swarm cell differentiation and multicellular migration by Proteus mirabilis. Mol Microbiol. 1991;5:1975–1982. doi: 10.1111/j.1365-2958.1991.tb00819.x. [DOI] [PubMed] [Google Scholar]
- 2.Arima K, Kakinuma A, Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun. 1968;31:488–494. doi: 10.1016/0006-291x(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 3.Clark J D, Maaløe O. DNA replication and the cell cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 4.Cosby W M, Vollenbroich D, Lee O H, Zuber P. Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control. J Bacteriol. 1998;180:1438–1445. doi: 10.1128/jb.180.6.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 6.Eberl L, Christiansen G, Molin S, Givskov M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhDC master operon. J Bacteriol. 1996;178:554–559. doi: 10.1128/jb.178.2.554-559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberl L, Winson M K, Sternberg C, Stewart G S A B, Christensen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 8.Gaisser S, Hughes C. A locus coding for putative non-ribosomal peptide/polyketide synthase functions is mutated in a swarming-defective Proteus mirabilis strain. Mol Gen Genet. 1997;253:415–427. doi: 10.1007/s004380050339. [DOI] [PubMed] [Google Scholar]
- 9.Gerard J, Lloyd R, Barsby T, Haden P, Kelly M T, Andersen R. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J Nat Prod. 1997;60:223–229. doi: 10.1021/np9606456. [DOI] [PubMed] [Google Scholar]
- 10.Givskov M, Eberl L, Christiansen G, Benedik M J, Molin S. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhDC. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 11.Givskov M, Eberl L, Molin S. Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol Lett. 1997;148:115–122. [Google Scholar]
- 12.Givskov M, Östling J, Eberl L, Lindum P W, Christensen A B, Christiansen G, Molin S, Kjelleberg S. Two separate systems participate in the control of swarming motility of Serratia liquefaciens. J Bacteriol. 1998;180:742–745. doi: 10.1128/jb.180.3.742-745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gocht M, Marahiel M A. Analysis of core sequences in the d-Phe activating domain of the multifunctional peptide synthetase TycA by site-directed mutagenesis. J Bacteriol. 1994;176:2654–2662. doi: 10.1128/jb.176.9.2654-2662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain D K, Thompson D K C, Lee H, Trevors J T. A drop-collapsing test for screening surfactant producing microorganisms. J Microbiol Methods. 1991;13:271–279. [Google Scholar]
- 15.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 16.Kleinkauf H, von Döhren H. Biosynthesis of peptide antibiotics. Annu Rev Microbiol. 1987;41:259–289. doi: 10.1146/annurev.mi.41.100187.001355. [DOI] [PubMed] [Google Scholar]
- 17.Kleinkauf H, von Döhren H. Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem. 1990;192:1–15. doi: 10.1111/j.1432-1033.1990.tb19188.x. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipmann F. Bacterial production of antibiotic polypeptides by thiol-linked synthesis on protein templates. Adv Microbiol Physiol. 1980;21:227–266. doi: 10.1016/s0065-2911(08)60357-4. [DOI] [PubMed] [Google Scholar]
- 20.Matsuyama T, Fujita M, Yano I. Wetting agent produced by Serratia marcescens. FEMS Microbiol Lett. 1985;28:125–129. [Google Scholar]
- 21.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol. 1992;174:1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama T, Bhasin A, Harshey R M. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol. 1995;177:987–991. doi: 10.1128/jb.177.4.987-991.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuyama T, Nakagawa Y. Surface-active exolipids: analysis of absolute chemical structures and biological functions. J Microbiol Methods. 1996;25:165–175. [Google Scholar]
- 24.Mendelson N H, Salhi B. Patterns of reporter gene expression in the phase diagram of Bacillus subtilis colony forms. J Bacteriol. 1996;178:1980–1989. doi: 10.1128/jb.178.7.1980-1989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootz H D, Marahiel M A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neu T R, Hartner T, Poralla K. Surface active properties of viscosin—a peptidolipid antibiotic. Appl Microbiol Biotechnol. 1990;32:518–520. [Google Scholar]
- 27.Neu T R. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev. 1996;60:151–166. doi: 10.1128/mr.60.1.151-166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pospiech A, Cluzel B, Bietenhader J, Schupp T. A new Myxococcus xanthus gene cluster for the biosynthesis of the antibiotic saframycin Mx1 encoding a peptide synthetase. Microbiology. 1995;141:1793–1803. doi: 10.1099/13500872-141-8-1793. [DOI] [PubMed] [Google Scholar]
- 29.Puskas A, Greenberg E P, Kaplan S, Schaefer A L. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stachelhaus T, Marahiel M A. Modular structures of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 31.Turgay K, Krause M, Marahiel M A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]