Abstract
Objective
The metabolic benefits of GLP-1 receptor (GLP-1R) agonists on glycemic and weight control are well established as therapy for type 2 diabetes and obesity. Glucagon's ability to increase energy expenditure is well described, and the combination of these mechanisms-of-actions has the potential to further lower hepatic steatosis in metabolic disorders and could therefore be attractive for the treatment for non-alcoholic steatohepatitis (NASH). Here, we have investigated the effects of a dual GLP-1/glucagon receptor agonist NN1177 on hepatic steatosis, fibrosis, and inflammation in a preclinical mouse model of NASH. Having observed strong effects on body weight loss in a pilot study with NN1177, we hypothesized that direct engagement of the hepatic glucagon receptor (GCGR) would result in a superior effect on steatosis and other liver related parameters as compared to the GLP-1R agonist semaglutide at equal body weight.
Methods
Male C57Bl/6 mice were fed a diet high in trans-fat, fructose, and cholesterol (Diet-Induced Obese (DIO)-NASH) for 36 weeks. Following randomization based on the degree of fibrosis at baseline, mice were treated once daily with subcutaneous administration of a vehicle or three different doses of NN1177 or semaglutide for 8 weeks. Hepatic steatosis, inflammation and fibrosis were assessed by immunohistochemistry and morphometric analyses. Plasma levels of lipids and liver enzymes were determined, and hepatic gene expression was analyzed by RNA sequencing.
Results
NN1177 dose-dependently reduced body weight up to 22% compared to vehicle treatment. Plasma levels of ALT, a measure of liver injury, were reduced in all treatment groups with body weight loss. The dual agonist reduced hepatic steatosis to a greater extent than semaglutide at equal body weight loss, as demonstrated by three independent methods. Both the co-agonist and semaglutide significantly decreased histological markers of inflammation such as CD11b and Galectin-3, in addition to markers of hepatic stellate activation (αSMA) and fibrosis (Collagen I). Interestingly, the maximal beneficial effects on above mentioned clinically relevant endpoints of NN1177 treatment on hepatic health appear to be achieved with the middle dose tested. Administering the highest dose resulted in a further reduction of liver fat and accompanied by a massive induction in genes involved in oxidative phosphorylation and resulted in exaggerated body weight loss and a downregulation of a module of co-expressed genes involved in steroid hormone biology, bile secretion, and retinol and linoleic acid metabolism that are also downregulated due to NASH itself.
Conclusions
These results indicate that, in a setting of overnutrition, the liver health benefits of activating the fasting-related metabolic pathways controlled by the glucagon receptor displays a bell-shaped curve. This observation is of interest to the scientific community, due to the high number of ongoing clinical trials attempting to leverage the positive effects of glucagon biology to improve metabolic health.
Keywords: GLP1R/GCGR agonist, Hepatic steatosis, Fibrogenesis, Transcriptome profiling
Highlights
-
•
Glucagon/glucagon-like peptide-1 receptor co-agonism shows efficacy on several liver related endpoints.
-
•
NN1177 is a glucagon/GLP-1 receptor co-agonist investigated for weight loss.
-
•
NN1177 administration resulted in dose-dependent reduction of liver fat.
-
•
NN1177 resulted in a reduction of inflammatory and fibrosis relevant biomarkers.
1. Introduction
With the growing incidence of type 2 diabetes, obesity, and the associated diseases, there is an increased focus on the liver related disorder coined non-alcoholic fatty liver (NAFL). NAFL is considered as the hepatic manifestation of the metabolic syndrome and is characterized by enlarged liver size (hepatomegaly), and increased levels of lipids, stored as triglycerides and to a lesser degree free cholesterol, in the hepatocytes (hepatosteatosis) [1]. In some individuals, NAFL progresses to a more severe form of the disease, non-alcoholic steatohepatitis (NASH). NASH is diagnosed clinically via histological assessment of the presence and degree of steatosis, lobular inflammation and hepatocyte ballooning which is combined into the NAFLD Activity Score (NAS). Additionally, the progression of disease is determined by a fibrosis score ranging from absence of fibrosis (F0) to de-compensated cirrhosis (F4d) or end-stage liver disease [2].
The estimated global prevalence of NASH is between 1.5% and 6.5% [3] and is higher in people with type 2 diabetes mellitus (T2D) and obesity reaching ∼22% and ∼12%, respectively [4]. The epidemiology and pathophysiology of NASH are not well understood, but genetic predisposition as well as the presence of insulin resistance are recognized to be key factors for disease development [5]. Only a proportion of patients with NASH will progress to advanced liver fibrosis and end-stage liver disease. The risk factors for progression include weight gain, diabetes and hypertension [6].
Currently, no drugs are approved for the treatment of NASH. First-line treatment is lifestyle intervention to achieve weight loss, and treatment of comorbidities (e.g., hyperlipidemia, hypertension, and diabetes) [7].
Clinical evaluation of the GLP-1 receptor (GLP-1R) agonist semaglutide in a double-blind phase 2 trial involving people with biopsy-confirmed NASH showed significant efficacy with respect to NASH resolution without worsening of fibrosis [8]. GLP-1R agonism results in body weight reduction by more than 10% and improved glycemic control, and consequently these data suggest great potential as treatment of patients with NASH by addressing the underlying metabolic disorders.
Glucagon and engineered glucagon receptor (GCGR) agonists have well-defined clinical benefits in people living with type 1 diabetes (T1D) for acute treatment of insulin induced hypoglycemia [9]. Glucagon agonism raises blood glucose through activation of hepatic glucagon receptors, mediating glycogenolysis and release of glucose. Glucagon also affects energy expenditure, protein handling and overall liver health which is less well described.
During the early 2000, numerous glucagon receptor antagonists were identified and developed for improving glucose control in people living with T2D. As predicted, the inhibition of hepatic glucose release resulted in lowering of fasting glucose and HbA1c in clinical trials [10], however, despite the beneficial effect on glucose homeostasis most glucagon antagonist programs were halted due to side effects related to liver function (ALT/AST elevation), LDL cholesterol increase, and body weight gain. The elevation of liver enzymes was accompanied by an increase in hepatic fat and demonstrated a role for glucagon in maintaining healthy liver function [11].
Clinical data from the glucagon antagonists showing adverse effects on liver health suggested a potential benefit of glucagon agonism in the treatment of NAFL. However, for activation of this pathway to be a viable treatment strategy for a people living with NAFL where T2D is a highly prevalent comorbidity, the glucagon-induced stimulation of hepatic glucose production and resulting impairment in blood glucose control needed to be mitigated, for instance via GLP-1 mediated insulin release. Substantial preclinical data have been generated in diet-induced mouse models suggesting that the combination of glucagon and GLP-1 receptor agonism can be balanced to mitigate the negative effects of glucagon on glucose homeostasis while maintaining its beneficial effects on liver steatosis and enhancing the GLP-1 mediated body weight loss [[12], [13], [14], [15]]. Oxyntomodulin is a natural occurring peptide with GLP-1R and GCGR agonistic properties [16]. Acute administration of oxyntomodulin suppresses food intake and results in body weight loss in obese subjects [17]. These data have encouraged pharmacological development on similar dual acting compounds with a range of potency ratios between the two receptors, as well as different pharmacokinetic properties. Some are currently being evaluated in clinical phase 1 and 2 studies in patients with obesity, type 2 diabetes, and NASH [18]. Cotadutide, a protracted, once-daily peptide agonist with dual GLP-1 and glucagon receptor activity was evaluated in people living with T2D. Parameters of glucose homeostasis [19] and hepatic health, such as liver enzymes [20], suggested a beneficial drug profile and prompted for progression to evaluation in people with NASH in a Phase 2b/3 trial. Here we report the preclinical findings from studies of the GLP-1/glucagon receptor dual agonist NN1177 [14], in order to contribute to the learnings and understanding of the physiological effects of combining activation of these metabolically opposing pathways in a setting of overnutrition and NASH.
1.1. Aim
The aim of this study was to evaluate the efficacy of the GLP-1/glucagon receptor agonist NN1177 with dual receptor affinities, with a ratio of 1–3, meaning 3 times higher affinity for the GLP1 receptor than the Glucagon receptor [14]) and improved pharmacokinetic properties compared to the native molecule oxyntomodulin [14] in a diet-induced NAFLD/NASH mouse model (DIO-NASH). Semaglutide was included in a clinically meaningful dose and served as a bodyweight matched control to the high dose of NN1177 to investigate the additional benefits introduced by NN1177 treatment.
This animal model has been extensively used in the preclinical investigation of pharmacotherapies that have progressed to clinical studies [12,21] because this model reflects a number of human-relevant disease markers [12,13,15,22]. To elucidate the affected biological pathways, the effect of NN1177 on the transcriptome from the DIO-NASH mice livers was investigated and compared to human liver transcriptomic data generated from patients with confirmed NASH [23,24].
2. Methods
2.1. Mouse model
The DIO-NASH study was performed at Gubra Aps, Denmark, and approved by The Danish Animal Experiments Inspectorate using permission 2013-15-2934-00784. C57BL/6JRj mice (5 weeks old) were obtained from Janvier Labs (Le Genest Saint Isle, France) and housed in a controlled environment (12 h light/dark cycle, 21 ± 2.0 °C, humidity 50 ± 10 %). C57BL/6J mice had ad libitum access to AMLN diet (DIO-NASH mice) or chow for 39 weeks prior to treatment start and for the duration of the treatment period. DIO-NASH mice were fed AMLN diet containing 40 kcal% fat, 40% carbohydrates (20% fructose) and 2% cholesterol (AMLN diet, 4.49 kcal/g, #D09100301, Research Diets, New Brunswick, NJ). A group of mice was maintained on regular rodent chow (2.85 kcal/g, Altromin 1324, Brogaarden, Hoersholm, Denmark) for comparison. A liver biopsy was sampled from the mice 3 weeks before treatment start, as described in detail previously [21].
Only mice with steatosis score 2 or higher and fibrosis stage ≥1 as outlined by [2] were included in the studies. Included DIO-NASH animals were then randomised into treatment groups based on mean baseline Col1 area% and body weight one week before dose start.
2.2. Study drug
Compound was delivered from Novo Nordisk to the test site Gubra as lyophilized protein and vehicle stocks. Every 14 days of the study, a vial of 100 nmol NNC9204-0000-1177-6C (NN1177) was dissolved in 1 ml of standard vehicle number 9042. The 100 nmol/ml stock solution was then further diluted to nominal concentrations of 0.8 (0.81), 0.4 (0.37) and 0.15 (0.13) nmol/ml (measured concentrations in parenthesis).
2.3. Intervention
All animals received subcutaneous (s.c.) dosing with active compound or vehicle once daily, using an insulin syringe and a dose volume of 5 ml/kg, daily dosing frequency was based on previous studies in rodents [14] and predicted dosing frequency in humans is once weekly (Friedrichsen et al. accepted for publication in MOLMET-D-23-00453R1). Body weight was measured daily during the intervention period. Animals were terminated in week 8 (day 56–58) in a non-fasting state. Latest drug dose was administered ∼24 h before termination. Animals were put in anaesthesia (isoflurane) and cardiac blood obtained for collection of terminal plasma. Upon necropsy, whole liver was collected and weighed. A piece (100–200 mg) of the left lateral lobe was fixed in 4% PFA for histology. The medial lobe was divided into pieces and snap frozen in liquid nitrogen for biochemical analysis in RNA later for RNA isolation and gene expression analysis.
2.4. Tissue homogenization
1 ml 5% NH-40/ddH2O solution (ab142227, Abcam) was added to the FastPrep tube. The tubes were placed in a FastPrep homogenizer and shaked for 2 × 60 s. After homogenization, the samples were slowly heated to 80–100 °C in a heating block for 3 min. After being cooled to room temperature the heating step was repeated. The samples were centrifuged for two minutes at top speed using a microcentrifuge to remove any insoluble material. The supernatant was stored at −80 °C until usage.
2.4.1. Hepatic triglyceride, cholesterol, and glycogen
Triglycerides and total cholesterol content in liver homogenates were measured in single determinations using autoanalyzer Cobas C-111 with commercial kits (Roche Diagnostics, Germany) according to manufacturer's instructions.
2.5. Plasma lipids, ALT, and AST
100 μl blood was collected into Lithium-Heparin tubes. Plasma was separated and samples were stored at 4 °C for one day prior to analysis. ALT, AST, TG, and total cholesterol were measured in single determinations using commercial kits (Roche Diagnostics, Germany) on the Cobas™ C-111 autoanalyzer according to the manufacturer's instructions.
2.6. Immunohistochemistry and histochemistry
Liver tissue was fixed in 4% PFA and paraffin embedded. Sections (3 μm each) were stained according to standard protocols using Meyer's haematoxylin (Dako) and Eosin Y solution (Sigma–Aldrich) for hematoxylin and eosin (HE) staining. Immunohistochemical (IHC) staining for CD11b (AbCam, cat. No. ab1333357), galectin-3 (Gal-3) (Biolegend, Cat. # 125402), alpha smooth muscle actin (αSMA) (AbCam, cat. No. ab124964), and Type I collagen (collagen 1a1) (Southern Biotech, Cat. 1310-01) were performed using standard procedures. Briefly, after antigen retrieval and blocking of endogenous peroxidase activity, slides were incubated with primary antibody. The primary antibody was detected using a linker secondary antibody followed by amplification using a polymeric HRP-linker antibody conjugate. Next, the primary antibody was visualized with DAB as chromogen. Finally, sections were counterstained in haematoxylin and cover slipped.
2.7. Morphometric quantification of immunohistochemistry and histochemistry by Visiopharm Integrator System
Image analysis using the Visiomorph software (Visiopharm, Denmark) was applied for quantification of steatosis and fibrosis on HE and sirius red stained slides, respectively, and for quantification of markers of inflammation and fibrogenesis on immunohistochemically stained slides. Slides were scanned at 20× using the Nanozoomer 2.0 HT System (Hamamatsu). The quantitative estimates of steatosis were calculated as an area fraction (see calculation below). Determination of steatosis score and fibrosis stage for inclusion criteria was done using the Kleiner scoring system [2] on slides stained with HE and sirius red, respectively.
For evaluation of treatment effects on hepatic inflammation and fibrogenesis, area fractions of the IHC staining for CD11b, Gal-3, αSMA, and collagen 1a1 were calculated as percent of steatosis-free tissue area (see calculation below).
2.8. RNA-sequencing sample preparation and sequencing
Lysis of tissue from the medial lobe was performed at Gubra using a MP FastPrep system. Briefly, 15–20 mg liver biopsy was homogenized and used for RNA extraction on NucleoSpin Plus RNA columns (Macherey–Nagel) as recommended by the supplier. The quantity of the RNA was assessed using a NanoDrop 2000 spectrophotometer (ThermoScientific). The purified RNA was stored at −80 °C. The NeoPrep (Illumina) was used to generate libraries for sequencing which were sequenced on a NextSeq 500 (Illumina). After sequencing, the raw data (fastq files) were provided by Gubra for analyses.
2.9. Sequencing data processing and analyses
RNA-sequencing data was processed with Trim Galore! (v. 0.6.4_dev; adaptor trimming) before alignment with Salmon (v. 1.9.0) [25] using decoy-aware indexes (GRCm38, release M23 for the mouse data; GRCh38, release 38 for the human data). Quality control and alignment statistics were obtained with MultiQC (v. 1.11) [26]. All subsequent data processing and analyses were performed in R (v. 4.2.0) using the tidyverse packages (v. 2.0.0). The RNA-seq data was loaded with tximeta (v. 1.14.1) [27] and DESeq2 (v. 1.36.0, default parameters) [28] was used for the quality control and differential expression analysis. Only genes that were expressed in at least half of the samples of one of the groups tested were kept to test for differential expression. Log-fold changes were adjusted using DESeq2's function lfcshrink (type = normal) and p-values adjusted for multiple testing using the Benjamini and Hochberg method. For the gene-set enrichment analysis, clusterProfiler's (v. 4.7.1.002) compareCluster and enrichKEGG functions were used [29].
For the mouse dataset, one outlier sample was excluded after inspection of the clustering results of the variance-stabilised transformed transcriptome data (DEseq2's vst() function) (Average euclidean distance to the other samples further than 3 standard deviations away from the mean distances across samples). As a result, all experimental groups included 8 samples, apart from the chow + vehicle group that included 7 samples. After this exclusion, the RNA-seq dataset had an average number of mapped reads of 16M reads (s.d. 4.5), with an average mapping rate of 91.5% (s.d. 4.5%).
To obtain an overview of the transcriptome differences and similarities between samples, a principal component analysis (PCA) was first done on the vst-transformed data using the 500 most variable genes. PCA projections were further plotted as densities for the first 2 components, and the distributions between groups were compared using t-tests (Bonferroni-corrected within each principal component for multiple comparison testing).
Gene co-expression modules were determined using a set of n = 11537 significantly differentially expressed (p adj ≤ 0.05) in any pairwise-comparison between sample groups. Correlated gene pairs with Pearson r > 0.8 were retained and used for graph-based clustering using the Leiden algorithm implementation of the igraph package (v.1.5.1) [30] ]. Only modules with at least 100 genes were kept for pathway enrichment analyses. To determine if the expression of genes within each module was different between sample groups, module expression scores were obtained with the GSVA package (v.1.44.5) [31] and the resulting scores were compared with t-tests (Bonferroni-corrected within each module for multiple comparison testing).
The clinical relevance of the gene expression changes due to NN1177 in NASH in our mouse model was investigated using two publicly available datasets of human NASH liver samples obtained from the Gene Expression Omnibus (GEO) [32]: GSE162694 (SRP295965) [24] and GSE130970 (SRP19735) [23]. The sex of the samples in each dataset was determined based on the expression levels of the X chromosome gene XIST and the 10 most variable Y chromosomes genes in each dataset. Samples with mismatches between the expression-based sex and the metadata-provided sex were excluded from further analyses. Furthermore, clusterings of the vst-transformed datasets were performed on the expression data to exclude outlier samples (Average euclidean distance to the other samples further than 3 standard deviations away from the mean distances across samples). Finally, only samples with information about both the fibrosis score and the NAFLD activity scores were kept for analyses. The number of samples included in the analyses were: SRP197353: n = 71 (fibrosis scores: F_0 = 21, F_1 = 27, F_2 = 9, F_3 = 12, F_4 = 2; NAFLD activity score: NAS_0 = 4, NAS_1 = 4, NAS_2 = 8, NAS_3 = 16, NAS_4 = 16, NAS_5 = 16, NAS_6 = 7); SRP295965: n = 112 (fibrosis scores: F_0 = 54, F_1 = 25, F_2 = 25, F_3 = 5, F_4 = 3; NAFLD activity score: NAS_0 = 29, NAS_1 = 12, NAS_2 = 9, NAS_3 = 10, NAS_4 = 13, NAS_5 = 18, NAS_6 = 12, NAS_7 = 9). After these exclusions, these RNA-seq datasets had a respective average number of reads of 29.3M and 36.1M (s.d. 6.6 and 3.8), with an average mapping rate of 52.8% and 65% (s.d. 7% and 4.2%), respectively. For each dataset, differential expression analyses were performed to find genes associated with 2 variables: the NAFLD activity score and the fibrosis score, accounting for age, sex and either fibrosis score or NAFLD activity score, respectively. This allowed to find genes independently associated with either of these two variables. Then, meta-analyses were performed to integrate the results of these two datasets together and increase the statistical power and robustness of the findings (associations with NAS or fibrosis). For this, the R package metafor (v. 4.2-0) [33] was used, based on the log2-fold changes and their standard deviations, using the REML (restricted maximum likelihood estimator) random-effect model. Datasets were unweighted in the meta-analyses. For between-species comparisons, Biomart (v. 2.52.0) [34] was used to obtain one-to-one orthologues pairs from the Ensembl data (release 102) [35], and only orthologues detected in both species included in analyses.
For plotting, the following packages were used: ggplot2 (v. 3.4.3), ggrepel (v. 0.9.3), RColorBrewer (v. 1.1-3) and cowplot (v. 1.1.1). Supplementary Tables were generated using openxlsx (v. 4.2.5.2).
2.10. Statistical methods
Test for treatment effect vs vehicle: P-values where indicated were generated by one way ANOVA with Bonferroni correction for multiple comparisons to test. In cases of significantly different standard deviations on Brown–Forsythe tests, Welsh correction ANOVA with Dunnett's T3 correction was used instead. Superiority to semaglutide tested using the above statistical methods. Chow group was excluded from ANOVA tests. Test pre-treatment biopsy vs post-treatment biopsy: P-values where indicated were generated by 2-tailed paired Students t-test. Tested against DIO-NASH vehicle or pre-vs post treatment: ∗∗∗ = p < 0.001, ∗∗ = P < 0.01, ∗ = p < 0.05 Tested against semaglutide 30 nmol/kg: ¤¤¤ = p < 0.001, ¤¤ = p < 0.01, ¤ = p < 0.05 NA = not applicable NT = not tested NS = not significant.
2.11. Data availability
The mouse data generated in this paper is available on the online repository Gene Expression Omnibus (GEO) under the accession GSE220519. The publicly available human data analysed in this paper is available under the accessions GSE162694 and GSE130970. The code used to generate the results is available in the GitHub repository https://github.com/novonordisk-research/NN1177_liver_RNAseq.
3. Results
Male C57Bl/6 mice were fed a fructose and high fat rich diet for 36 weeks to induce the NASH phenotype (DIO-NASH) and animals with steatosis score of minimum 2 and fibrosis score of minimum 1 were randomized based on baseline collagen 1a1 area% to receive either once daily s.c. administration of vehicle (NASH vehicle) or three different doses of NN1177 (0.75 nmol/kg, 2 nmol/kg or 4 nmol/kg) for 8 weeks. In addition, a reference group of animals was treated with the GLP-1 analogue semaglutide (30 nmol/kg). A vehicle-dosed group of animals fed with a normal chow diet (Chow vehicle) was also included. 12 mice were randomised to each treatment, and 10 mice for chow vehicle.
Randomisation based on biopsy reduces heterogeneity of the treatment cohorts and allows measurement of changes in histological liver features from baseline to termination. Hepatic steatosis, inflammation, and fibrogenesis were assessed from liver biopsies by morphometric analyses of immunohistochemical and histochemical stained slides. Total RNA sequencing was performed, and plasma levels of lipids, cholesterol, and liver enzymes were also determined.
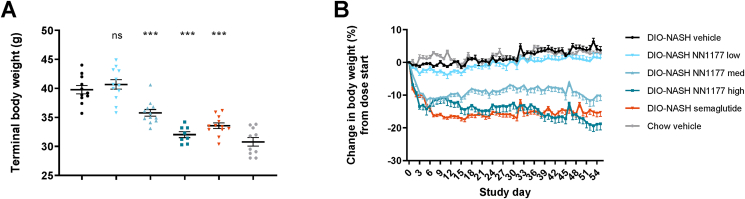
NN1177 dose-dependently reduced body weight of 10- and 20% in the medium and high dose groups, respectively, as compared to vehicle treatment, while semaglutide induced a body weight loss of 16% (Table 1, Figure 1). During the intervention period, four mice were euthanized in the group receiving the highest dose of NN1177 due to excessive weight loss. Per our protocol, these mice were euthanised when they approached 30% weight loss from their individual starting weight or if appearing lethargic or showing signs of malaise. The culled mice were rapidly losing weight over multiple days despite substitution with intraperitoneal saline and softened food. There were no clinical signs of adverse effects other than excessive weight loss, and ALT as a signal for liver injury was reduced in all groups showing weight loss.
Table 1.
Overview of 8 weeks DIO-NASH study data. Data presented as mean per group (n = 8–12), standard deviation (SD) and tested for statistical significance against DIO-NASH vehicle or semaglutide using one-way ANOVA with Bonferroni correction, chow vehicle excluded from tests. Tested vs DIO-NASH vehicle: ∗∗∗∗ = p < 0.0001, ∗∗∗ = p < 0.001, ∗∗ = P < 0.01, ∗ = p < 0.05 Tested vs semaglutide 30 nmol/kg, ¤¤¤¤ = p < 0.0001 NA = not applicable NT = not tested NS = not significant.
| DIO-NASH vehicle n = 11 |
DIO-NASH NN1177 low (0.75 nmol/kg) n = 12 |
DIO-NASH NN1177 med (2 nmol/kg) n = 12 |
DIO-NASH NN1177 high (4 nmol/kg) n = 8 |
DIO-NASH semaglutide (30 nmol/kg) n = 12 |
Chow vehicle n = 9–10 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Stat. Sign. vs veh | Stat. Sign. vs sema | Mean | SD | Stat. Sign. vs veh | Stat. Sign. vs sema | Mean | SD | Stat. Sign. vs veh | Stat. Sign. vs sema | Mean | SD | Stat. Sign. vs veh | Mean | SD | Stat. Sign. | |
| Baseline body weight | 38.4 | 2.4 | 40.2 | 2.6 | NS | NS | 39.9 | 2.7 | NS | NS | 39.5 | 2.1 | NS | NS | 39.7 | 2.1 | NS | 29.9 | 2.2 | NT |
| Terminal BW (g) | 39.8 | 2.4 | 40.7 | 2.8 | NS | ¤¤¤ | 35.8 | 2 | ∗∗∗ | NS | 32 | 1.4 | ∗∗∗ | NS | 33.6 | 1.7 | ∗∗∗ | 30.8 | 2.4 | NT |
| Cumulative food intake Day 0–13 (g GAN diet) | 39.1 | 2.8 | 41.0 | 3.4 | NS | ¤¤¤ | 37.6 | 5.6 | NS | ¤¤¤ | 39.1 | 6.0 | NS | ¤¤¤ | 29.8 | 2.9 | ∗∗∗ | N/A | N/A | N/A |
| Liver weight (g) | 3.5 | 0.6 | 3.3 | 0.6 | NS | ¤¤¤ | 1.9 | 0.3 | ∗∗∗ | NS | 1.5 | 0.1 | ∗∗∗ | ¤ | 2.1 | 0.4 | ∗∗∗ | 1.3 | 0.1 | NT |
| Liver index (% of BW) | 8.9 | 1.2 | 8.2 | 1.3 | NS | ¤¤¤ | 5.2 | 0.8 | ∗∗∗ | NS | 4.7 | 0.4 | ∗∗∗ | ¤¤ | 6.2 | 1 | ∗∗∗ | 4.1 | 0.5 | NT |
| ALT (U/L) | 215.3 | 60.7 | 200 | 78.7 | NS | ¤¤¤ | 56.4 | 18.3 | ∗∗∗ | NS | 54 | 17.1 | ∗∗∗ | NS | 52.3 | 24.8 | ∗∗∗ | 25.4 | 7.2 | NT |
| AST (U/L) | 183.1 | 43.8 | 271 | 90.9 | ∗∗∗ | ¤¤¤ | 168.4 | 55.8 | NS | ¤¤ | 157.8 | 36.6 | NS | ¤ | 93.1 | 23.6 | ∗∗∗ | 48.4 | 17.4 | NT |
| Plasma Triglyceride (mmol/L) | 0.5 | 0.2 | 0.5 | 0.1 | NS | NS | 0.6 | 0.2 | NS | NS | 0.6 | 0.2 | NS | NS | 0.4 | 0.2 | NS | 0.8 | 0.3 | NT |
| Plasma total cholesterol (mmol/L) | 7.6 | 1.5 | 6.4 | 1.5 | NS | NS | 3.6 | 0.6 | ∗∗∗ | ¤¤¤ | 2.9 | 0.2 | ∗∗∗ | ¤¤¤ | 5.4 | 1.1 | ∗∗ | 2.4 | 0.3 | NT |
| Liver TG (mg/g) | 95.8 | 19.5 | 86.9 | 11.5 | NS | ¤¤ | 33.1 | 11 | ∗∗∗ | ¤¤¤ | 17.9 | 4.1 | ∗∗∗ | ¤¤¤ | 68.6 | 14.4 | ∗∗ | 9 | 3.1 | NT |
| Liver TC (mg/g) | 8.6 | 2.8 | 8.9 | 1.2 | NS | NS | 5.2 | 1.1 | ∗∗∗ | ¤¤¤ | 5.2 | 1.2 | ∗∗∗ | ¤¤¤ | 8.8 | 1.8 | NS | 3.6 | 0.5 | NT |
| Steatosis area% post biopsy | 29.6 | 2.17 | 22.7 | 3.97 | ∗∗ | ¤¤¤ | 6.88 | 3.33 | ∗∗∗ | ¤¤ | 2.68 | 1.24 | ∗∗∗ | ¤¤¤ | 12.2 | 3.8 | ∗∗∗ | 0.943 | 0.223 | NT |
| Liver glycogen (mg/g) | 8.2 | 5.6 | 13.5 | 5.2 | NS | NS | 15.6 | 4.9 | ∗ | ¤ | 17.1 | 10.5 | ∗ | ¤¤ | 7.8 | 4.1 | NS | 7.6 | 4.9 | NT |
| CD11b fat-free area% | 0.95 | 0.30 | 0.74 | 0.16 | NS | NS | 0.51 | 0.44 | ∗∗ | NS | 0.64 | 0.20 | NS | NS | 0.49 | 0.31 | ∗∗ | 0.34 | 0.41 | NT |
| Gal-3 fat-free area% | 6.9 | 1.09 | 4.89 | 0.75 | ∗ | NS | 3.45 | 1.18 | ∗∗∗ | NS | 3.7 | 0.75 | ∗∗ | NS | 3.54 | 1.28 | ∗∗∗ | 1.71 | 0.92 | NT |
| aSMA fat-free area% | 7.25 | 1.79 | 4.81 | 0.72 | ∗∗∗ | ¤¤¤ | 2.19 | 1.05 | ∗∗∗ | NS | 1.85 | 0.8 | ∗∗∗ | NS | 2.76 | 1.03 | ∗∗∗ | 0.64 | 0.27 | NT |
| Col1a1 fat-free area% | 14.8 | 4.81 | 12.0 | 2.66 | NS | NS | 10.3 | 2.87 | ∗ | NS | 9.99 | 2.99 | ∗ | NS | 12.8 | 3.38 | NS | 2.32 | 0.77 | NT |
Figure 1.
Mice body weight.A. Terminal body weight day 55. B. Change in body weight (%) during dose-phase.
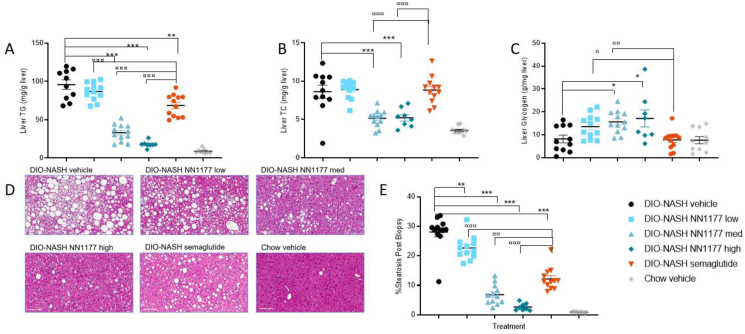
Compared with semaglutide, NN1177 reduced hepatic steatosis to a greater extent at equal body weight loss, as demonstrated by two independent methods: biochemical assessments of liver cholesterol (TC) and triglycerides (TG), and morphometric quantification of steatosis. Liver TG was significantly reduced by the two highest doses of NN1177 and by semaglutide. The greatest magnitude of reduction in TGs was observed in the highest NN1177 dose group, with both 2 and 4 nmol/kg providing significantly greater reduction compared with semaglutide (p < 0.0001) (Table 1, Figure 2A). Liver cholesterol was significantly reduced by NN1177 at the two highest doses compared with DIO-NASH vehicle, which was not achieved with semaglutide. However, plasma cholesterol was significantly reduced by both NN1177 and semaglutide treatment (Table 1, Figure 2B). The differences in TG levels as determined by biochemical analysis of liver fat were also evident from the liver histology. Morphometric quantification of steatosis induced by the DIO-NASH diet was noticeably reduced in the 2- and 4 nmol/kg NN1177-treated groups, compared with either the DIO-NASH vehicle or semaglutide treatment (Table 1, Figure 2D–E). The more pronounced effect of the middle dose of NN1177 on steatosis compared to semaglutide was obtained despite a larger reduction in body weight in the semaglutide treated group, indicating a contributing effect from the glucagon agonism of the combination molecule. The hepatic glycogen content was likewise measured, and the dual pharmacology of glucagon receptor activation and GLP1 mediated insulin release resulted in a small, but significant increase in glycogen storage with the mid and high doses of NN1177 (Table 1, Figure 2C).
Figure 2.
Hepatic triglyceride, cholesterol, and glycogen and liver steatosis at end-of-treatment.A. Liver triglyceride (mg/g wet tissue); B. Liver total cholesterol (mg/g wet tissue); C. Liver glycogen (mg/g wet tissue); D. Representative images of liver morphology at the end of treatment period (magnification 20×, scale bar = 100 μm); E. Morphometric quantification of liver lipid content on HE stained sections.
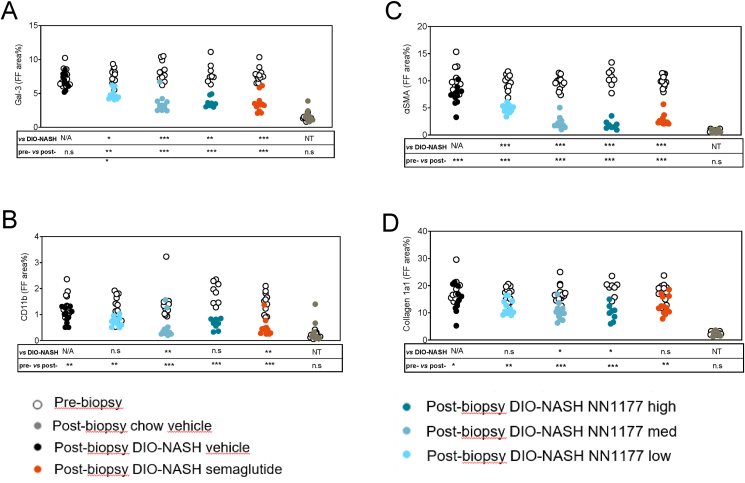
As shown in Table 1 and depicted in Figure 3, the histological markers of inflammation (CD11b and Gal-3) and fibrogenesis (αSMA and collagen 1a1) were all markedly upregulated in livers from DIO-NASH vehicle compared to chow vehicle mice and significantly decreased by all doses of NN1177 and semaglutide.
Figure 3.
Quantitative change in Gal-3, CD11b, aSMA, and Collagen 1a1 staining from Pre-treatment biopsy to end-of-treatment biopsy normalized to fat free area.
Together, these data demonstrate that despite NN1177 inducing a larger reduction in liver steatosis than semaglutide, this only has little impact on the histological endpoints of inflammation and fibrosis in the DIO-NASH model. To further investigate the effect of NN1177 on fibrotic livers, we profiled the hepatic transcriptome of the different treatment groups in the study and investigated the gene expression programs associated with the observed phenotypes.
We obtained high-quality transcriptome profiles from liver samples across the experimental groups. After quality control, the sample number per group was DIO-NASH vehicle (n = 8), DIO-NASH semaglutide (n = 8), DIO-NASH NN1177 low (low dose, n = 8), DIO-NASH NNCC1177 med (medium dose, n = 8), DIO-NASH NNCC1177 high (high dose, n = 8), chow vehicle (n = 7).
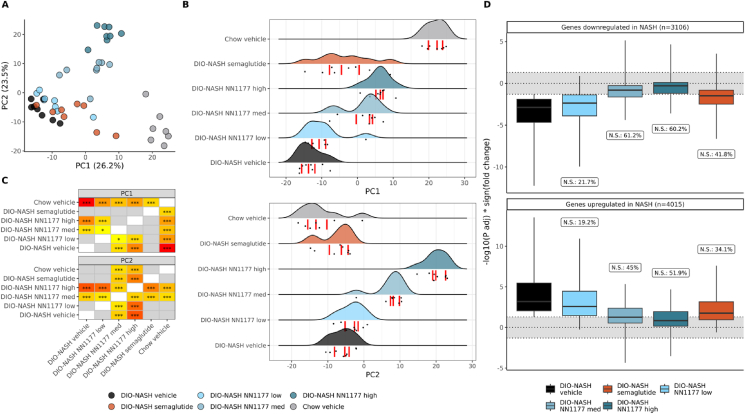
The induction of the disease phenotype DIO-NASH explains most of the hepatic gene expression variance. Principal component analysis of the top 500 variables genes across all samples display the most extensive global changes in transcriptome between chow + vehicle mice, DIO-NASH + vehicle mice (Figure 4A–C). The transcriptomes from mice treated with NN1177 or semaglutide cluster between the chow vehicle and DIO-NASH vehicle groups demonstrate these compounds' positive effect in ameliorating the experimental NASH phenotype. Interestingly, the DIO-NASH samples from the medium and high doses of NN1177 were significantly distinct from the DIO-NASH vehicle samples on this axis (PC1, Figure 4B–C). The primary genes contributing to the PC1 component include well-described markers of liver dys-metabolism and damage, such as proteins involved in lipid metabolism (Cidec, Apoa4, Cidea) inflammation (Ly6d, AnxA2, Hamp) and extracellular matrix formation (Serpina1e, Col3a1, Col1a1, Col6a3, Col1a2, Adamtsl2, Spp1) (Suppl. Figure 1). An almost equal proportion of the variance in the data (23.5%, PC2) reflects a dose-dependent NN1177-specific effect. PC2 differences are mainly driven by the major urinary protein (Mup) genes, which are found in rodents but not in humans, as well as Cyp genes, involved in cholesterol, bile, and lipid metabolism. Cyp gene isoforms and their role in detoxification and metabolism pathways are highly species specific between mouse and man, so the translational value of this observation is uncertain.
Figure 4.
NN1177 effects on the transcriptome of diet-induced NASH in mice, compared to semaglutide.A. Principal component analysis of the top 500 most variable genes in the dataset (VST-normalised data). The top 30 genes driving the first two principal components are shown in Suppl. Figure 1. B. Sample distributions on the first two principal components. Points represents each sample, the curves represent the sample distribution per group and the red bars represents the first, second and third quartiles. C. Statistical differences in principal components values between groups. Colours represent statistical significance from less significant (yellow) to most significant (red). T-test results with p > 0.1 are shown in grey, results with 0.1 < p < 0.05 are shown in yellow with no annotation. Annotations: ∗: p ≤ 0.05; ∗∗: p ≤ 0.01; ∗∗∗: p ≤ 0.001 (Bonferroni-corrected within modules). D. Reversal of the NASH-induced transcriptome changes with semaglutide and NN1177. Grey zone: genes not statistically differently expressed from the control group (chow + vehicle diet). N.S. % = percentage of genes significantly different between the chow vehicle group and the DIO-NASH vehicle group but not significantly differentially expressed after treatment. Outlier genes are not represented in the plot.
We then evaluated the ability of NN1177 and semaglutide to reverse the NASH transcriptome phenotype. For this, we estimated the fraction of the DIO-NASH perturbed genes compared to the chow vehicle control that does not appear to be perturbed in NN1177 or semaglutide treatment compared to the chow-vehicle control (Figure 4D). After treatment with semaglutide, 41.8% and 34.1% of the genes respectively down-regulated and up-regulated in NASH were no longer statistically different from the control group (chow + vehicle). Comparatively, the low dose of NN1177 seems to have a lesser effect on these genes (21.7%/19.2%), while the medium and high doses had a higher effect on the NASH-affected genes (medium dose: 61.2%/45%; high dose: 60.2%/51.9%). These results suggest a dose-dependent effect of NN1177 in reversing the expression of approximately half of the genes perturbed by the NASH phenotype.
To investigate the specific gene expression programs affected by NN1177 treatment, we build a gene co-expression network (Pearson r > 0.8) with the genes associated with NASH induction and the treatments and identified six co-expressed gene modules. We scored the expression activity of these modules for each sample. Figure 5A shows the expression patterns and magnitude of perturbations between the different treatment groups in the six gene modules identified. Module 1 represent a group of genes that are significantly repressed due to the NASH induction (5A–B). Module 1 expression is rescued by the highest dose of NN1177 (Figure 5B). Module 1 comprises pathways involved in metabolic homeostasis, such as retinol metabolism, steroid biogenesis, amino acid metabolism and oxidative phosphorylation (Figure 5C column 1). Module 2 (M2) consists of pathways involved in focal adhesion and ECM receptors, typically associated with fibrogenesis which is upregulated by the NASH diet (5A). This M2 expression is partially rescued by treatment with both semaglutide and the different doses of NN1177, both the medium and high dose of NN1177 achieves a significant effect, with the highest dose essentially normalising expression. Module 3 encompasses a strong overrepresentation of biological pathways of ribosomal biosynthesis and oxidative phosphorylation. The expression activity of this module trends towards being downregulated by the NASH diet and it is partially rescued by the treatment with semaglutide or the two lower doses of NN1177. The high dose of NN1177 induces expression of this gene significantly set to a level that overshoots the normal levels seen in the chow vehicle control mice (Figure 5A). Module 4 ([4]) is enriched in pathways involved in steroid and steroid hormone biogenesis in addition to retinol and linoleic acid metabolism and bile secretion. The expression activity of this module is repressed in the DIO-NASH vehicle group. While semaglutide treatment move the module expression activity towards normalisation, NN1177 seems to have a dose-dependent effect leading to an increased repression of these pathways. The pathways found in module 5 are related to inflammation, phagocytosis and immune modulation. This module is upregulated in the NASH model and all treatments tested induce a trend towards lower levels, although the effect did not increase further with the higher doses of NN1177. Finally, module 6 ([6]) contains pathways that are upregulated in the NASH vehicle animals but are not affected by either NN1177 or semaglutide treatment. This category contains genes involved in ECM and focal adhesion which may be part of the pro-fibrotic response and overlap with the pathways from module 2. This could suggest that the beneficial impact of semaglutide and NN1177 treatment on hepatic fibrosis are mediated through effects on a subset of the molecular mechanisms involved in regulating the equilibrium between fibrosis build up and degradation.
Figure 5.
Co-expression modules and pathway enrichment analyses.A. Co-expression modules. Modules corresponds to sets of genes whose expression is strongly correlated (Pearson r > 0.8). For each module, gene-set variation analysis (GSVA) was performed to generate scores representing expression levels of genes within each module per sample, relative to the other samples. B. Statistical differences in GSVA scores between groups. Colours represent statistical significance from less significant (yellow) to most significant (red). T-test results with p > 0.1 are shown in grey, results with 0.1 < p < 0.05 are shown in yellow with no annotation. Annotations: ∗: p ≤ 0.05; ∗∗: p ≤ 0.01; ∗∗∗: p ≤ 0.001 (Bonferroni-corrected within modules). C. Over-representation enrichment analyses (KEGG pathways) for genes modules and genes significantly associated with NASH and/or NN1177 (ordinal regression analysis NASH vehicle < NN1177 low < NN1177 med < NN1177 high). The pathways represented are selected from the top 5 pathways per gene-sets. Each gene-set tested is identified by a number in bracket and referenced with this number in the results. The numbers of genes in each gene sets are shown in parenthesis. The variations in number of genes between (A) and (C) are due to differences in gene ID mappings between Ensembl IDs used for expression analyses (A) and Entrez IDs used for enrichment analyses (C). Full differential expression and enrichment results are presented in Suppl. Table 1.
To determine the dose-dependent effect of NN1177, an ordinal regression analyses was done to look at genes associated with increasing doses of NN1177 (NASH + vehicle < NASH + low NN1177 < NASH + med. NC1177 < NASH + high NN1177) or decreasing doses of NN1177. Enrichment analyses were then also run on sets of genes for which significant changes in expression were observed in the same or opposite direction in NASH and under the effect of NN1177, as well on genes only associated with NASH or NN1177 (Figure 5C right panel). The right panel of Figure 5C visualises how the KEGG pathways overrepresented in the NASH phenotype with or without NN1177 treatment overlaps and are regulated in either the same direction or the opposite (and therefor supposedly curative direction). The column [7] (“Increased in NN1177”) shows that treatment with NN1177 induces gene expression of pathways involved in amino acid metabolism, the coagulation and complement cascade, ribosome biosynthesis, and oxidative phosphorylation. The column [8] (“Decreased in NASH”) shows that these same pathways, in addition to steroid and steroid hormone biosynthesis, retinol and linoleic acid metabolism and bile secretion are downregulated by the NASH phenotype. The column [9] (“Decreased by NN1177”) shows that NN1177 decreases various pathways involved in inflammation, ECM, focal and cell adhesion in addition to retinol metabolism. The column [10] (“Increased in NASH”) shows majority of these pathways are increased by the DIO-NASH diet. Column [11] (“Opposite direction”) shows the pathways that are enriched when considering genes that are affected in an opposite way between the effect of NN1177 on DIO-NASH mice versus the effect of the DIO-NASH diet itself (intersection of the genes from columns [7] and [8] + intersection of the genes from columns [9] and [10]). These pathways, where NN1177 can potentially revert the effect of the DIO-NASH diet, are mainly around amino-acid metabolism, oxidative phosphorylation, ribosomal pathways and cell adhesion. This highlight that this could be the main effects of NN1177. On the other hand, the column [12] (“Same direction”) shows the genes and pathways that are perturbed in the same direction by NASH diet and further so with treatment with NN1177 (intersection of the genes from columns [8] and [9] + intersection of the genes from columns [7] and [10]). These are pathways involved in steroid hormone biosynthesis, retinol and linoleic metabolism, bile secretion and chemical carcinogenesis such as DNA adducts. The latter may be a consequence of the increased oxidative stress induced by the glucagon mediated increase in oxidative phosphorylation and high fatty acid content in the NASH diet. Column [13] (“Only signif. with NN1177”) contains genes that are only significantly regulated in the NN1177 treated animals and seem to be mainly related to ribosomal genes. Finally, column [14] (“Only signif. in NASH”) contains the genes and their associated pathways that were significantly regulated in the NASH model and not affected by NN1177 treatment. These genes map notably to steroid biosynthesis, pancreatic secretion and natural-killer cell mediated cytotoxicity.
Treatment with NN1177 is associated with substantial body weight loss, to the point where some of the animals treated with the highest dose had to be euthanised. For this reason, some of the observed transcriptome changes may be driven by the changes in body weight rather than by NN1177. In order to ensure that the effects observed in Figure 5 are not only driven by body weight loss, the pathway enrichment analysis was repeated after correcting for this variable in the differential gene expression analyses (Suppl. Figure 2 and Suppl. Table 2). While the number of significantly differentially expressed genes is reduced after this adjustment, as anticipated, this analysis shows that the well described direct effects of hepatic glucagon receptor activation such as induction of oxidative phosphorylation and amino acid metabolism remained significantly induced. Interestingly, a subset of genes involved in ECM and focal adhesion also remains significantly downregulated by NN1177 following adjustment for bodyweight. Several pathways involved in inflammation and amino acid metabolism also remain significantly affected in the NASH animals independent of bodyweight, likely demonstrating the rerouting of hepatic metabolism necessary to manage the low nutritious value and pro-inflammatory effects of this high-fat and high-sucrose diet.
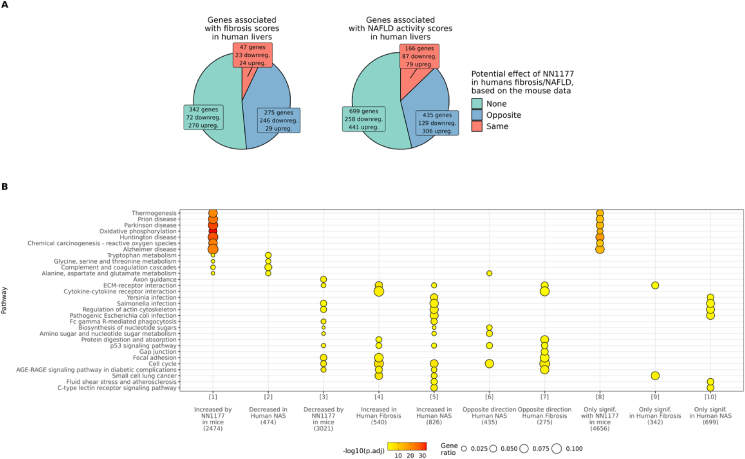
To evaluate if the positive effects of NN1177 treatment on the hepatic health of the DIO-NASH mouse model could have potential to be translatable into a treatment for the human condition we compared the genes that are significantly associated with fibrosis score and NAFLD activity scores in human livers to the genes that are significantly affected by NN1177 in this murine model of NASH. For this purpose, we analysed two publicly available datasets, GSE162694 [24] and GSE130970 [23]. We used this RNA sequencing data from human NASH liver to find which gene perturbations associated independently with either fibrosis stages or NAFLD activity scores, and subsequently performed a meta-analysis to combine the results from the two datasets. Figure 6A shows the overlap between genes significantly associated with fibrosis (left panel) or NAFLD activity score (right panel) in humans and their corresponding regulation in NN1177 treated DIO-NASH mouse (analyses were restricted to genes with human/mice homologues). When exploring the assumption that NASH improvement corresponds to NN1177 inducing an opposite effect on gene expression in mice than how the gene is perturbed in association with fibrosis/NAFLD in human livers, while worsening corresponds to NNC117 having a similar effect in mice and fibrosis/NAFLD in human livers, we find that NN1177 has the potential to reverse a large fraction of the dysregulated hepatic gene expression seen in people living with NAFLD and NASH. To further understand which biology and specific pathways may be targeted by this co-agonist in humans, a pathway enrichment analysis was also performed on these comparisons (Figure 6B). Interestingly, as opposed to what was seen in the mice, we did not find significant repression of the oxidative phosphorylation pathways in human NAFLD/hepatic fibrosis in the datasets analysed. This challenges the translatability of the mouse model with respect to energy expenditure/energy handling and suggests that in humans, the main pathways that potentially could be rescued with NN1177 treatment are those related to the inflammatory and pro-fibrotic pathways.
Figure 6.
Potential effect of NN1177 on the transcriptome in human NASH.A. Genes significantly associated with fibrosis score and NAFLD activity scores in human livers are grouped by direction of effect of NN1177 in mice. Opposite: NN1177 has an opposite effect in mice than fibrosis/NAFLD in human livers (for opposite genes, upregulated = upregulated in mice data, downregulated in human data, and inversely). Same: NNC117 has a similar effect in mice than fibrosis/NAFLD in human livers. B. Over-representation enrichment analyses (KEGG pathways) genes significantly associated with NASH in human and/or NN1177 in mice (ordinal regression analysis NASH vehicle < NN1177 low < NN1177 med < NN1177 high). The pathways represented are selected from the top 5 pathways per gene-sets. Each gene-set tested is identified by a number in bracket and referenced with this number in the results. The numbers of genes in each gene sets are shown in parenthesis. The following gene-sets tested had no significant enrichments and are not represented in the plot: “Decreased in Human Fibrosis” (n = 124), “Opposite direction in human NAS” (n = 435), “Same direction Human NAS” (n = 166), “Same direction Human Fibrosis” (n = 47). Full differential expression and enrichment results are presented in Suppl. Table 3.
4. Discussion
Here we have evaluated the dose-dependent effects of the dual GLP-1 and glucagon receptor agonist NN1177 in a DIO-NASH mouse model. Similar to the GLP-1 receptor agonist semaglutide, NN1177 induced substantial body weight reduction (Figure 1) and mediated an even more profound effect on hepatic steatosis and cholesterol content (Figure 2) at equal body weight loss, confirming that NN1177 has maintained a strong glucagon mediated effect on hepatic lipid oxidation. The effect on feeding behaviour has previously been evaluated in DIO mouse studies and the effect on food intake was comparable to a GLP1 reference molecule, but the overall body weight loss was superior to the reference suggesting increased energy expenditure due to the glucagon agonism [14]. In this study the cumulative food intake in the NN1177 groups were significantly higher than in the semaglutide group, suggesting a compensatory eating behaviour due to the increased energy expenditure One of the observed effects that was specific to the dual GLP-1 and glucagon receptor agonist is the induction of hepatic glycogen content (Figure 2C). The accumulation of glycogen is not observed with semaglutide, and is a probably a consequence of simultaneous GLP-1 receptor mediated insulin release, counteracting the glucagon mediated hepatic glucose output.
NN1177 and semaglutide showed significant effects on several liver related parameters, such as ALT, inflammatory biomarkers; CD11b and Gal-3, as well as Col1. Unlike the effect on steatosis, the efficacy on ALT and inflammatory markers and fibrosis markers obtained with NN1177 was not superior to the efficacy obtained with semaglutide. Neither of these hepatic improvements achieved with either compound translated into an effect on the histological fibrosis endpoints after 8 weeks of treatment.
In the research field pursuing development of a pharmaceutical treatment for NASH, it is widely debated how soon regression of deposited collagen fibers can be expected to be observed following improvement of hepatocyte metabolic health. Clinical studies of hepatitis C patients suggest that even after fully removing the underlying cause by eliminating the virus it takes several years to see improvement in the fibrosis stage in a large fraction of the patients evaluated [36]. For this reason, many of the NASH clinical trials aiming at improving hepatocyte metabolic health have a duration of a year or more [37,38] to increase the probability of meeting the fibrosis regression endpoint. Similarly, there are also uncertainties and discussions around the optimal duration of preclinical studies to demonstrate decreased fibrosis progression or actual regression.
Subsequent in-house investigations into the timeline for regression of hepatic fibrosis by switching the mice from DIO-NASH to chow diet have shown that longer study duration is necessary to demonstrate changes in SR area% (data not shown). Therefore, we proceeded with gene expression and bioinformatic analysis to investigate if NN1177 treatment induced transcriptional changes that pointed to an accelerated effect on inflammation and/or fibrosis resolution. Interestingly, NN1177 seem to have a dose-specific effect on the expression of a subset of genes associated with fibrogenesis and oxidative phosphorylation, trending toward higher corrections compared to what could be achieved with semaglutide in this study (Figure 5, gene module 2 and 3). Previously the concerns with respect to glucagon agonism have been focused on glucose homeostasis, extensively described for NN1177 in high fat diet induced obese (DIO) mouse model [14]. Here, mice develop glucose intolerance over time as determined by an acute glucose challenge test. Administration of NN1177 improved glucose handling but not to the same extend than observed by administration of the GLP1 reference alone [14]. In this study using the metabolic DIO-NASH model with comparable glucose intolerance to the DIO model [22], none of the pathways induced with NN1177 seem directly related to glucose homeostasis. This could suggest that the added benefit of the GLP-1 mediated insulin release is able to maintain glucose control in this setting, although further experiments would be warranted to confirm this. On the other hand, NN1177 activation of pathways such as oxidative phosphorylation and other KEGG pathways indicating the presence of mitochondrial overactivity, were very apparent in this study. The overexpression of genes related to oxidative phosphorylation and increased mitochondrial respiration have previously been described as a direct effect of glucagon agonism [39] but also by GLP1/GCGR agonism [15]. While oxidative phosphorylation pathways were found to be downregulated in the DIO-NASH mouse model, no significant changes in oxidative phosphorylation pathways were observed in the study of hepatic gene expression from NASH patients relative to healthy controls (however this pathway enrichment analysis is restricted to genes with human/mouse homologues for comparison purposes and might miss human-specific signals). This could suggest that these pathways either are differentially regulated among rodents and humans or that they are temporarily dysregulated following the sub-chronic switch to a high fat diet in the rodent DIO-NASH model but normalises during the decades of western diet intake that usually supersedes the development of NASH in humans. Another interesting observation is the strong NN1177 mediated induction of the ribosomal biosynthesis pathway, which is the most significantly affected pathway as shown in Figure 5C. Insulin signalling, with its anabolic activity, is well-known to increase ribosomal biosynthesis [40]. Deregulation of this pathway has not previously been associated with NAFLD, or glucagon agonism, suggesting that this could be a unique feature arising from the dual GLP-1R/GCGR pharmacology of NN1177.
Figure 5A–C is also pointing to pathways where NN1177 appears to further exacerbate some of the effects of the DIO-NASH diet-induced phenotype (Gene module 4). These pathways are involved in steroid and steroid hormone biosynthesis in addition to retinol and linoleic acid metabolism and bile secretion and are repressed in the DIO-NASH vehicle group. The biological processes and expression of the enzymes in these pathways are known to be modulated by high fat and high cholesterol intake [41]. The observation that semaglutide treatment does not further repress these pathways and that they remain significantly repressed after correcting for body weight loss points towards this effect being partly mediated by the glucagon component of NN1177. It is difficult to interpret the downstream biological consequences of this observation in this complex setting of high glucagon signalling in an animal being fed a high-fat, high-sucrose, and high-cholesterol diet while undergoing massive weight loss. However, fundamentally the biological role of glucagon is to safeguard blood glucose levels by making alternative substrates available to the organism to consume for energy production. Therefore, it is plausible to speculate that in any metabolic setting, very high levels of glucagon agonism will result in the associated fasting related gene programme switching metabolic fluxes towards a severe catabolic state that may lead to an inhibition of important anabolic processes including steroid biosynthesis and conversions of retinols and essential fatty acids. Perhaps the explanation for the downregulation of the gene programmes in module 4 in the DIO-NASH vehicle control and even further in the higher dose of NN1177 is that while the former is a result of high levels of cholesterol and lipids exerting negative feedback inhibition on the expression of the enzymes producing these metabolites, the latter is a consequence of glucagon agonism inducing a severe catabolic state.
A high degree of overlap was identified in genes dysregulated by NN1177 in the DIO-NASH rodent model and by NAFLD-associated fibrosis in human (Figure 6), suggesting that if the mechanism of action of NN1177 is conserved in humans, it could be a potential treatment for NASH achieving fibrosis regression. In particular, Figure 5A Module 2 points towards NN1177 decreasing the expression of genes involved in fibrogenesis, including ECM receptors and focal adhesion molecules. Importantly, this activity appears to be partly independent of body weight loss (Supp. Figure 2). The observation that NN1177 also strongly activates pathways involved in oxidative phosphorylation and ribosomal biosynthesis, signals that NN1177 treatment induces an adaptation to increased energy expenditure and protein turn over. While glucagon receptor agonism may mediate increases in energy expenditure which is predicted to additively accelerate the GLP-1 mediated body weight loss, this effect could indirectly exacerbate the minor increase in heart rate observed with GLP-1R agonists. Glucagon has also been reported to have direct chronotropic effects on the heart [42] and NN1177 have been shown to effect heart rate in early clinical testing (Friedrichsen et al. accepted for publication in MOLMET-D-23-00453R1).
Taken together, the substantial amount of preclinical and clinical data published on GLP-1/glucagon co-agonism demonstrates that incorporating the glucagon component in these molecules has the potential to significantly increase the efficacy of treating metabolic disease, in particular NAFL and NASH. Even though, the preclinical model has translational limitations, the data points towards the antifibrotic effects of NN1177. However, there appears to only be a narrow window for balancing the potency of the glucagon and GLP-1 agonism to achieve the maximum beneficial effects of both while staying below the threshold of glucagon pharmacology that may lead to unwanted metabolic changes and chronotrophic effect on the heart. Several of the compounds tested in clinical trials so far, has failed to achieve a significant body weight loss [20]. However, there is further innovation on-going in the field with the development of triple agonism molecules, including the GLP-1/GIP/glucagon tri-agonist Retatruvide, with promising results on weight loss and resolution of hepatic steatosis and acceptable glycaemic control and effects on heart rate.
5. Conclusion
The GLP-1/GCG receptor agonist NN1177 was evaluated in the DIO-NASH mouse model. Dose-related efficacy was obtained on clinically relevant parameters, such as steatosis, but also on relevant biomarkers for inflammation and fibrogenesis. Transcriptomic analyses identified several relevant hepatic pathways directly regulated towards normalisation by NN1177, including pathways relevant to human NASH. We also identified pathways which could result in additional downstream effects, which may be dose limiting in the clinical situation. When compared to semaglutide, NN1177 is superior on liver fat reduction, whereas a comparable efficacy is obtained on inflammatory (CD11b and GAL-3) and fibrosis related biomarkers (aSMA and Col1).
The above presented preclinical data as well as the human transcriptomic analysis suggest that balanced GLP-1R/GCGR co-agonism could improve clinically relevant biological pathways which are specific to people living with NASH and that this effect could be obtained without additional body weight loss when compared to GLP-1R agonism alone. The potential for achieving the clinical endpoints of NASH resolution and fibrosis reduction still remains to be evaluated in clinical trials and on-going and future Phase 2b studies will shed more lights on the therapeutic potential of dual GLP-1R/GCGR agonists in people living with NASH.
CRediT author statement
Jenny Norlin: Data curation, Methodology, Writing – original draft, Writing – review & editing. Anne Bugge: Writing – original draft, Writing – review & editing. Thomas Monfeuga: Formal analysis, Writing – original draft, Writing – review & editing. Michael Feigh: Writing – review & editing. Dorte Holst: Conceptualization, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. Sanne S. Veidal: Writing – review & editing. Pia S. Petersen: Writing – review & editing. Cesar A. Prada-Medina: Formal analysis, Writing – review & editing. Markus Latta: Writing – original draft, Writing – review & editing. Elisabeth D. Gaalsgaard: Data curation, Methodology, Writing – original draft.
Declaration of competing interest
Thomas Monfeuga, Jenny Norlin, Anne Bugge, Elisabeth D. Gaalsgaard, Cesar A. Prada-Medina, Markus Latta, Sanne S. Veidal and Dorte Holst are employees of Novo Nordisk A/S and shareholders in Novo Nordisk A/S. Pia S. Petersen is currently employee at Ascendis Pharma A/S and Michael Feigh is employee at Gubra A/S.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101850.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Malhotra P., Gill R.K., Saksena S., Alrefai W.A. Disturbances in cholesterol homeostasis and non-alcoholic fatty liver diseases. Front Med (Lausanne) 2020;7:467. doi: 10.3389/fmed.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Stepanova M., Lawitz E.J., Reddy K.R., Wai-Sun Wong V., Mangia A., et al. Patients with nonalcoholic steatohepatitis experience severe impairment of health-related quality of life. Am J Gastroenterol. 2019;114(10):1636–1641. doi: 10.14309/ajg.0000000000000375. [DOI] [PubMed] [Google Scholar]
- 4.Williams C.D., Stengel J., Asike M.I., Torres D.M., Shaw J., Contreras M., et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140(1):124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 5.Dongiovanni P., Valenti L. Genetics of nonalcoholic fatty liver disease. Metabolism. 2016;65(8):1026–1037. doi: 10.1016/j.metabol.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Singh S., Allen A.M., Wang Z., Prokop L.J., Murad M.H., Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol. 2015;13(4) doi: 10.1016/j.cgh.2014.04.014. p. 643–654.e1-9;quiz e39-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Younossi Z.M., Marchesini G., Pinto-Cortez H., Petta S. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: implications for liver transplantation. Transplantation. 2019;103(1):22–27. doi: 10.1097/TP.0000000000002484. [DOI] [PubMed] [Google Scholar]
- 8.Newsome P.N., Sejling A.S., Sanyal A.J. Semaglutide or placebo for nonalcoholic steatohepatitis. Reply. N Engl J Med. 2021;385(2):e6. doi: 10.1056/NEJMc2106921. [DOI] [PubMed] [Google Scholar]
- 9.Isaacs D., Clements J., Turco N., Hartman R. Glucagon: its evolving role in the management of hypoglycemia. Pharmacotherapy. 2021;41(7):623–633. doi: 10.1002/phar.2534. [DOI] [PubMed] [Google Scholar]
- 10.Scheen A.J., Paquot N., Lefebvre P.J. Investigational glucagon receptor antagonists in Phase I and II clinical trials for diabetes. Expert Opin Investig Drugs. 2017;26(12):1373–1389. doi: 10.1080/13543784.2017.1395020. [DOI] [PubMed] [Google Scholar]
- 11.Guzman C.B., Zhang X.M., Liu R., Regev A., Shankar S., Garhyan P., et al. Treatment with LY2409021, a glucagon receptor antagonist, increases liver fat in patients with type 2 diabetes. Diabetes Obes Metab. 2017;19(11):1521–1528. doi: 10.1111/dom.12958. [DOI] [PubMed] [Google Scholar]
- 12.Henderson S.J., Konkar A., Hornigold D.C., Trevaskis J.L., Jackson R., Fritsch Fredin M., et al. Robust anti-obesity and metabolic effects of a dual GLP-1/glucagon receptor peptide agonist in rodents and non-human primates. Diabetes Obes Metab. 2016;18(12):1176–1190. doi: 10.1111/dom.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdecantos M.P., Pardo V., Ruiz L., Castro-Sánchez L., Lanzón B., Fernández-Millán E., et al. A novel glucagon-like peptide 1/glucagon receptor dual agonist improves steatohepatitis and liver regeneration in mice. Hepatology. 2017;65(3):950–968. doi: 10.1002/hep.28962. [DOI] [PubMed] [Google Scholar]
- 14.Simonsen L., Lau J., Kruse T., Guo T., McGuire J., Jeppesen J.F., et al. Preclinical evaluation of a protracted GLP-1/glucagon receptor co-agonist: translational difficulties and pitfalls. PLoS One. 2022;17(3) doi: 10.1371/journal.pone.0264974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann T., Thomas L., Baader-Pagler T., Haebel P., Simon E., Reindl W., et al. BI 456906: discovery and preclinical pharmacology of a novel GCGR/GLP-1R dual agonist with robust anti-obesity efficacy. Mol Metab. 2022;66 doi: 10.1016/j.molmet.2022.101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldissera F.G., Holst J.J., Knuhtsen S., Hilsted L., Nielsen O.V. Oxyntomodulin (glicentin-(33-69)): pharmacokinetics, binding to liver cell membranes, effects on isolated perfused pig pancreas, and secretion from isolated perfused lower small intestine of pigs. Regul Pept. 1988;21(1–2):151–166. doi: 10.1016/0167-0115(88)90099-7. [DOI] [PubMed] [Google Scholar]
- 17.Wynne K., Park A.J., Small C.J., Patterson M., Ellis S.M., Murphy K.G., et al. Subcutaneous oxyntomodulin reduces body weight in overweight and obese subjects: a double-blind, randomized, controlled trial. Diabetes. 2005;54(8):2390–2395. doi: 10.2337/diabetes.54.8.2390. [DOI] [PubMed] [Google Scholar]
- 18.Hope D.C.D., Vincent M.L., Tan T.M.M. Striking the balance: GLP-1/glucagon co-agonism as a treatment strategy for obesity. Front Endocrinol (Lausanne) 2021;12 doi: 10.3389/fendo.2021.735019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambery P., Parker V.E., Stumvoll M., Posch M.G., Heise T., Plum-Moerschel L., et al. MEDI0382, a GLP-1 and glucagon receptor dual agonist, in obese or overweight patients with type 2 diabetes: a randomised, controlled, double-blind, ascending dose and phase 2a study. Lancet. 2018;391(10140):2607–2618. doi: 10.1016/S0140-6736(18)30726-8. [DOI] [PubMed] [Google Scholar]
- 20.Nahra R., Wang T., Gadde K.M., Oscarsson J., Stumvoll M., Jermutus L., et al. Effects of cotadutide on metabolic and hepatic parameters in adults with overweight or obesity and type 2 diabetes: a 54-week randomized phase 2b study. Diabetes Care. 2021;44(6):1433–1442. doi: 10.2337/dc20-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolbol K.S., Kristiansen M.N., Hansen H.H., Veidal S.S., Rigbolt K.T., Gillum M.P., et al. Metabolic and hepatic effects of liraglutide, obeticholic acid and elafibranor in diet-induced obese mouse models of biopsy-confirmed nonalcoholic steatohepatitis. World J Gastroenterol. 2018;24(2):179–194. doi: 10.3748/wjg.v24.i2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen H.H., HM A.E., Oro D., Evers S.S., Heeboll S., Eriksen P.L., et al. Human translatability of the GAN diet-induced obese mouse model of non-alcoholic steatohepatitis. BMC Gastroenterol. 2020;20(1):210. doi: 10.1186/s12876-020-01356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang S.A., Oseini A., Feaver R.E., Cole B.K., Asgharpour A., Vincent R., et al. Gene expression predicts histological severity and reveals distinct molecular profiles of nonalcoholic fatty liver disease. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-48746-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantano L., Agyapong G., Shen Y., Zhuo Z., Fernandez-Albert F., Rust W., et al. Molecular characterization and cell type composition deconvolution of fibrosis in NAFLD. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-96966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewels P., Magnusson M., Lundin S., Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Soneson C., Hickey P.F., Johnson L.K., Pierce N.T., Shepherd L., et al. Tximeta: reference sequence checksums for provenance identification in RNA-seq. PLoS Comput Biol. 2020;16(2) doi: 10.1371/journal.pcbi.1007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csárdi G., Nepusz T. 2006. The igraph software package for complex network research. [Google Scholar]
- 31.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36(3):1–48. [Google Scholar]
- 34.Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., et al. BioMart--biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmona I., Cordero P., Ampuero J., Rojas A., Romero-Gomez M. Role of assessing liver fibrosis in management of chronic hepatitis C virus infection. Clin Microbiol Infect. 2016;22(10):839–845. doi: 10.1016/j.cmi.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Sanyal A.J., Chalasani N., Kowdley K.V., McCullough A., Diehl A.M., Bass N.M., et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med. 2010;362(18):1675–1685. doi: 10.1056/NEJMoa0907929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loomba R., Abdelmalek M.F., Armstrong M.J., Jara M., Kjaer M.S., Krarup N., et al. Semaglutide 2.4 mg once weekly in patients with non-alcoholic steatohepatitis-related cirrhosis: a randomised, placebo-controlled phase 2 trial. Lancet Gastroenterol Hepatol. 2023;8(6):511–522. doi: 10.1016/S2468-1253(23)00068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breton L., Clot J.P., Baudry M. Effects of glucagon on basal metabolic rate and oxidative phosphorylation of rat liver mitochondria. Horm Metab Res. 1983;15(9):429–432. doi: 10.1055/s-2007-1018747. [DOI] [PubMed] [Google Scholar]
- 40.Proud C.G. Regulation of protein synthesis by insulin. Biochem Soc Trans. 2006;34(Pt 2):213–216. doi: 10.1042/BST20060213. [DOI] [PubMed] [Google Scholar]
- 41.Vidon C., Boucher P., Cachefo A., Peroni O., Diraison F., Beylot M. Effects of isoenergetic high-carbohydrate compared with high-fat diets on human cholesterol synthesis and expression of key regulatory genes of cholesterol metabolism. Am J Clin Nutr. 2001;73(5):878–884. doi: 10.1093/ajcn/73.5.878. [DOI] [PubMed] [Google Scholar]
- 42.Hernandez-Cascales J. Does glucagon have a positive inotropic effect in the human heart? Cardiovasc Diabetol. 2018;17(1):148. doi: 10.1186/s12933-018-0791-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse data generated in this paper is available on the online repository Gene Expression Omnibus (GEO) under the accession GSE220519. The publicly available human data analysed in this paper is available under the accessions GSE162694 and GSE130970. The code used to generate the results is available in the GitHub repository https://github.com/novonordisk-research/NN1177_liver_RNAseq.