Abstract
Myosin binding protein-C (MyBP-C) is a multidomain protein that regulates muscle contraction. Mutations in MYBPC3, the gene encoding for the cardiac variant (henceforth called cMyBP-C), are amongst the most frequent causes of hypertrophic cardiomyopathy. Most mutations lead to a truncated version of cMyBP-C, which is most likely unstable. However, missense mutations have also been reported, which tend to cluster in the central domains of the cMyBP-C molecule. This suggests that these central domains are more than just a passive spacer between the better characterized N- and C-terminal domains. Here, we investigated the potential impact of four different missense mutations, E542Q, G596R, N755K, and R820Q, which are spread over the domains C3 to C6, on the function of MyBP-C on both the isolated protein level and in cardiomyocytes in vitro. Effect on domain stability, interaction with thin filaments, binding to myosin, and subcellular localization behavior were assessed. Our studies show that these missense mutations result in slightly different phenotypes at the molecular level, which are mutation specific. The expected functional readout of each mutation provides a valid explanation for why cMyBP-C fails to work as a brake in the regulation of muscle contraction, which eventually results in a hypertrophic cardiomyopathy phenotype. We conclude that missense mutations in cMyBP-C must be evaluated in context of their domain localization, their effect on interaction with thin filaments and myosin, and their effect on protein stability to explain how they lead to disease.
Keywords: ATPase, cardiomyopathy, cell culture, cytoskeleton, heart, microscopic imaging, mutant, protein stability
Hypertrophic cardiomyopathy (HCM) is a hereditary disease that is caused by mutations mainly in genes that encode for proteins of the sarcomere, the basic unit of the myofibrils, which mediates muscle contraction. It is assumed to have a frequency of 1:500 in the human population, and most mutations are seen in the genes encoding for beta-myosin heavy chain (MYH7) and the cardiac isoform of myosin binding protein-C, cMyBP-C (MYBPC3) (1). cMyBP-C is a multidomain protein of the A-band that was reported to interact with all three filament systems of the sarcomere: thin (actin), thick (myosin), and titin (Fig. 1A; for recent review, see Ref. (2)). The cardiac isoform is composed of eight immunoglobulin and three fibronectin type three-like domains (termed C0 to C10) with several interspersed regions of a less well-defined structure (3) (Fig. 1B). The N-terminal third of the molecule is thought to bind to actin, myosin (subfragments S1 and S2), and the regulatory light chain of myosin (reviewed in Ref. (4, 5)), whilst the C-terminal third was shown to bind to titin and myosin (LMM) and to be required for A-band targeting (6, 7). cMyBP-C is involved in the regulation of heart muscle contractility via regulatory interactions with both the thin and thick filaments (8, 9). Whilst 60 to 90% of HCM mutations in MYBPC3 lead to truncated protein that is not detectable in patient heart samples (10, 11), the remaining are missense mutations, some of which can be detected in patient hearts as well as in inbred cat strains that are prone to HCM (12, 13). The majority of these missense mutations are found in the middle part of the MyBP-C molecule (domains C3C6; (11, 12, 14)), the function of which is not very well understood. Previous studies have shown that cMyBP-C with a missense mutation fails to restore contractile parameters in engineered heart tissue from cMyBP-C knockout background despite being incorporated normally into the A-band, suggesting it may play a role akin to a poison peptide (15). In feline HCM, fairly normal incorporation of mutant cMyBP-C (A31P) into the A-bands was demonstrated with a mutation-specific antibody (12). On the other hand, studies on patient hearts see reduced expression levels down to 80% also of missense cMyBP-C similar to truncated cMyBP-C mutants (E542Q; R502W; and E258K), making haploinsufficiency a possibility (10). However, transgenic expression of as little as 40% of cMyBP-C was able to rescue the phenotype in a cMyBP-C knockout mouse strain (16), suggesting that the situation might be more complex and species and mutation site dependent.
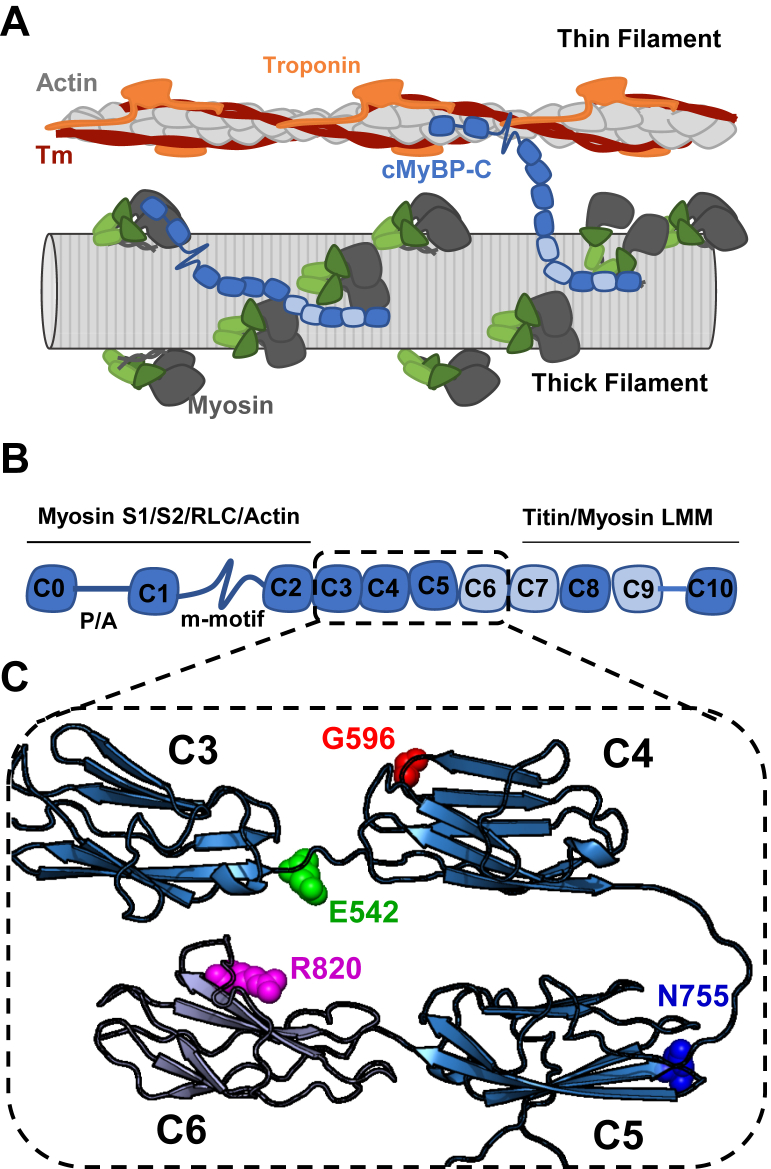
Figure 1.
Schematic representation of cardiac MyBP-C (cMyBP-C).A, cartoon representation of potential interactions of cMyBP-C (blue) with thick and thin filaments. B, domain organization of cMyBP-C composed of eight Ig- domains (dark blue) and three Fn3 domains (light blue, interspersed by linker regions of low complexity). Identified protein interactions are indicated accordingly. C, AlphaFold2 model of the structure of C3C6 with the investigated missense mutations indicated in color. cMyBP-C, cardiac isoform of myosin binding protein-C; Tm, tropomyosin.
In the current study, we decided to investigate the structural and functional consequences of four HCM-associated variants (E542Q, G596R, N755K, and R820Q) that are situated in the central domains C3C6. They were identified using the ClinVar database and had an allelic frequency of less than 0.01% according to the gnomAD database (17, 18). Multidomain constructs were expressed recombinantly in Escherichia coli and subjected to biophysical and biochemical analysis. Multidomain and full-length GFP-tagged versions were also expressed in primary cultures of neonatal rat cardiomyocytes (NRCs) and analyzed for targeting behavior to the sarcomere as well as for potential disruptive effects on sarcomere structure. Our results show that these four missense mutations have an effect on cMyBP-C behavior, but that this effect is mutation site specific. The expected functional consequence of all of them is that cMyBP-C fails to regulate contraction properly, which will eventually lead to HCM.
Results
Biophysical characterization of C3C6 WT and missense variants
To analyze the effect of HCM-causing missense mutations in the central domains of cMyBP-C on domain stability and folding, we generated bacterial expression constructs for the C3C6 domains (WT) and four variants (E542Q, G596R, N755K, and R820Q; for modeling of mutated residues on the domains, see Fig. 1C). All C3C6 constructs were solubly expressed in E. coli, suggesting that the missense mutations did not completely disrupt domain folding, which would lead to protein aggregation in the insoluble fraction (“inclusion bodies”) (Fig. 2A).
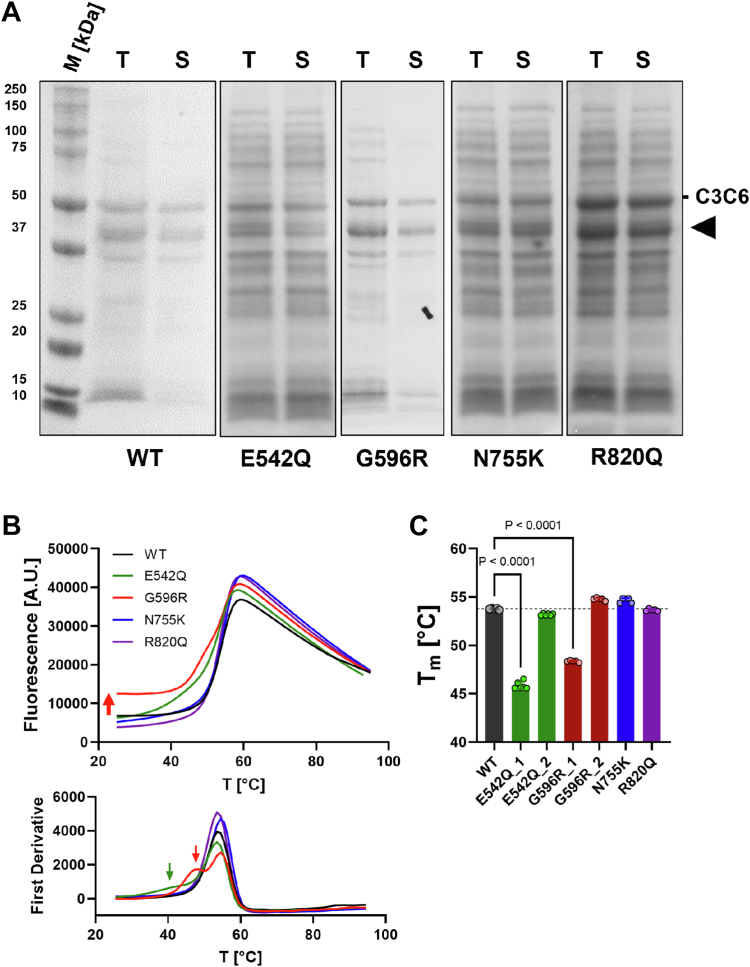
Figure 2.
Effect of missense mutations on C3C6 domain stability.A, expression profiles of WT and HCM variants of C3C6 in bacterial systems. C3C6 is indicated accordingly. Black arrowhead indicates degradation product. B, top, differential scanning fluorimetry unfolding curves for WT and HCM variants of C3C6. Please note that G596R shows increased background fluorescence at low temperature (red arrow). Bottom, first derivative of unfolding curves shown above. G596R and E542Q show an additional transition at lower temperatures (indicated by green and red arrow, respectively). C, summary of measured Tms. Data are presented as means ± SD, n = 5. Statistical significance between WT and HCM variants was assessed with a one-way ANOVA followed by Tukey’s post hoc test. HCM, hypertrophic cardiomyopathy; S, soluble fraction of bacterial lysate; T, total bacterial lysate.
To assess protein stability, differential scanning fluorometry (DSF) experiments were carried out (Fig. 2B), which revealed that the melting curves for WT and the N755K and R820Q variants showed a single transition (Fig. 2B) with almost identical Tm of about 54 °C, suggesting that domain stability is not affected by these substitutions. In contrast, E542Q and G596R showed two transitions in the DSF curves, one with a Tm identical to those measured for the WT protein (Tm of about 54 °C) and a second transition with a markedly reduced Tm of 45 °C and 47 °C, respectively (quantified in Fig. 2C). This suggests that the C3C6 variants E542Q and G596R likely exist in at least two distinct conformations, or that individual domains in the multidomain C3C6 construct are not correctly folded in the presence of the substitutions. Moreover, the high background fluorescence in the G596R variant (red trace in Fig. 2B) further indicates that one of the conformations exists in a partially unfolded state under physiologically relevant temperatures (≥37 °C).
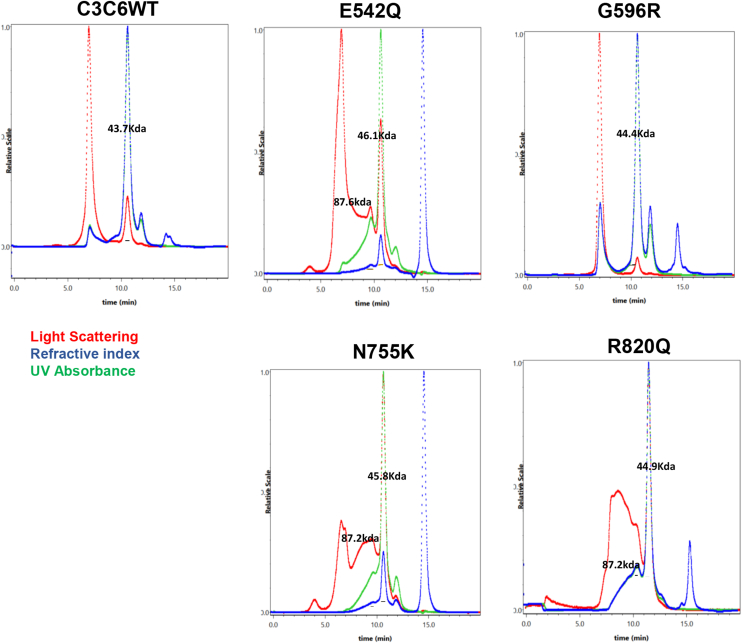
Next, we tested for the effects of the variants on protein aggregation using size-exclusion chromatography–multiangle light scattering (SEC–MALS, Figs. 3 and S1). WT C3C6 shows a single peak in SEC–MALS with a calculated molecular weight of about 44 kDa, which is in good agreement with the calculated molecular weight of about 48 kDa for the C3C6 monomer. In contrast, the R820Q, N755K, and E542Q variants cause increased aggregation, as indicated by additional shoulders preceding the main peak in the SEC chromatogram. Integration of the UV absorbance trace suggests that the aggregated species correspond to about 37%, 24%, and 34% of the total protein for R820Q, N755K, and E542Q, respectively. The major aggregate species had a molecular weight of about 87 kDa, consistent with the molecular weight of the dimer (Fig. S1). However, higher molecular weight aggregates could also be detected. Strikingly, although G596R likely exists in two conformations, of which at least one is partially unfolded according to the DSF experiments, the SEC–MALS analysis shows a single predominant species with a molecular weight corresponding to the monomer and very little to no protein aggregation. Although proteins were equilibrated at room temperature (RT) for about 2 h prior to SEC–MALS analysis, time-dependent increase in aggregation cannot be excluded by the present results.
Figure 3.
Effect of missense mutations on protein aggregation. WT and HCM variants of C3C6 were analyzed by size-exclusion chromatography–multiangle light scattering (SEC–MALS). Traces for light scattering, refractive index, and UV absorbance are shown for each variant in red, blue, and green, respectively. HCM, hypertrophic cardiomyopathy.
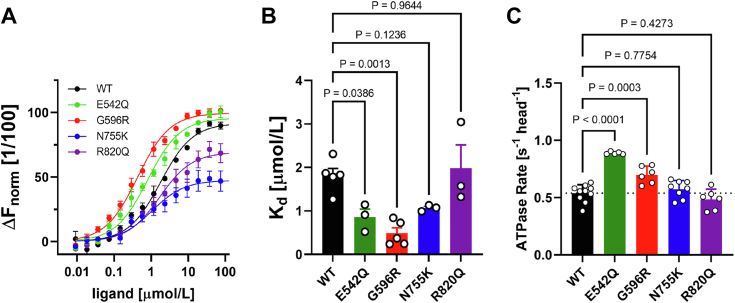
We have previously shown that the central domains of cMyBP-C can interact with the isolated myosin head domain (19), and we therefore performed microscale thermophoresis (MST) binding experiments of either the WT C3C6 or the four variants and the isolated bovine beta-cardiac myosin S1 fragment (Fig. 4A). In good agreement with previously published results, the C3C6 region of cMyBP-C binds bovine beta-cardiac myosin S1 with high affinity, as indicated by a steady-state dissociation constant (Kd) in the low micromolar range (Fig. 4B). Neither N755K nor R820Q had a significant effect on this interaction. In contrast, E542Q and G596R increased the affinity for myosin S1 as indicated by decreased Kd values.
Figure 4.
Effect of missense mutations on binding to myosin motor domain and actomyosin ATPase activity.A, microscale thermophoresis (MST) curves for WT and HCM variants of C3C6 binding to isolated bovine beta-cardiac myosin S1. B, summary of determined steady-state dissociation constants (Kd). C, normalized F-actin-stimulated bovine cardiac myosin S1 ATPase activity in the presence of WT (control) or in the presence of variant C3C6 fragments. Data are presented as means ± SD, n = 3 to 9. Statistical significance between WT and HCM variants was assessed with a one-way ANOVA followed by Tukey’s post hoc test. HCM, hypertrophic cardiomyopathy.
We then assessed the functional effects of HCM variants by measuring the ATPase activity of isolated bovine beta-cardiac myosin S1 in the presence of F-actin and C3C6 variants (Fig. 4C). In excellent agreement with the MST data, both N755K and R820Q showed no significant change in the actomyosin ATPase activity compared with WT C3C6 at a concentration of 10 μmol/l with an acto-myosin ATPase rate of about 0.5 s−1 head−1. In contrast, adding either E542Q or G596R significantly increased the ATPase activity by about 50% and 30%, respectively, suggesting these mutants might act as a less effective brake.
Cellular characterization of C3C6 WT and missense variants
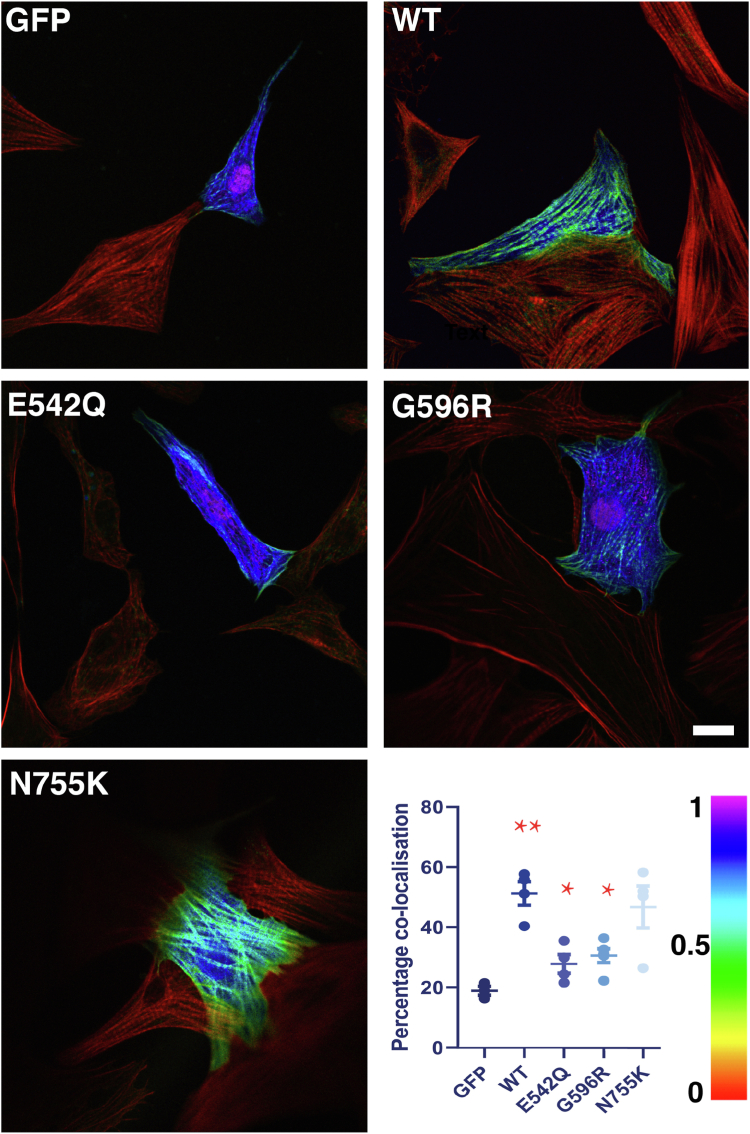
We then wanted to investigate the behavior of WT and missense C3C6 in a eukaryotic cellular environment and therefore generated GFP-tagged expression constructs for all of them. Initial analysis of expression in transiently transfected COS-1 cells revealed that the constructs were distributed mostly diffusely throughout the cells, with some association with actin filaments (data not shown) and that for WT, E542Q, G596R, and N755K bands with the expected molecular weight could be detected by immunoblotting (Fig. S2). To our surprise, no signal could be detected for the R820Q mutant, despite it expressing well in the bacterial system. Since COS-1 cells are a simplified experimental system, we then went on to study these GFP-tagged multidomain constructs in transiently transfected primary cultures of NRCs. Again, no transfected cells were seen for R820Q, whilst all the other constructs showed a mostly diffuse GFP signal, which seemed in part more intense along the myofibrils (Fig. S3). Previously, it was shown by others that an interaction site with the thin filament might sit in the central domains of MyBP-C (20). In order to evaluate a potential effect of the variants on this more precisely, we analyzed transiently transfected NRC followed by labeling of the F-actin cytoskeleton using phalloidin and then quantified the overlap of the GFP and F-actin signal on confocal micrographs using an algorithm written in Mathematica (Wolfram). The ratiometric images of these experiments are presented in Figure 5 and showed interesting differences between the different missense variants. Whilst N755K behaved exactly like WT C3C6 and showed association with the F-actin signal that was clearly more intense than the signal detected for GFP on its own (green signal in ratiometric images), less signal overlap was detected for E542Q and G596R, with levels only slightly higher than GFP on its own. This suggests that these two missense variants may have a decreased ability to interact with the thin (actin) filaments.
Figure 5.
Some missense mutations can compromise the association with cardiac thin filaments. GFP-tagged C3C6 constructs were transiently transfected into primary cultures of neonatal rat cardiomyocytes that were then counterstained for F-actin with phalloidin. The images shown are ratiometric (as quantified using a Mathematica algorithm), which means that high overlap of the GFP signal with F-actin is depicted in green, whilst low overlap is shown in blue/purple (see lookup table in bottom right corner; original images are shown in Fig. S3). WT C3C6 and the N755K variant display high spatial overlap with F-actin, which is significantly decreased for E542Q and G596R, which are similar to GFP on its own (see quantification). Scale bar equals 10 μm.
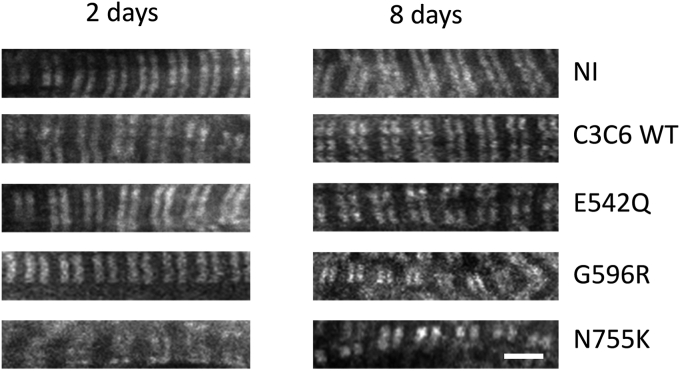
The aforementioned experiments already revealed mutation site-specific effects on C3C6 behavior. We then went on to study short-term and long-term effects of the expression of these constructs on myofibril integrity in NRC, as revealed by staining for endogenous cMyBP-C (using an antibody recognizing C1C2) followed by super-resolution microscopy using stimulated emission depletion (STED) (Fig. 6). Also at long term, the GFP signal for C3C6 remained mostly diffuse with increased signal overlapping with the myofibrils, and there was no evidence of aggregates (data not shown), as is for example seen for missense mutations in some isolated titin domains (21). However, overexpressing C3C6 appeared to cause some myofibril disarray in the transfected cells, and endogenous cMyBP-C doublets were more disorganized in the STED microscope. For N755K, this was already evident at 2 days, and the slight disarray became even more prominent at 8 days, when also some disarray was seen in the case of the G596R construct (for quantification, see Fig. S4).
Figure 6.
Expression of mutant GFP-tagged C3C6 can affect myofibril structure. STED images of primary cultures of neonatal rat cardiomyocytes transiently transfected with GFP-tagged C3C6 constructs. The images only show the signal for endogenous MyBP-C, as visualized by antibodies against C1 and C2 in the STED mode (the GFP constructs localized in a completely diffuse fashion). Whilst the WT construct has no effect, some of the mutant versions (G596R; N755K) lead to myofibril disarray of the endogenous cMyBP-C doublets, which becomes especially evident after 8 days in culture (for quantification, see Fig. S4). Scale bar equals 2 μm. cMyBP-C, cardiac isoform of myosin binding protein-C; STED, stimulated emission depletion.
Most of the full-length GFP-tagged cMYBP-C mutants incorporate normally into sarcomeres
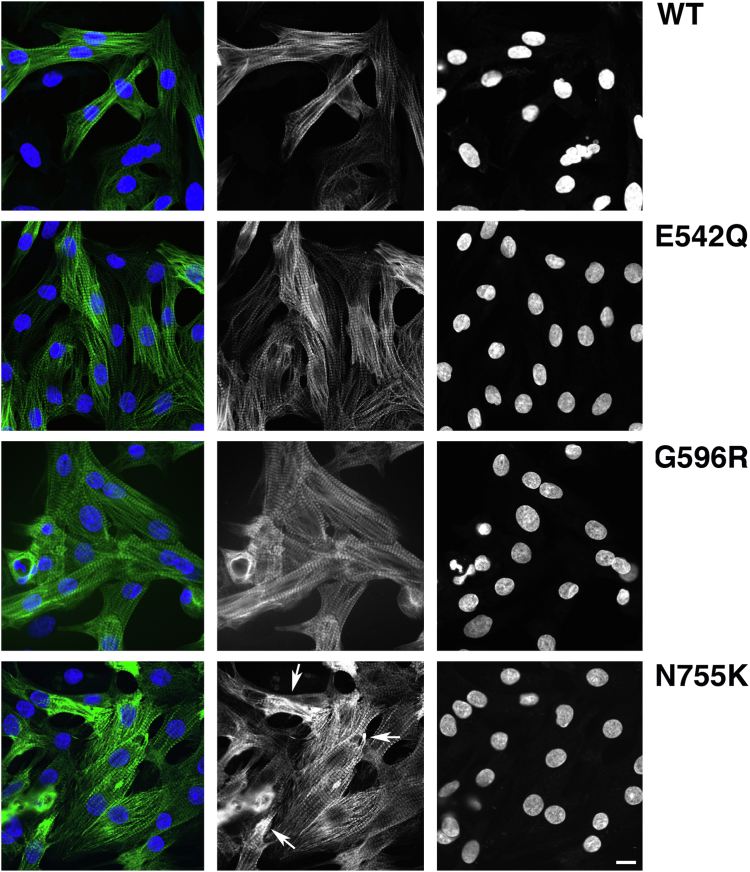
As the C3C6 constructs lack the C-terminal domains that are necessary for integration into the myofibrils (6, 7), we expressed full-length versions of N-terminally GFP-tagged cMyBP-C. Since primary cardiomyocytes do not transfect efficiently, we generated adenoviral constructs that expressed full-length WT or mutant cMyBP-C and used them to infect the NRC (Fig. S5). Usually, the stoichiometry of sarcomeric proteins is under tight control, which results in downregulation of the expression of endogenous proteins when GFP-tagged versions are expressed with adenovirus (11, 22). This appeared to be similar for most of our constructs. Only the N755K variant displayed aberrant behavior, with widely varying expression levels between different cardiomyocytes and also some subcellular areas that indicated protein aggregation (Fig. 7). Again, there was no signal detected for the adenovirus that expressed the full-length GFP-tagged R820Q variant (Fig. S5), suggesting that a failure to express this variant as a stable protein is a general phenomenon in eukaryotic cells and was not just an artifact of the multidomain construct. Quantification of the GFP signal in infected cells gave similar results to the transiently transfected cells, with evidence for overexpression and reduced striated signal for the N755K, whilst the WT and the other two variants, E542Q and G596R, were mostly present in a striated pattern (quantified in Fig. S6).
Figure 7.
With the exception of N755K, all full-length mutant GFP-tagged cMyBP-C versions integrate normally into the A-bands in short-term cultures. Confocal micrographs of primary cultures of neonatal rat cardiomyocytes that were infected with adenovirus encoding for full-length GFP-tagged WT or mutant cMyBP-C. GFP signal is shown in the middle column and in green in the overlay, DAPI (4′,6-diamidino-2-phenylindole) signal is shown in the right hand column and in blue in the overlay. Whilst WT, E542Q, and G592R show a homogeneous incorporation into the myofibrils with perfect doublets, accumulation in the GFP signal and inconsistent expression levels between cells are evident for the N755K mutant (for quantification, see Fig. S6). Scale bar equals 10 μm. cMyBP-C, cardiac isoform of myosin binding protein-C.
In conclusion, our experiments demonstrate that the different missense variants have distinct effects on cMyBP-C function, with some shifting the balance from thin filament interaction toward myosin binding (E542Q and G596R), whilst others affect sarcomeric targeting and myofibril structure (N755K) or stability of cMyBP-C protein in eukaryotic cells (R820Q).
Discussion
In this work, we show that different missense variants in the central domains of cMyBP-C have distinct effects on the behavior and the stability of the molecule. As the modeling in Figure 1C shows, these mutations sit in different positions on the Ig and Fn3 domains. The E542Q mutation is located in the linker region between C3 and C4, whilst both G596R and N755K reside in terminal loops between two beta-sheets in C4 and C5, respectively (23). NMR spectroscopy and unfolding experiments suggested that N755K may have a destabilizing effect on the loop it sits in, because the lysine is a bulkier and charged residue (23). The R820Q mutation sits on the surface on top of the Fn domain C6 and does not seem to have an effect on the structure of this domain as analyzed by NMR and small-angle X-ray scattering (24). None of these missense mutations is expected to dramatically affect the folding of the Ig domains or Fn3 domains, which is the likely explanation for why it was possible to express the recombinant multidomain proteins solubly. Whilst no effects on thermostability were detected for N755K and R820Q, the E542Q and G596R variants clearly showed two populations, one with a Tm close to WT, whilst the Tm of the other population was reduced by about 10 °C, suggesting partially impaired stability. This is somewhat different to missense mutations characterized for titin Ig and Fn3 domains that were analyzed by Rees et al. (21), which were usually found to be insoluble and to have a thermal stability that was 10 °C lower than the WT for all the variant proteins. The MyBP-C variants described here thus seem to have a much milder phenotype as far as protein folding is concerned. Another indication for compromised domain folding is the presence of aggregates that are often clumped around the Z-disc, when GFP-tagged versions of multidomain or single domain constructs are expressed in NRC (21). For most of the missense variants analyzed here, there was no evidence for pronounced aggregation in cells and the multidomain constructs localized mostly in a diffuse fashion with increased signal at the myofibrils, whilst full-length constructs incorporated as a doublet into the A-band, indistinguishable from endogenous MyBP-C. Constructs (either multidomain or full length) bearing the R820Q mutation failed to express at all in eukaryotic cells. This is puzzling at the moment, since the modeling suggests that the mutation is unlikely to have any dramatic effect on domain folding. It sits on the domain surface and probably affects protein–protein interaction (24). Currently, the potential interaction partner of cMyBP-C at C6 is unknown, but a closer investigation of the recent cryo-EM structure of human cardiac thick filaments from the Craig laboratory suggests that this residue may interact with the sentinel myosin (25). Impaired protein stability for a missense mutation in the central domains was reported previously by Smelter et al. (26) for W792R. Another intriguing possibility is that the R820Q variant causes mRNA destabilization. In good agreement, previous reports have shown that about a third of investigated variants cause protein depletion via alterations in mRNA splicing (27).
For both E542Q and G596R mutations, we see similar effects on the protein behavior, which might be due to their physical proximity on the three-dimensional structure of the molecule. There is a slight impairment of thermal stability and also a shift from thin filament interaction, which appears to be less efficient, toward increased myosin binding. C3C6 on its own does bind myosin in vitro (19) but does not integrate into myofibrils in doublets in cardiomyocytes as the full-length version would. This is probably because of the lack of C7C10, which were previously shown to be essential for proper sarcomeric targeting (6, 7). It was always assumed that the targeting is due to an interaction of C7C10 with both titin filaments and the myosin tails; however, very recent structural evidence on human cardiac thick filaments from the Craig laboratory shows that actually there is no direct interaction between the C terminus of MyBP-C and titin (25). The same structure also suggests that C4 might interact with a blocked myosin head, which could be the interaction that is affected in the E542Q and G596R variants.
Judging from our short-term experiments, the majority of the variants appear to integrate normally into the myofibrils and do not lead to massive disarray, similar to results reported by Helms et al. (11) for mutations in the central domains (R495Q; R502W; F503L; W792R; and R810H), whilst mutations in C10 (L1238P; N1257K) failed to incorporate into sarcomeres. Only the N755K variant displayed slightly impaired integration and a minor tendency to aggregate, which was also seen previously when this mutation was analyzed by NMR (23).
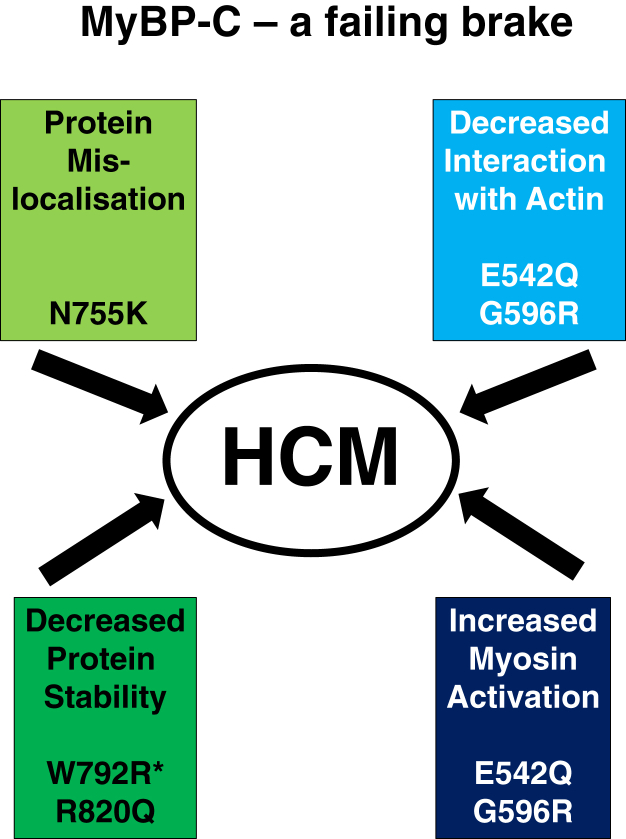
Taken together, our results suggest that missense mutations in the central domains of cMyBP-C have site-specific effects on the protein’s behavior in cells and its function. Whilst some (E542Q, G596R) appear to shift the balance of cMyBP-C interaction from actin (thin filament) toward myosin, another leads to a tendency for protein mislocalization and overexpression (N755K), whilst others cause decreased protein stability (R820Q; W792R; (26)), thereby causing haploinsufficiency (Fig. 8). All these changes can provide an explanation for cMyBP-C’s failure to work as a brake and to regulate contraction properly (8), which will eventually lead to a HCM phenotype. However, our data also indicate that additional missense mutations in MYPBC3 will have to be analyzed carefully for their effects on protein function and stability and that the prediction of disease-causing variants will only be straightforward in cases that have obvious consequences for domain folding like demonstrated for some of the titin domain mutants (21, 28).
Figure 8.
Proposed mechanisms for how different missense mutations in cMyBP-C lead to HCM. Depending on their position along the molecule, missense mutations in MyBP-C can either shift the balance of interaction with thin filaments and myosin (e.g., E542Q; G596R), lead to protein mislocalization and aggregation (N755K), or decrease protein stability (R820Q in this study; W792R in Ref. (26), indicated with an asterisk). cMyBP-C, cardiac isoform of myosin binding protein-C; HCM, hypertrophic cardiomyopathy.
Experimental procedures
Recombinant protein expression
A pET6a-His-TEV plasmid containing the C3C6 insert was transformed into BL21(DE3)RIPL E. coli cells (Agilent), liquid cultures were grown in Terrific Broth (Sigma), protein expression was induced by adding 0.1 mM of IPTG (VWR chemicals), and purification was carried out using a HisTrap FF column (GE Life Sciences). Protein expression was checked by immunoblot using a mouse anti-His antibody (Novagen). Protein containing fractions were pooled, and tobacco etch virus protease was added at a concentration of 1:50 to remove the His tag and was then dialyzed against a solution of 25 mM Hepes, 200 mM NaCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, overnight at 4 °C.
Generation of eukaryotic expression constructs
The C3C6 domain of cMyBP-C was amplified from adult human heart complementary DNA using forward (TTTCTCGAGTGAGCCCCCTGTGCTCATCACG) and reverse (TTTGGATCCACGCGTTTAACCGATAGG CATGAAGGG) primers (Sigma–Aldrich) and cloned into the XhoI and BamH1 restriction sites of a pEGFP-C2 vector (Clontech).
Site-directed mutagenesis of C3C6 constructs
HCM-linked mutations within the C3C6 region were identified using the ClinVar database (17), and the following primers were designed to generate these mutations: E542Q: reverse (CTCCAGCTTCTTTTGCTGCACAATGAGC); G596R: reverse (GGTGTCCCACATCAGGCGGGTCCACAAACTG); N755K: forward (GGTCACAGTGAAGAAACCTGTGGGCGAGG); R820Q: reverse (CAGCCGCATCCACTGGTAGCTCTTCTTC). The PCR was used to transform ultracompetent E. coli cells (ThermoFisher Scientific). Isolated DNA was sent off for sequencing in order to verify the correct mutation had occurred (Source Bioscience).
DSF
DSF experiments were performed as described previously (19). Briefly, C3C6 constructs at 1 mg/ml were mixed with 20X SYPRO Orange (Thermo Fisher Scientific) in PBS containing 1 mmol/l DTT, and 20 μl were transferred in individual wells of Bio-Rad quantitative PCR plates. Plates were heated from 25 to 95 °C at a rate of 1 °C/min in an MX3005p quantitative PCR machine (Agilent Technologies). Fluorescent emission at 610 nm following excitation at 492 nm was measured, with the resulting curve defined in Excel, and the Tm was calculated in GraphPad Prism (GraphPad Software, Inc).
Acto-myosin ATPase activity measurements
Bovine cardiac myosin S1 was prepared from bovine ventricle as previously described (29). Rabbit skeletal F-actin was purchased from Cytoskeleton, Inc and prepared for experiments according to the manufacturer’s instructions.
About 20 μl of enzyme mix in buffer A (composition in mmol/l: 10 Mops, 0.1 EGTA, 1 DTT, pH 7) containing 500 nmol/l bovine cardiac myosin S1, 40 U/ml lactate dehydrogenase, and 400 U/ml pyruvate kinase were dispensed into a black 96-well half area plate (Greiner). C3C6 fragments were added to each well to a final concentration of 10 μmol/l, and plates were incubated on a plate shaker at 30 °C for 10 min at 2000 rpm. Reactions were started by adding 20 μl substrate mix in buffer B (composition in mmol l−1: 4 Mops, 9.1 EGTA, 2 MgCl2, 3 NaN3, 1 DTT, pH 7.0) containing 10 μmol/l F-actin, 440 mmol/l NADH, 4 mmol/l 2-phosphoenolpyruvate, and 4 mmol/l ATP. Plates were briefly mixed by shaking at 5000 rpm on a plate shaker and spun down at 3000g for 10 s. NADH fluorescence intensity was measured for each well using a ClarioStar Plate Reader for 10 min every 30 s at 30 °C with the following settings: excitation at 380 nm with a 10 nm bandwidth and emission at 470 nm with a 24 nm bandwidth. ATPase activity was extracted by linear regression to changes in fluorescence intensity and converted to rates by using standards with defined ADP concentrations.
MST experiments
MST experiments were performed on a Monolith NT.115 instrument (NanoTemper) in interaction buffer containing 20 mmol/l Mops (pH 7), 1 mmol/l MgCl2, 50 mmol/l KCl, 1 mmol/l DTT, and 0.05% (v/v) Tween-20. Myosin S1 was labeled with Alexa 647-NHS (Molecular Probes, Inc; Thermo Fisher Scientific) according to the manufacturer's instructions. All proteins were either gel-filtered into and/or extensively dialyzed against interaction buffer. Titration experiments were performed with a fixed concentration of 50 nmol/l of Alexa 647-labeled myosin S1 in premium capillaries.
SEC–MALS
SEC–MALS experiments were performed in PBS on an Agilent 1260 Infinity II HPLC coupled to a SEC Analytical Column 5 μm 500 Å (WYATT Technologies) and DAWN MALS detector (WYATT Technologies).
COS-1 cells
COS-1 cells (30) were negative for mycoplasma, grown as described previously and transiently transfected in 35 mm culture dishes using Escort IV (Sigma) (31). For immunoblots, COS-1 cells were lysed in loading buffer (3.7 M urea, 134.6 mM Tris [pH 6.8], 5.4% SDS, 2.3% NP-40, 4.45% beta-mercaptoethanol, 4% glycerol, and 6 mg/ml bromophenol blue) following a PBS wash. The cell lysate was then boiled for 2 min and run on a precast 4 to 20% SDS-PAGE gel (Generon). After overnight wet blot transfer onto a nitrocellulose membrane (GE Life Sciences) and Ponceau Red (Sigma) staining, the membrane was blocked for 1 h on a rotator at RT in 5% skimmed milk (Sainsbury’s) in low salt (LS) buffer (0.9% NaCl; 0.1% Tween-20; 9 mM Tris base [pH 7.4]). The membrane was then incubated in primary antibody solution (Mouse anti GFP; Roche or Rabbit anti all actin isoforms; Sigma) in 3% milk/LS for 1 h at RT on a shaker. The membrane was washed three times for 5 min in 3% milk/LS and then incubated in secondary antibody solution (horseradish peroxidase–rabbit antimouse immunoglobulins; DAKO; horseradish peroxidase–goat anti-Rabbit immunoglobulins; Calbiochem) on a shaker at RT for 1 h. The membrane was washed three times 5 min in LS and afterward incubated in Clarity Western Enhanced Chemiluminescence solution (ECL, Bio-Rad). The membrane was then developed using a Bio-Rad gel imaging machine.
NRCs
Primary cultures of NRCs were isolated and cultured as described previously (31). For immunofluorescence staining, NRCs were washed with PBS and then fixed with 4% paraformaldehyde/PBS (Agar Scientific) for 10 min. Cells were then permeabilized with 0.2% Triton X-100 in PBS for 5 min, followed by a primary monoclonal mouse antibody against myomesin (32) and afterward incubation with counterstain solution consisting of Cy3-conjugated goat anti-mouse immunoglobulins (Jackson Immunochemicals), 4′,6-diamidino-2-phenylindole (Sigma), and Alexa Fluor 633 phalloidin (ThermoFisher) diluted in 1% bovine serum albumin/Tris-buffered saline in a humid chamber on a tilting table at RT for 1 h. Cells were washed three times for 5 min in PBS and then mounted using Lisbeth’s mounting medium (0.1 M Tris–HCl/glycerol and 50 mg/ml n-propyl-gallate at pH 9.5). Samples were imaged on a SP5 confocal microscope (Leica) using a 63/1.4 numerical aperture oil immersion lens and 405 blue diode, argon, and helium neon lasers as described previously (31).
Adenovirus constructs and infection
A pAd vector containing the full-length human cMyBP-C tagged at the N terminus with GFP was generated by VectorBuilder. Human embryonic kidney 293 cells were transiently transfected with the pAd full-length cMyBP-C DNA using Lipofectamine 3000 (ThermoFisher). The infection was allowed to continue until 80% cytopathic effect was seen and then adenovirus-containing cells were harvested and then underwent three times freeze–thaw cycles (−80 °C for 30 min followed by 37 °C for 15 min). The cell lysate was then centrifuged at 3000 RPM for 15 min, and the supernatant was taken and stored at −80 °C. To generate missense mutants, the full-length human cMyBP-C gene was amplified out of the pAd vector using forward (caattctctaaggaaatacttaaccATGGTGAGCAAGGGCGAG) and reverse (gtctagatatctcgacggatcctaTCACTGAGGCACTCGCAC) primers (Sigma–Aldrich) and cloned into a pENTR vector (ThermoFisher). Site-directed mutagenesis was then carried out on the pENTR full-length cMyBP-C construct using the primers described previously. Mutant pAd full-length cMyBP-C constructs were then propagated in human embryonic kidney 293 cells. For adenovirus titration, the AdEasy viral titer kit (Agilent Technologies) was used according to the manufacturer. NRCs were infected with WT adenovirus at a dilution of 1:100, and mutant constructs were added at an equivalent dilution based on the infectious units calculated via titration. Cells were incubated for 24 h at 37 °C, 5% CO2, and then either SDS samples were made for immunoblot analysis to determine protein expression levels (as aforementioned), or cells were fixed and immunostained for confocal microscopy as described previously.
STED microscopy
NRCs were transiently transfected with GFP-tagged C3C6 constructs using Escort III (Sigma) as described previously (31) and then fixed and permeabilized as aforementioned. Immunostaining was carried out using primary antibodies against mouse antisarcomeric alpha-actinin (Sigma) and rabbit anti MyBP-C (a generous gift from Prof Mathias Gautel, Kings College London) and secondary antibody/counterstain solution (Cy3-goat anti-mouse, catalog no.: 115-165-146; Jackson Immunochemicals) and Atto 647N-goat anti-rabbit (Sigma). Cells were mounted using 70% glycerol/PBS according to the antibody manufacturer instructions. Samples were imaged using a STEDYCON (Abberior) attached to a Leica TCS SP5 confocal microscope equipped with 594 nm and 640 nm excitation lasers and a 775 nm depletion laser using a 100×/numerical aperture 1.4 oil immersion lens. Images were recorded at a pixel size of 20 nm.
STED image analysis
STED images along with their respective metadata-containing text files were imported into Mathematica 12.2. The images were then transformed using a fast Fourier transform and converted to absolute magnitude to give a 2D spectrogram. The original image was multiplied by a Gaussian kernel with the same image dimensions to reduce edge artifacts. An inverse fast Fourier transform was then applied to this to produce a 2D cepstrogram. This shows a linear series of bands emanating from the center of the spectrogram with a spacing that is the same as the original image. The cepstrogram was then rotated so that the major axis of the signal was horizontal. The cepstrogram was then further transformed using a radon transform, which shows distance of signal versus angle/direction. This has the benefit of smearing any background noise to give a cleaner signal. A line scan through the middle of the radon transform was then taken to visualize intensity as a profile plot. These profiles were used to calculate the average sarcomere length (the major nonzero peak in the profiles). These plots were then normalized to take account of slight variations in sarcomere length and the mean and standard deviations calculated for each mutant and control. Images of cells displaying good MyBP-C localization produced profiles that had two further intensity peaks corresponding to the MyBP-C signal positioned between the two major peaks in the profile. This was accentuated by removing the baseline signal using the Difference of Gaussians approach (similar to background subtraction as commonly used in image processing). It was then possible to extract the intensity of the first MyBP-C peak as a measure of signal integrity. It was found that this peak was the best indicator for this.
Image processing and image analysis
Figures were compiled using Adobe Photoshop software. NRCs were transiently transfected with either GFP alone or a GFP-tagged WT C3C6 containing plasmid, immunostained for F-actin, and images were taken using a confocal microscope. Mathematica 11.3 software was then used to calculate the arctangent of the intensities in the red channel compared with those in the green channel and normalized to a linear scale of 0 to 1, with values close to zero (red pixels) indicating F-actin alone, values close to one (purple/dark blue) indicating the GFP-tagged C3C6 containing construct alone, and values around 0.5 (green/cyan) indicating the presence of both with similar intensities (A). ImageJ was then utilized to determine the percentage of the cell that was green.
Data availability
All data are within the article (main figures and supporting information); original files (data measurements; full-length gels and blots; Mathematica algorithm) are available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Amy Pearce was a participant of the BHF MRes/PhD programme at the School of Cardiovascular Medicine and Sciences at King's College London (FS/16/57/32733). Rylan Beckingham is a PhD student funded by the BHF Centre of Research Excellence at King's College London (RE/18/2/34213). We are grateful to Prof Mathias Gautel for the donation of the rabbit anti-MyBP-C antibody and to all laboratory members for support and scientific discussions. Work in the Ehler laboratory was supported by UKRI-MRC (grant no.: MR/R017050/1) and by the BHF. Work in the Kampourakis laboratory was supported by a British Heart Foundation Intermediate Basic Science Research Fellowship (grant no.: FS/16/3/31887). The SEC-MALS work was carried out at the Centre for Biomolecular Spectroscopy at King's College London on an instrument that had been funded by the BBSRC (BBSRC 20ALERT BB/V01966X/1).
Author contributions
E. E. conceptualization; M. R. H. software; M. R. H. and T. K. formal analysis; A. P., S. P., R. B., and A. L. K. investigation; E. E. writing–original draft; A. P., M. R. H., T. K., and E. E. writing–review & editing; T. R., T. K., and E. E. supervision; T. K. and E. E. project administration; T. K. and E. E. funding acquisition.
Reviewed by members of the JBC Editorial Board. Edited by Enrique De La Cruz
Supporting information
SEC-MALS chromatograms for WT and mutant C3C6 constructs. Light scatter and refractive index are shown in blue and red, respectively. Calculated molar masses (black squares) are shown superimposed onto the chromatograms.
Expression of GFP-tagged C3C6 constructs in COS-1 cells. COS-1 cells were transiently transfected with eukaryotic expression plasmids and SDS samples were subjected to SDS-PAGE and immunoblotting with anti GFP antibodies. A, antibodies against all actin isoforms were used as a loading control. B, the first lane are not transfected (NT), followed by GFP on its own, WT C3C6, and the missense mutants (bottom). All constructs yielded a signal of the expected molecular weight, except for GFP-tagged R820Q, where hardly any signal was detected despite similar amounts of loaded protein as evident from the actin band below. Molecular weight markers are indicated on the left.
Confocal micrographs of GFP-tagged C3C6 constructs after transient expression in NRC. Original data for the ratiometric images shown in Figure 5. Transiently transfected NRC (GFP in green) were stained for DAPI (blue signal in overlay) and counterstained for the M-band myomesin to validate their cardiomyocyte identity (red signal in overlay). The scale bar represents 10 μm.
Quantification of the STED signal for endogenous cMyBP-C in NRC that were transiently transfected with different GFP-tagged C3C6 constructs. Short-term (ST) cultures (2 days) are shown on the left and long-term (LT; 8 days) on the right. An algorithm written in Mathematica, described under Experimental procedures, was used for the quantification. The plotted cepstrograms show a series of peaks. The middle peak and those to the far left and right (annotated with green arrows) are derived from the distance between adjacent sarcomeres based on gross cMYBP-C organization. The four other minor peaks (light blue arrows) are derived from the position and integrity of the local cMyBP-C signal. The more clear and obvious that these peaks are represents greater structural integrity. Less clear peaks, as seen with some mutants, represent less structural integrity (G596R; N755K; W792R), and lower peaks represent less protein (W792R in LT). Dark blue curves represent the averaged data, while red curves represent the standard deviation.
Adenoviral expression of GFP-tagged cMyBP-C in NRC. NRC were infected with adenovirus encoding for full-length GFP-tagged cMyBP-C (WT and missense variants), and SDS samples were subjected to SDS-PAGE and immunoblotting with anti-MyBP-C (top) and GFP antibodies (middle). Ponceau Red is shown as a loading control (bottom). The first lane is not infected NRC (1), followed by WT (2), E542Q (3), G596R (4), N755K (5), and R820Q (6). Except for GFP-tagged R820Q, bands of the expected molecular weight were detected for all exogenous constructs (for the MyBP-C antibody above the endogenous cMyBP-C). Molecular weight markers are indicated on the left.
Quantification of localization behavior of full-length GFP-tagged cMyBP-C in NRC. Adenovirus encoding for the different variants were used to infect primary cultures of neonatal rat cardiomyocytes, and the GFP signal was quantified on confocal micrographs using the classifications shown on the right. The N755K mutant showed impaired integration into sarcomeres compared to WT and the other two mutants.
References
- 1.Frey N., Luedde M., Katus H.A. Mechanisms of disease: hypertrophic cardiomyopathy. Nat. Rev. Cardiol. 2012;9:91–100. doi: 10.1038/nrcardio.2011.159. [DOI] [PubMed] [Google Scholar]
- 2.Heling L.W.H.J., Geeves M.A., Kad N.M. MyBP-C: one protein to govern them all. J. Muscle Res. Cell Motil. 2020;41:91–101. doi: 10.1007/s10974-019-09567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gautel M., Zuffardi O., Freiburg A., Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfuhl M., Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J. Muscle Res. Cell Motil. 2012;33:83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 5.Craig R., Lee K.H., Mun J.Y., Torre I., Luther P.K. Structure, sarcomeric organization, and thin filament binding of cardiac myosin-binding protein-C. Pflugers Arch. 2014;466:425–431. doi: 10.1007/s00424-013-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert R., Kelly M.G., Mikawa T., Fischman D.A. The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J. Cell Sci. 1996;109:101–111. doi: 10.1242/jcs.109.1.101. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert R., Cohen J.A., Pardo S., Basu A., Fischman D.A. Identification of the A-band localization domain of myosin binding proteins C and H (MyBP-C, MyBP-H) in skeletal muscle. J. Cell Sci. 1999;112:69–79. doi: 10.1242/jcs.112.1.69. [DOI] [PubMed] [Google Scholar]
- 8.Previs M.J., Beck Previs S., Gulick J., Robbins J., Warshaw D.M. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337:1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampourakis T., Yan Z., Gautel M., Sun Y.B., Irving M. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18763–18768. doi: 10.1073/pnas.1413922112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marston S., Copeland O., Gehmlich K., Schlossarek S., Carrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J. Muscle Res. Cell Motil. 2012;33:75–80. doi: 10.1007/s10974-011-9268-3. [DOI] [PubMed] [Google Scholar]
- 11.Helms A.S., Thompson A.D., Glazier A.A., Hafeez N., Kabani S., Rodriguez J., et al. Spatial and functional distribution of MYBPC3 pathogenic variants and clinical outcomes in patients with hypertrophic cardiomyopathy. Circ. Genom Precis Med. 2020;13:396–405. doi: 10.1161/CIRCGEN.120.002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris S.P., Lyons R.G., Bezold K.L. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ. Res. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kittleson M.D., Meurs K.M., Harris S.P. The genetic basis of hypertrophic cardiomyopathy in cats and humans. J. Vet. Cardiol. 2015;17 Suppl 1:S53–S73. doi: 10.1016/j.jvc.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Geist J., Grogan A., Hu L.R., Kontrogianni-Konstantopoulos A. Thick filament protein network, functions, and disease association. Compr. Physiol. 2018;8:631–709. doi: 10.1002/cphy.c170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Lange W.J., Grimes A.C., Hegge L.F., Spring A.M., Brost T.M., Ralphe J.C. E258K HCM-causing mutation in cardiac MyBP-C reduces contractile force and accelerates twitch kinetics by disrupting the cMyBP-C and myosin S2 interaction. J. Gen. Physiol. 2013;142:241–255. doi: 10.1085/jgp.201311018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadayappan S., Gulick J., Osinska H., Martin L.A., Hahn H.S., Dorn G.W., 2nd, et al. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ. Res. 2005;97:1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponnam S., Kampourakis T. Microscale thermophoresis suggests a new model of regulation of cardiac myosin function via interaction with cardiac myosin-binding protein C. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2021.101485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inchingolo A.V., Previs S.B., Previs M.J., Warshaw D.M., Kad N.M. Revealing the mechanism of how cardiac myosin-binding protein C N-terminal fragments sensitize thin filaments for myosin binding. Proc. Natl. Acad. Sci. U. S. A. 2019;116:6828–6835. doi: 10.1073/pnas.1816480116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rees M., Nikoopour R., Fukuzawa A., Kho A.L., Fernandez-Garcia M.A., Wraige E., et al. Making sense of missense variants in TTN-related congenital myopathies. Acta Neuropathol. 2021;141:431–453. doi: 10.1007/s00401-020-02257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michele D.E., Albayya F.P., Metzger J.M. Thin filament protein dynamics in fully differentiated adult cardiac myocytes: toward a model of sarcomere maintenance. J. Cell Biol. 1999;145:1483–1495. doi: 10.1083/jcb.145.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idowu S.M., Gautel M., Perkins S.J., Pfuhl M. Structure, stability and dynamics of the central domain of cardiac myosin binding protein C (MyBP-C): implications for multidomain assembly and causes for cardiomyopathy. J. Mol. Biol. 2003;329:745–761. doi: 10.1016/s0022-2836(03)00425-x. [DOI] [PubMed] [Google Scholar]
- 24.Nadvi N.A., Michie K.A., Kwan A.H., Guss J.M., Trewhella J. Clinically linked mutations in the central domains of cardiac myosin-binding protein C with distinct phenotypes show differential structural effects. Structure. 2016;24:105–115. doi: 10.1016/j.str.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Dutta D., Nguyen V., Campbell K.S., Padrón R., Craig R. Cryo-EM structure of the human cardiac myosin filament. Nature. 2023;623:853–862. doi: 10.1038/s41586-023-06691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smelter D.F., de Lange W.J., Cai W., Ge Y., Ralphe J.C. The HCM-linked W792R mutation in cardiac myosin-binding protein C reduces C6 FnIII domain stability. Am. J. Physiol. Heart Circ. Physiol. 2018;314:H1179–H1191. doi: 10.1152/ajpheart.00686.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suay-Corredera C., Pricolo M.R., Herrero-Galan E., Velazquez-Carreras D., Sanchez-Ortiz D., Garcia-Giustiniani D., et al. Protein haploinsufficiency drivers identify MYBPC3 variants that cause hypertrophic cardiomyopathy. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laddach A., Gautel M., Fraternali F. TITINdb-a computational tool to assess titin's role as a disease gene. Bioinformatics. 2017;33:3482–3485. doi: 10.1093/bioinformatics/btx424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohde J.A., Thomas D.D., Muretta J.M. Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1796–E1804. doi: 10.1073/pnas.1611698114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 31.Iskratsch T., Lange S., Dwyer J., Kho A.L., dos Remedios C., Ehler E. Formin follows function: a muscle specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 2010;191:1159–1172. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grove B.K., Kurer V., Lehner C., Doetschman T.C., Perriard J.C., Eppenberger H.M. Monoclonal antibodies detect new 185,000 dalton muscle M-line protein. J. Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SEC-MALS chromatograms for WT and mutant C3C6 constructs. Light scatter and refractive index are shown in blue and red, respectively. Calculated molar masses (black squares) are shown superimposed onto the chromatograms.
Expression of GFP-tagged C3C6 constructs in COS-1 cells. COS-1 cells were transiently transfected with eukaryotic expression plasmids and SDS samples were subjected to SDS-PAGE and immunoblotting with anti GFP antibodies. A, antibodies against all actin isoforms were used as a loading control. B, the first lane are not transfected (NT), followed by GFP on its own, WT C3C6, and the missense mutants (bottom). All constructs yielded a signal of the expected molecular weight, except for GFP-tagged R820Q, where hardly any signal was detected despite similar amounts of loaded protein as evident from the actin band below. Molecular weight markers are indicated on the left.
Confocal micrographs of GFP-tagged C3C6 constructs after transient expression in NRC. Original data for the ratiometric images shown in Figure 5. Transiently transfected NRC (GFP in green) were stained for DAPI (blue signal in overlay) and counterstained for the M-band myomesin to validate their cardiomyocyte identity (red signal in overlay). The scale bar represents 10 μm.
Quantification of the STED signal for endogenous cMyBP-C in NRC that were transiently transfected with different GFP-tagged C3C6 constructs. Short-term (ST) cultures (2 days) are shown on the left and long-term (LT; 8 days) on the right. An algorithm written in Mathematica, described under Experimental procedures, was used for the quantification. The plotted cepstrograms show a series of peaks. The middle peak and those to the far left and right (annotated with green arrows) are derived from the distance between adjacent sarcomeres based on gross cMYBP-C organization. The four other minor peaks (light blue arrows) are derived from the position and integrity of the local cMyBP-C signal. The more clear and obvious that these peaks are represents greater structural integrity. Less clear peaks, as seen with some mutants, represent less structural integrity (G596R; N755K; W792R), and lower peaks represent less protein (W792R in LT). Dark blue curves represent the averaged data, while red curves represent the standard deviation.
Adenoviral expression of GFP-tagged cMyBP-C in NRC. NRC were infected with adenovirus encoding for full-length GFP-tagged cMyBP-C (WT and missense variants), and SDS samples were subjected to SDS-PAGE and immunoblotting with anti-MyBP-C (top) and GFP antibodies (middle). Ponceau Red is shown as a loading control (bottom). The first lane is not infected NRC (1), followed by WT (2), E542Q (3), G596R (4), N755K (5), and R820Q (6). Except for GFP-tagged R820Q, bands of the expected molecular weight were detected for all exogenous constructs (for the MyBP-C antibody above the endogenous cMyBP-C). Molecular weight markers are indicated on the left.
Quantification of localization behavior of full-length GFP-tagged cMyBP-C in NRC. Adenovirus encoding for the different variants were used to infect primary cultures of neonatal rat cardiomyocytes, and the GFP signal was quantified on confocal micrographs using the classifications shown on the right. The N755K mutant showed impaired integration into sarcomeres compared to WT and the other two mutants.
Data Availability Statement
All data are within the article (main figures and supporting information); original files (data measurements; full-length gels and blots; Mathematica algorithm) are available upon request.