Abstract
A gene (estA) encoding a 42-kDa cell-bound esterase, EstA, was found to be located 75 bp upstream of the cyclophilin A gene (cypA) of Streptomyces chrysomallus. Western blot analysis revealed the presence of EstA (42 kDa) in cell extracts of S. chrysomallus X2 and Streptomyces lividans. EstA specifically hydrolyzes short-chain p-nitrophenyl esters. EstA formation starts at the end of growth phase, and its activity level remains constant throughout stationary phase. Expression of estA from the melanin (mel) promoter in plasmid pIJ702 led to a substantial increase of total esterase activity in streptomycetes.
Streptomycetes are filamentous soil bacteria possessing a large repertoire of extracellular enzymes for the degradation of biopolymeric material in their natural habitats (13). The presence of these enzymatic activities is often substrate regulated but can also depend on a specific stage in the life cycle (21). The life cycle of the streptomycetes is characterized by the development of a vegetative substrate mycelium from which aerial mycelium and later spores are formed. Production of antibiotics and other secondary metabolites is a characteristic feature of streptomycetes and usually starts at the onset of the stationary growth phase (2). The regulation of formation of lipases and esterases in streptomycetes has attracted particular interest. Stationary-phase-dependent formation of lipases has been demonstrated in several cases, e.g., with lipA from Streptomyces exfolians (20). In addition, interesting regulation systems involving coexpression of activator proteins for Streptomyces lipase or esterase gene expression have been described previously (1, 20). Esterases hydrolyze water-soluble or emulsified esters with short-chain carboxylic acids, whereas lipases are specific for emulsified substrates with fatty acyl chains. Esterase sequences show the conserved motif G-X1-S-X2-G containing the active-site serine representing the nucleophilic residue in the catalytic triad of these enzymes (4, 13). This triad normally consists of an additional histidine and an additional aspartate located at specific positions in the polypeptide chains, which, however, are not obviously the same ones in the various enzymes (3–5). Interestingly, two previously identified Streptomyces esterases lack the classical G-X-S-X-G/G-X-S motif and may contain a novel variant of the catalytic triad (22).
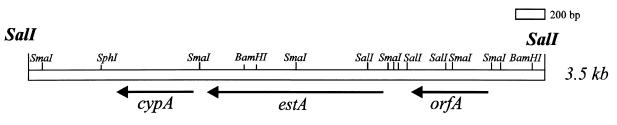
Sequencing of the upstream region of cypA from Streptomyces chrysomallus.
Plasmid pMS104 (17) contains a 9-kb XhoI fragment of the S. chrysomallus X2 (10) chromosome carrying the cyclophilin gene cypA. About 2.4 kb of the immediate upstream region of cypA was sequenced (EMBL accession no. Z15137), and sequence analysis revealed two open reading frames (ORFs) (estA and orfA; 1,170 and 435 bp, respectively). The identified ORFs are separated by 268 bp and are in same orientation as that of cypA (Fig. 1); estA starts with a GTG codon, and the TGA stop codon is 74 bp in front of cypA. The noncoding regions upstream of orfA and estA did not conspicuously show conserved promoter structures. The G+C content of estA is 74.4% (of orfA, 74.8%), and the codon usage is typical for a streptomycete gene with an average G+C content of 93.5% (orfA, 87.1%) in the third codon position. From the deduced amino acid sequence, it is apparent that estA encodes a protein of 41,178 Da and orfA encodes one of 15,441 Da. Southern hybridization analyses showed that estA is present as a single copy on the chromosome of S. chysomallus.
FIG. 1.
Organization of the estA gene region of S. chrysomallus X2. A ca 2.4-kb fragment of genomic S. chrysomallus X2 DNA is shown. The ORFs are represented by arrows. cypA encodes cyclophilin A, estA encodes cell-bound esterase A, and orfA is of unknown function.
estA encodes an esterase.
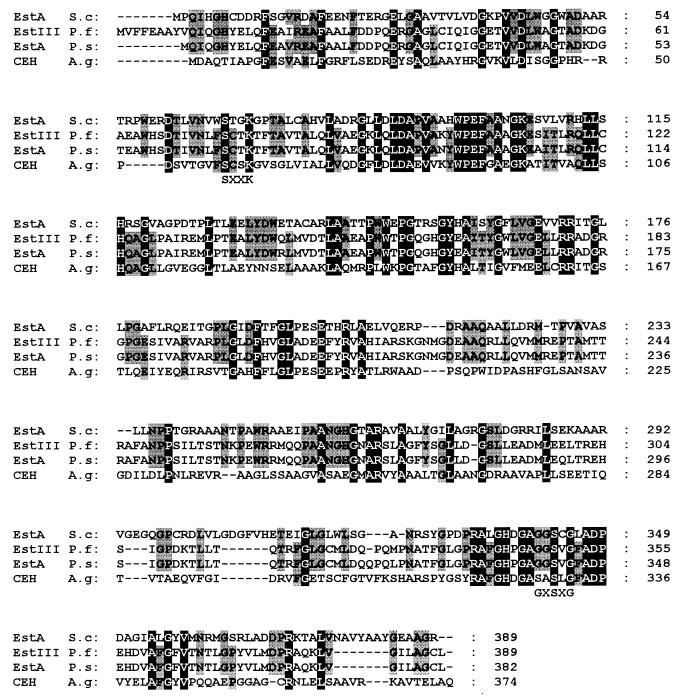
Database searches with the complete protein sequence deduced from the estA nucleotide sequence revealed the highest similarities to two cell-bound esterases from psychrotrophic pseudomonads (11, 14) (43% identity to both) and to the ethyl chrysanthemate esterase from Arthrobacter globiformis (15) (38% identity) (Fig. 2). Like these proteins, EstA does not contain a signal sequence, which indicates that it is a cell-bound protein. EstA contains the sequence GGS343CG, which agrees with the consensus sequence surrounding the active-site serine, G-X1-S-X2-G, observed in a large number of serine esterases, including lipases and serine proteases (4). Interestingly, the N-terminal part of EstA (amino acids 30 to 100) revealed additional sequence similarity to β-lactamases, e.g., to cephalosporinase from Escherichia coli (8) (50% identity) and to β-lactamase from Citrobacter freundii (16) (44% identity), with the sequence featuring the conserved S-X-X-K motif which forms part of the catalytic center of β-lactamases (16).
FIG. 2.
Amino acid sequence alignment of esterase A from S. chrysomallus X2. Sequences for esterase A from S. chrysomallus X2 (EstA S.c), esterase III from Pseudomonas fluorescens (EstIII P.f), esterase A from Pseudomonas spp. (EstA P.s), and carboxylic ester hydrolase from Arthrobacter globiformis (CEH A.g) are shown.
Heterologous expression of estA in E. coli.
estA was modified by PCR with oligonucleotides A (5′-AGGGAGGCCGCATGCCGCAGATCCAC-3′) and B (5′-AACTGCAGTCACCTCCCGGCGGCCTC-3′). The PCR fragment was cloned into the E. coli expression plasmid pQE32 (Qiagen), which generates pQEBOX321 encoding an N-terminal six-His-tagged EstA. In crude extracts of E. coli transformants, a 42-kDa protein was detectable. Only a small amount of the protein was soluble, and it was tested with p-nitrophenylbutyrate as the substrate. The specific esterase activity was four to five times higher than that in the extract from the control strain (data not shown). Only the denatured soluble protein (in 6 M urea or 8 M guanidine hydrochloride) binds to the Ni-chelate matrix, indicating that the six-His tag is buried in the native form. To generate antibodies, the denatured form was eluted from the Ni-chelate matrix and further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Overexpression of estA and characterization of its gene product in S. chrysomallus and Streptomyces lividans.
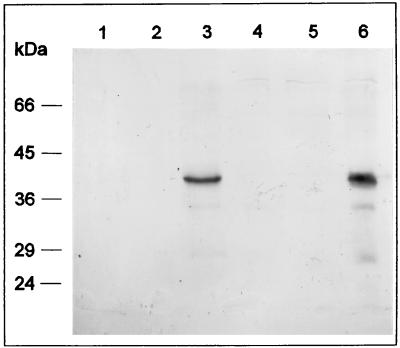
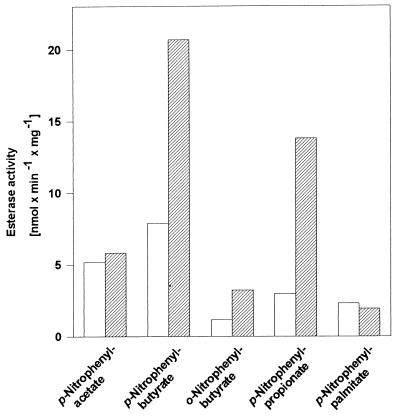
For overexpression of estA in streptomycetes, two PCR-generated estA derivatives were cloned as SphI-SstI fragments behind the melanin promoter in plasmid pIJ702 (7, 9), which generates pBOX9 and pBOX19. Primers A and B (see above) were used to generate the wild-type gene (pBOX9), primers C (5′-GGAGGGCCGCATGCACCACCACCACCACCACTGCGACGACCGCTTCAG-3′) and B were used to generate an estA derivative coding for a six-His-tagged EstA (pBOX19). Both PCR fragments were first subcloned as SphI-PstI fragments into pTZ18 (Pharmacia) to obtain a 3′ SstI site, necessary for cloning into pIJ702. S. chrysomallus X2 and S. lividans transformants carrying plasmids pBOX9, pBOX19, and pIJ702 as well as the nontransformed strains were then grown in complete medium (2-day-old mycelium was used), and protein extracts derived from these strains were analyzed with respect to the presence of EstA and their lipolytic activities. In Western analyses, strains transformed with pBOX9 (Fig. 3, lanes 3 and 6) or pBOX19 (data not shown) revealed the presence of EstA. By contrast, EstA was hardly detectable in strains transformed with pIJ702 (Fig. 3, lanes 2 and 5) and nontransformed strains (Fig. 3, lanes 1 and 4). Attempts to purify six-His-tagged EstA expressed from the pBOX19 construct in S. chrysomallus X2 failed, as in the case of the very similar construct in E. coli, indicating the inaccessibility of the six-His tag in this protein. Therefore, esterase and lipase activity was determined in crude extracts by using p-nitrophenyl acetate (pNPAc), p-nitrophenyl propionate (pNPPr), o- or p-nitrophenyl butyrate (oNPBu or NPBu, respectively), and p-nitrophenyl palmitate (pNPPa) as substrates. Total esterase activity in S. chrysomallus X2 transformed with pBOX9 (Fig. 4) or pBOX19 (data not shown) greatly exceeds that of the untransformed strain (Fig. 4) or that harboring pIJ702. Increases in specific activities in the cleavage of pNPPr and pNPBu are observable, whereas the low basal activities for pNPPa and pNPAc hydrolysis remain unchanged. This unambiguously identifies the estA gene product as an esterase. Furthermore, the plasmid-encoded esterase activity was inhibited by 80% within 20 min of incubation at 37°C in the presence of 30 μM phenylmethylsulfonyl fluoride. Phenylmethylsulfonyl fluoride covalently binds to the active-site serines of many serine proteases and lipases. Measurements of esterase activity in cell extracts of untransformed S. lividans (with pNPBu as the substrate) revealed a ca. 10-fold-higher basal esterase activity than that in S. chrysomallus X2, which indicates that S. lividans may contain additional esterases distinct from EstA, because in Western blot analyses (see below) the EstA levels in S. lividans and S. chrysomallus X2 are nearly the same.
FIG. 3.
Overexpression of estA in streptomycetes. The Western blot shows SDS-PAGE separations of total cellular protein of sonicated cells from S. chrysomallus X2 (lane 1), S. chrysomallus X2 harboring pIJ702 (lane 2), S. chrysomallus X2 harboring pBOX9 (lane 3), S. lividans (lane 4), S. lividans with pIJ702 (lane 5), and S. lividans harboring pBOX9 (lane 6). Each lane contains about 50 μg of protein. All streptomycetes were grown for 30 h in complete medium.
FIG. 4.
Cell-bound lipolytic activity and substrate specificity of EstA. The substrate specificity of EstA encoded by pBOX9 as measured by hydrolysis of p-nitrophenyl carboxylic acid esters is shown. The plasmid-bearing strain (hatched bars) shows elevated levels of hydrolysis of pNPBu and pNPPr, while no increases in hydrolysis of pNPAc and pNPPa were observed. Results for Nontransformed S. chrysomallus X2 (open bars) are also shown. S. chrysomallus X2 was grown to stationary phase, and 100 μg of total cellular protein was used in the test.
estA is expressed in the stationary phase.
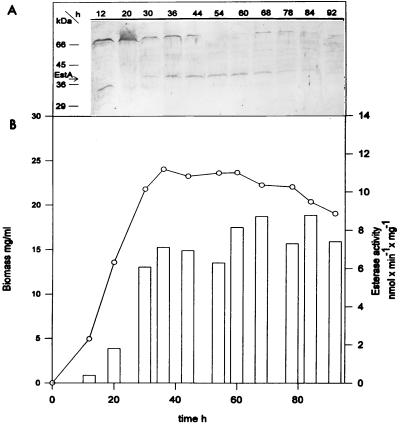
Immunoblot analyses of protein extracts from S. chrysomallus X2 mycelium harvested at different times of cultivation revealed that formation of EstA begins at the end of the growth phase (Fig. 5). For extracts from all stages of cultivation, there was an additional cross-reacting immunoreactive band of 66 kDa. The nature of this protein is not known. Esterase activity determined with pNPBu is correlated with the appearance of EstA. Protein level and enzymatic activity remained constant during the stationary phase of cultivation (Fig. 5). A very similar picture was obtained when S. chrysomallus X2 was grown in chemically defined media (data not shown). In these media, the total esterase activity was not affected by the choice of carbon sources, such as glucose, maltose, glycerol, acetate, or the esterase substrate triacetin. Results of Western analyses with S. lividans mycelium obtained from the growth and stationary phases show a similar pattern to that of S. chrysomallus X2: the appearance of a 42-kDa protein was clearly visible in stationary-phase cultures, while it was hardly detectable in younger cultures (less than 2 days of growth).
FIG. 5.
Growth-phase-dependent expression of estA in S. chrysomallus X2. (A) SDS-PAGE separations of soluble protein extracts derived from sonicated cells of S. chrysomallus X2 from various times of cultivation in complete medium were analyzed by Western blotting. (B) Esterase activity in protein extracts derived from S. chrysomallus X2 grown for the indicated times was determined. Vertical bars represent esterase activity with pNPBu as the substrate; mycelial growth (wet weight) (○) is also shown.
Conclusion.
Previous reports have shown that streptomycetes possess various lipolytic enzymes classified as extracellular esterases or lipases (6, 18–21). In this report, we describe for the first time a cell-bound esterase from a streptomycete. Previously described cell-bound esterases from other organisms are not essential for growth, and thus their functions are not clear. With S. chrysomallus X2, a detailed investigation of EstA function was not possible because this organism is not amenable to gene disruption due to insufficient frequencies of transformation and integration. The gene estA lies immediately 5′ to the previously described cyclophilin A gene (cypA) of S. chrysomallus X2. Interestingly, the gene encoding the cell-bound esterase in Acinetobacter calcoaceticus BD 413 also lies upstream of a cyclophilin gene in the same orientation (12).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant Ke 452/6-2).
We thank S. Lucania from Bristol-Myers Squibb for the gift of thiostrepton.
REFERENCES
- 1.Babcock M J, McGrew M, Schottel J L. Identification of a protein-binding sequence involved in expression of an esterase gene from Streptomyces scabies. J Bacteriol. 1992;174:4287–4293. doi: 10.1128/jb.174.13.4287-4293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb M J. The regulation of antibiotic production in Streptomyces coelicolor A3. Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 3.Blow D. More of the catalytic triad. Nature. 1990;351:694–695. doi: 10.1038/343694a0. [DOI] [PubMed] [Google Scholar]
- 4.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley R, Turkenburg J P, Christiansen L, Huge-Jensen B, Norskov L, Thim L, Menge U. A serine protease triad forms the catalytic center of a triacyl glycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 5.Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988;334:491–494. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 6.Cruz H, Pérez C, Wellington E, Castro C, Servín-González L. Sequence of the Streptomyces albus G lipase-encoding gene reveals the presence of a prokaryotic lipase family. Gene. 1994;144:141–142. doi: 10.1016/0378-1119(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 8.Jaurin B, Grundstrom T. AmpC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz E, Thompson C J, Hopwood D A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983;129:2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- 10.Keller U, Krengel U, Haese A. Genetic analysis in Streptomyces chrysomallus. J Gen Microbiol. 1985;131:1181–1191. doi: 10.1099/00221287-131-5-1181. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y S, Lee H B, Choi K D, Park S, Yoo O J. Cloning of Pseudomonas fluorescens carboxylesterase gene and characterization of its product expressed in Escherichia coli. Biosci Biotechnol Biochem. 1994;58:111–116. doi: 10.1271/bbb.58.111. [DOI] [PubMed] [Google Scholar]
- 12.Kok R G, Christoffels V M, Vosman B, Hellingwerf K J. Growth-phase-dependent expression of the lipolytic system of Acinetobacter calcoaceticus BD413: cloning of a gene encoding one of the esterases. J Gen Microbiol. 1993;139:2329–2342. doi: 10.1099/00221287-139-10-2329. [DOI] [PubMed] [Google Scholar]
- 13.Korn-Wendisch F, Kutzner H J. The family Streptomycetaceae. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. New York, N.Y: Springer-Verlag; 1992. pp. 921–995. [Google Scholar]
- 14.McKay D B, Jennings M P, Godfrey E, MacRae I, Rogers P J, Beacham I R. Molecular analysis of an esterase-encoding gene from a lipolytic psychrotrophic pseudomonad. J Gen Microbiol. 1992;138:701–708. doi: 10.1099/00221287-138-4-701. [DOI] [PubMed] [Google Scholar]
- 15.Nishizawa M, Shimizu M, Ohkawa H, Kanaoka M. Stereoselective production of (+)-trans-chrysanthemic acid by a microbial esterase: cloning, nucleotide sequence, and overexpression of the esterase gene of Arthrobacter globiformis in Escherichia coli. Appl Environ Microbiol. 1995;61:3208–3215. doi: 10.1128/aem.61.9.3208-3215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oefner C, D’Arcy A, Daly J J, Gubernator K, Charnas R L, Heinze I, Hubschwerlen C, Winkler F K. Refined crystal structure of β-lactamase from Citrobacter freundii indicates a mechanism for β-lactam hydrolysis. Nature. 1990;343:284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- 17.Pahl A, Ühlein M, Bang H, Schlumbohm W, Keller U. Streptomycetes possess peptidyl-prolyl cis-trans isomerases that strongly resemble cyclophilins from eukaryotic organisms. Mol Microbiol. 1992;6:3551–3558. doi: 10.1111/j.1365-2958.1992.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 18.Pérez C, Juarèz E, Garcia-Castells E, Söberon G, Servín-González L. Cloning, characterization and expression in Streptomyces lividans 66 of an extracellular lipase-encoding gene from Streptomyces sp. M11. Gene. 1993;123:109–114. doi: 10.1016/0378-1119(93)90548-h. [DOI] [PubMed] [Google Scholar]
- 19.Raymer G, Willard J M A, Schottel J L. Cloning, sequencing, and regulation of expression of an extracellular esterase gene from the plant pathogen Streptomyces scabies. J Bacteriol. 1990;172:7020–7026. doi: 10.1128/jb.172.12.7020-7026.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Servín-González L, Castro C, Pérez C, Rubio M, Valdez F. bldA-dependent expression of the Streptomyces exfoliatus M11 lipase gene (lipA) is mediated by the product of a continuous gene, lipR, encoding a putative transcriptional activator. J Bacteriol. 1997;179:7816–7826. doi: 10.1128/jb.179.24.7816-7826.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tesch C, Nikoleit K, Gnau V, Götz F, Bormann C. Biochemical and molecular characterization of the extracellular esterase from Streptomyces diastatochromogenes. J Bacteriol. 1996;178:1858–1865. doi: 10.1128/jb.178.7.1858-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Y, Schottel J L, Derewenda U, Swenson L, Patkar S, Derewenda Z S. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat Struct Biol. 1995;2:218–223. doi: 10.1038/nsb0395-218. [DOI] [PubMed] [Google Scholar]