Abstract
In the allosteric aspartate transcarbamylase (ATCase) from the hyperthermophilic eubacterium Thermotoga maritima, the catalytic and regulatory functions, which in class B ATCases are carried out by specialized polypeptides, are combined on a single type of polypeptide assembled in trimers. The ATCases from T. maritima and Treponema denticola present intriguing similarities, suggesting horizontal gene transfer.
The Thermotogales represent one of the deepest branches and most slowly evolving lineages within the bacterial domain of life (5). They consist exclusively of extreme thermophiles and hyperthermophiles of hydrothermal origin. The species Thermotoga grows from 50 to 90°C, with an optimum at 80°C. The metabolic enzymes from this species investigated so far show a very high intrinsic thermostability (3, 7, 8, 23).
Aspartate transcarbamylase (ATCase) catalyzes the condensation of l-aspartate and carbamylphosphate (CP) in the first step of pyrimidine biosynthesis. CP is a very thermolabile compound, and its degradation product, cyanate, is a toxic carbamylating agent (10). Therefore, organisms growing at high temperatures must have evolved strategies to protect CP from thermal degradation. Channeling of this metabolite was indeed observed in Pyrococcus furiosus (10), Pyrococcus abyssi (14), and Thermus aquaticus (17, 21).
In this paper, we show that Thermotoga maritima synthesizes a new type of ATCase. The catalytic and regulatory chains which, in the class B ATCases from members of the Enterobacteriaceae (22), Vibrio sp. (25), and Archaea (2, 4, 15), are encoded by separated genes are fused in a single polypeptide in Thermotoga.
Cloning in Escherichia coli and sequencing of the T. maritima ATCase gene.
Two types of cloning experiments were performed by complementation of an E. coli pyrBI deletion mutant with genomic DNA prepared from T. maritima as described elsewhere (11).
First, one single clone growing on a medium without uracil which contained a 2.5-kb HindIII insert in the pKK 223-3 vector was obtained (1). The corresponding plasmid pTM8 harbors a gene which expresses an unstable and unregulated enzyme (17, 19).
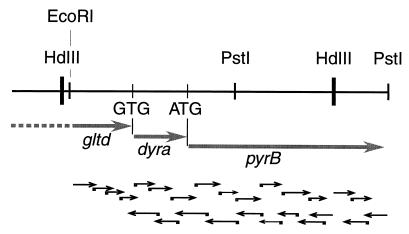
A second library of T. maritima prepared by ligation of genomic DNA partially digested by Sau3A in the λZAP Express vector linearized by BamHI (Stratagene) allowed isolation of a clone containing plasmid pTM14 with a 5.7-kb insert containing supplementary DNA downstream of the 2.5-kb HindIII fragment present in pTM8 (Fig. 1). A 0.5-kb HindIII-PstI fragment located immediately downstream of the sequence present in pTM8 was subcloned and found to contain the C-terminal part of the T. maritima ATCase gene. Analysis of the nucleotide sequence (16) of the 1,578-base open reading frame (ORF) encoding ATCase is preceded by a truncated gltD gene coding for the small chain of glutamate synthetase (EC 1.4.1.13.) and by a complete dyrA gene coding for dihydrofolate reductase (20). Two terminator-like structures were found upstream of the ATCase ORF. Therefore, it seems unlikely that the gene is expressed from the vector promoter. It seems more plausible that the ATCase gene is expressed from a Thermotoga putative promoter sequence located 40 nucleotides upstream of the ATG start codon (Fig. 2).
FIG. 1.
Sequencing strategy and genetic organization in the pyrBI region of T. maritima. Thick arrows, identified open reading frames (genes); thin black arrows, directions and extents of DNA sequencing.
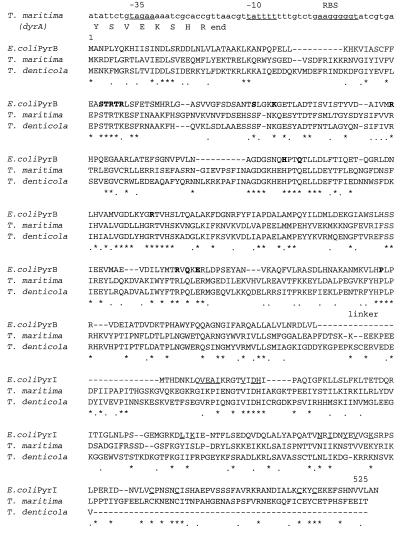
FIG. 2.
Alignment of the T. maritima ATCase sequence with the E. coli and T. denticola ATCases. Residues forming the catalytic site in E. coli are in boldface; those involved in the regulatory site as well as the four cysteines coordinated with the zinc atom are underlined. The nucleotide sequence upstream of the T. maritima pyrB::I gene is given, with the putative promoter sequence (−35 and −10) and the putative ribosome binding site (RBS). Asterisks, identities; dots, conservative substitutions.
This ORF is unusually long for an ATCase gene, i.e., 525 amino acids (aa). The N-proximal 335 aa show identity with other PyrB sequences (9). The C-proximal region (155 aa) is homologous to the PyrI regulatory chain from enteric bacteria (28.1 to 29.5% identity). A short linker (residues 336 to 370) separates the two regions. Moreover, deletion of the last 100 aa of this region results in the production of an unregulated and unstable enzyme (references 17 and 19 and this work). T. maritima ATCase thus exhibits a new type of structure in which the catalytic and regulatory regions appear to be combined in a single PyrBI polypeptide chain. Alignments with 31 ATCase genes (9) revealed that the T. maritima PyrBI protein presents the highest identity with T. denticola ATCase (51.7%). This identity spreads over the whole length of the published T. denticola ATCase sequence (475 aa) (6), including the linker region and the first half of the regulatory region (Fig. 2). Therefore, the PyrI region of T. denticola ATCase appears to be truncated and to lack the zinc-binding motive of the classical PyrI regulatory chains. However, if two frameshifts are introduced near the end of the T. denticola gene sequence, a 534-aa ORF with a full-length PyrI region is obtained, showing 50% identity with the PyrI region of the T. maritima enzyme and harboring the four cysteine residues coordinating the Zn atom in the canonical zinc-binding domain of class B ATCases. Nevertheless, the recombinant T. denticola enzyme did not appear to be regulated by CTP in crude extracts of the recombinant E. coli strain (6). This intriguing protein is worth further investigation, first of all to determine whether the frameshift brought to light by comparison with T. maritima is not due to a sequencing artifact. At any rate, when the widely different positions of Thermotoga and Treponema in the eubacterial phylogenetic tree are considered, the apparent kinship between these two ATCases is also intriguing. Such a situation suggests that horizontal transfer of an ATCase gene has occurred between organisms related to Thermotoga and Treponema (12).
In view of the position of T. maritima as a very ancient and slowly evolving lineage near the root of the universal phylogenetic tree, it is conceivable that the genetic organization pyrB:I is an ancestral one and that the separation between pyrB and pyrI occurred later in evolution.
Purification of T. maritima ATCase produced in E. coli.
T. maritima ATCase was purified to homogeneity from extracts of the E. coli strain containing the pTM14 plasmid. A 30-g amount of cells was resuspended in 120 ml of phosphate storage buffer (PSB [40 mM KH2PO4, 0.2 mM EDTA, 2 mM mercaptoethanol adjusted to pH 7.0 with KOH]), to which l-aspartate (10 mM) and DNase (50 μg · ml−1) were added. The cell suspension was sonicated for 5 min with a Raytheon sonicator and was centrifuged for 30 min at 20,000 × g to remove the cell debris. The extract was incubated at 78°C for 15 min and cleared of the precipitate formed by centrifugation (15 min at 20,000 × g). Ammonium sulfate was added to 70%, and the precipitate was collected by centrifugation. The pellet was resuspended in PSB, dialyzed against the same buffer, and concentrated by filtration on an Amicon membrane (PM30). The protein sample was loaded on a Pharmacia Mono Q ion-exchange column and was eluted by a phosphate gradient (40 to 600 mM). The fractions showing high-level activity were pooled, concentrated, and dialyzed against PSB. This sample was then loaded on a Pharmacia Mono S ion-exchange column and eluted by a salt gradient (0 to 500 mM KCl in PSB). The fractions having ATCase activity were treated as described above and loaded on a Pharmacia P12 molecular sieving column from which the pure enzyme was obtained by elution with PSB containing 150 mM NaCl. The overall yield was 15%, and the purification factor was 1,315. The pure enzyme eluted as a single peak corresponding to an Mr of approximately 200. The molecular mass of the monomeric unit calculated from the sequence was 60.5. Therefore, it appears that the native enzyme has a trimeric structure (c:r)3. Considering the fact that in Thermotoga the catalytic and regulatory gene equivalents form a single unit, this trimeric structure, (c:r)3, would be equivalent to one moiety (one hemisphere) of the 2c33r2 architecture of class B ATCases.
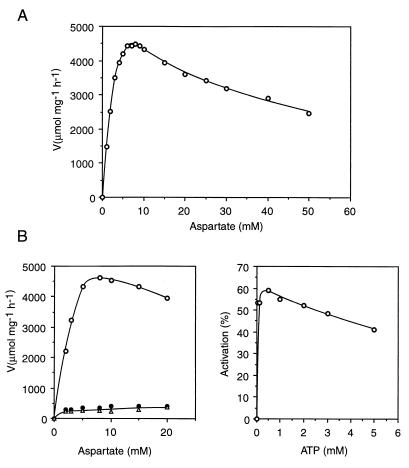
The enzymatic activity of the purified enzyme was measured by the colorimetric method (13) as described elsewhere (24). The activity proved stable up to 75°C (15 min), whereas E. coli ATCase lost most of its activity after such a treatment. The saturation curve by aspartate presented a marked inhibition by excess substrate (Fig. 3A). The curve was sigmoidal, and the kinetic parameters were calculated by fitting the data to the Hill equation were as follows: maximal velocity, 4,715 ± 135 μmol of carbamylaspartate (C-Asp) mg−1 h−1; aspartate concentration giving half-maximal velocity (S0.5Asp), 3.5 ± 0.3 mM; and Hill coefficient (nH), 1.8 ± 0.2. The regulatory properties were investigated by measuring the effect on the aspartate concentration curve of the addition of 2 mM nucleotides (ATP, CTP, or UTP) (Fig. 3B). CTP and UTP were potent inhibitors (up to 90% inhibition), whereas ATP had an activating effect. The ATP effect was investigated further by examining the effect of increasing concentrations (0 to 5 mM) in the presence of 3 mM aspartate and 5 mM CP (Fig. 3B).
FIG. 3.
Properties of the pure recombinant T. maritima ATCase. (A) Saturation by l-aspartate. (B) Regulation by nucleotide effectors. Left panel, saturation by l-aspartate in the absence of effectors (○) and in the presence of CTP (•) or UTP (▵); right panel, effect of increasing ATP concentration on activity at an aspartate concentration of S0.5 (3 mM).
In Thermotoga ATCase, all of the residues which were shown in E. coli to participate in the catalytic site are conserved (Fig. 2), and most of the residues known to interact with the nucleotides in E. coli are identical or similar. This suggests that the catalytic mechanism and the binding of the allosteric effectors in these enzymes are similar. When examining the two types of c-c interfaces described for E. coli ATCase, one observes that the residues connecting two catalytic chains within a trimer in E. coli are mostly conserved in T. maritima (9 of 13 are identical). This is also in keeping with the fact that the active site is shared between two adjacent c chains. On the other hand, the residues interacting at the c1-c4 type of interface (connecting two catalytic chains belonging to different trimers in class B ATCases) are much less conserved (only 3 of 10 are identical). When the E. coli r-c interfaces are considered, the corresponding residues are also more conserved (15 of 28) for the r1-c1 interface, which connects the two types of chains in the same half of the molecule, than for the r1-c4 interface, which connects a catalytic chain with a regulatory chain of the other hemisphere (only 3 of 15 are identical). Moreover, none of the residues belonging to the r1-r6 interface in the E. coli enzyme is identical in the Thermotoga ATCase. This suggests that T. maritima ATCase is similar to the basic trimer of other ATCases but that functional association of two T. maritima trimers is not possible, due to the absence of key residues at the appropriate location. We propose to designate this new type of ATCase class B′.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the EMBL Nucleotide Sequence database under accession no. Y10300.
Acknowledgments
This work was supported by a grant from the Flanders Foundation for Scientific Research and by a concerted action between the University and the Belgian State and the EEG-sponsored biotechnology program.
REFERENCES
- 1.Brosius J, Holy A. Regulation on ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geohaggen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelly J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1056–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Dams T, Bohm G, Auerbach G, Bader G, Schurig H, Jaenicke R. Homo-dimeric recombinant dihydrofolate reductase from Thermotoga maritima shows extreme intrinsic stability. Biol Chem. 1998;379:367–371. [PubMed] [Google Scholar]
- 4.Durbecq V, Thia-Toong T L, Charlier D, Roovers M, Legrain C, Glansdorff N. Aspartate carbamoyltransferase from the extremely thermophilic archaeon Sulfolobus solfataricus: gene cloning, sequence analysis and enzyme characterization. Arch Physiol Biochem. 1977;106:B6. [Google Scholar]
- 5.Hüber R, Langworthy T A, König H, Thomm M, Woese C R, Sleyter V W, Stetter K O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol. 1986;144:324–333. [Google Scholar]
- 6.Ishihara K, Ishihara M, Takazoe I, Okuda K. Cloning and expression of the aspartate carbamoyltransferase gene from Treponema denticola. Appl Environ Microbiol. 1992;58:3399–3403. doi: 10.1128/aem.58.10.3399-3403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeanicke R. Glyceraldehyde-3-phosphate dehydrogenase from Thermotoga maritima: strategies of protein stabilization. FEMS Microbiol Rev. 1996;18:215–224. doi: 10.1111/j.1574-6976.1996.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 8.Jeanicke R, Schurig H, Baucamp N, Ostendorp R. Structure and stability of hyperstable proteins: glycolytic enzymes from hyperthermophilic bacterium Thermotoga maritima. Adv Prot Chem. 1996;48:191–269. doi: 10.1016/s0065-3233(08)60363-0. [DOI] [PubMed] [Google Scholar]
- 9.Labedan, B., A. Boyen, M. Baetens, D. Charlier, P. Chen, R. Cunin, V. Durbecq, N. Glansdorff, G. Hervé, C. Legrain, Z.-Y. Liang, C. Purcarea, M. Roovers, R. Sanchez, T. L. Thia-Toong, M. Van de Casteele, F. Van Vliet, Y. Xu, and Y.-F. Zhang. The evolutionary history of carbamoyltransferases: a complete set of paralogous genes was already present in the last universal ancestor. J. Mol. Evol., in press. [DOI] [PubMed]
- 10.Legrain C, Demarez M, Glansdorff N, Piérard A. Ammonia-dependent synthesis and metabolic channeling of carbamoylphosphate in the hyperthermophilic archaeon Pyrococcus furiosus. Microbiology. 1995;141:1093–1099. doi: 10.1099/13500872-141-5-1093. [DOI] [PubMed] [Google Scholar]
- 11.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennisi E. Genome data shake tree of life. Science. 1998;280:672–674. doi: 10.1126/science.280.5364.672. [DOI] [PubMed] [Google Scholar]
- 13.Prescott M L, Jones M E. Modified methods for the determination of carbamyl aspartate. Anal Biochem. 1969;32:408–419. doi: 10.1016/s0003-2697(69)80008-4. [DOI] [PubMed] [Google Scholar]
- 14.Purcarea C. Etude des enzymes du métabolisme du carbamylphosphate chez l’archaebactérie marine hyperthermophile et barophile Pyrococcus abyssi. Ph.D. thesis. Orsay, France: Université de Paris-Sud; 1995. [Google Scholar]
- 15.Purcarea C, Hervé G, Ladjimi M M, Cunin R. Aspartate transcarbamylase form the deep-sea hyperthermophilic archaeon Pyrococcus abyssi: genetic organization, structure, and expression in Escherichia coli. J Bacteriol. 1997;179:4142–4157. doi: 10.1128/jb.179.13.4143-4157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van de Casteele M. The metabolic and genetic control of carbamoylation in extreme thermophilic eubacteria. Ph.D. thesis. Brussels, Belgium: Vrije Universiteit Brussel; 1994. [Google Scholar]
- 18.Van de Casteele M, Demarez M, Legrain C, Glansdorff N, Piérard A. Pathway of arginine biosynthesis in extreme thermophilic archaeo- and eubacteria. J Gen Microbiol. 1990;136:1177–1183. [Google Scholar]
- 19.Van de Casteele M, Desmarez L, Legrain C, Chen P G, Van Lierde K, Piérard A, Glansdorff N. Genes encoding thermophilic aspartate carbamoyltransferases of Thermus aquaticus ZO5 and Thermotoga maritima MSB8: modes of expression in Escherichia coli and properties of their products. Biocatalysis. 1994;11:165–179. [Google Scholar]
- 20.Van de Casteele M, Legrain C, Wilquet V, Glansdorff N. The dihydrofolate reductase-encoding gene dyrA in the hyperthermophilic bacterium Thermotoga maritima. Gene. 1995;158:101–105. doi: 10.1016/0378-1119(95)00090-s. [DOI] [PubMed] [Google Scholar]
- 21.Van de Casteele M, Legrain C, Desmarez L, Chen P G, Piérard A, Glansdorff N. Molecular physiology of carbamoylation under extreme conditions: what can we learn from extreme thermophilic microorganisms? Comp Biochem Physiol. 1997;118:463–473. doi: 10.1016/s0300-9629(97)00007-8. [DOI] [PubMed] [Google Scholar]
- 22.Wild J R, Wales M E. Molecular evolution and genetic engineering of protein domains involving aspartate transcarbamylase. Annu Rev Microbiol. 1990;44:193–218. doi: 10.1146/annurev.mi.44.100190.001205. [DOI] [PubMed] [Google Scholar]
- 23.Wilquet V, Gaspar J A, Van de Casteele M, Legrain C, Meiering E, Glansdorff N. Purification and characterization of recombinant Thermotoga maritima dihydrofolate reductase. Eur J Biochem. 1998;255:628–637. doi: 10.1046/j.1432-1327.1998.2550628.x. [DOI] [PubMed] [Google Scholar]
- 24.Xi X-G, Van Vliet F, Ladjimi M M, De Wannemaeker B, De Staercke C, Glansdorff N, Piérard A, Cunin R, Hervé G. Heterotropic interactions in Escherichia coli aspartate transcarbamylase: subunit interfaces involved in CTP inhibition and ATP activation. J Mol Biol. 1991;220:789–799. doi: 10.1016/0022-2836(91)90118-p. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Zhang Z-Y, Liang Y-F, Van de Casteele M, Legrain C, Glansdorff N. Aspartate carbamoyltransferase from a psychrophilic deep-sea bacterium, Vibrio strain 2693. Properties of the enzyme, genetic organization and synthesis in Escherichia coli. Microbiology. 1998;144:1435–1441. doi: 10.1099/00221287-144-5-1435. [DOI] [PubMed] [Google Scholar]