Abstract
The cell-associated β-d-fructosyltransferase of Streptococcus salivarius, which is devoid of the cell wall anchoring motif, LPXTG, is released on exposure to its substrate, sucrose. Deletions within the C terminus of the enzyme implicated both the hydrophobic and the proline-glycine-serine-threonine-rich wall-associated domain in stabilizing the enzyme on the cell surface.
The β-d-fructosyltransferases (Ftf’s) of oral streptococci and the levansucrases (SacB’s) of bacilli form a catalytically distinct family of proteins which polymerize the fructose moiety of sucrose into extracellular fructans (4, 6, 13, 23). Unlike the bacilli and mutans streptococci, which secrete their enzymes directly into the culture fluid (2, 3), the Ftf of Streptococcus salivarius is initially cell associated (14). The release of the S. salivarius Ftf from the surface of the cell occurs upon exposure to its substrate, sucrose (18). The initial cell surface binding and subsequent substrate-induced secretion appear to be unique.
It should be noted that since determining the sequence of the Ftf of S. salivarius (20), we have found that the signal sequence is cleaved at TQVKA⩵↓DQVTE to form the mature protein, not at the computer-predicted site, TLAFL⩵↓GATQV, previously published. This suggests that the predicted start site for translation is at Met-31, not at Met-52. As a result, the numbering of the amino acids has been modified by subtracting 52 from that previously published, giving rise to a predicted length of 917 amino acids for the cell-associated Ftf.
Previous deletion studies have shown that the C-terminal region of the Ftf directs surface attachment (20). This region displays high homology with the C termini of other gram-positive surface-bound polypeptides, such as the M protein of Streptococcus pyogenes (11, 19). In these proteins, the C terminus contains a cell wall sorting signal which consists of a consensus pentapeptide motif (LPXTG) followed by a C-terminal hydrophobic domain, ending with a positively charged tail (8). Secretion of these proteins is believed to be hindered by the presence of the charged tail, which allows the LPXTG consensus to be maintained in a position where it is proteolytically cleaved. In the case of Staphylococcus aureus, cleavage between the threonine and glycine results in cross bridge formation between the threonine and pentaglycine of the peptidoglycan (22). Such a consensus pentapeptide is absent in the Ftf of S. salivarius, raising the question of how the Ftf remains attached to the cell surface.
Besides the possible involvement of the hydrophobic domain in retention, another region common to surface-bound proteins lies directly N-terminal to the sorting signal. This so-called wall-associated domain is defined as a region spanning 50 to 125 residues with contents of proline-glycine and threonine-serine residues ranging from 15 to 32 and from 13 to 38%, respectively (8). The wall-associated domain of the Ftf of S. salivarius ATCC 25975 that directly precedes the hydrophobic C-terminal domain contains an extended proline-glycine-threonine-serine-rich domain spanning 178 amino acids from Pro-705 to Ser-882, with proline-glycine and threonine-serine contents of 17 and 22%, respectively. The wall-associated domain is separated from the catalytic domain of the Ftf (defined as that region between Gln-198 and Asp-631 possessing high homology with the SacB’s of bacilli [20]) by a spacer region of 73 amino acids (Fig. 1).
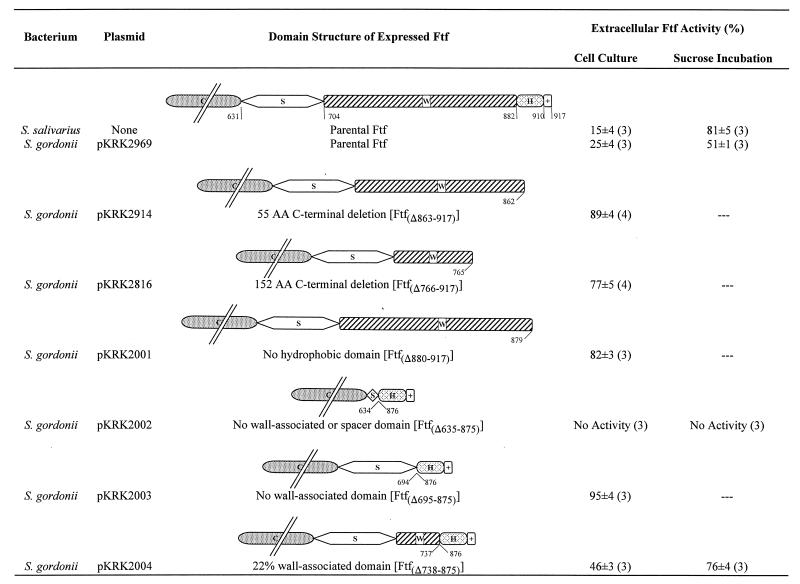
FIG. 1.
Comparison of the generalized domain structure of parental and mutated Ftf and their cellular location prior to and following incubation of washed cells with sucrose. C, catalytic domain; S, 73-amino-acid spacer; W, wall-associated domain; H, hydrophobic domain; +, positively charged amino acid C terminus. Numbers refer to the first or last amino acid present within a given or truncated domain.
In order to distinguish the roles of the hydrophobic and wall-associated domains in the attachment of the Ftf of S. salivarius ATCC 25975 to the surface of the cell, regions of the ftf gene coding for these domains were deleted and the recombinant proteins were expressed in the heterologous host Streptococcus gordonii LGR2 (20). S. gordonii LGR2, which does not produce an Ftf, was used as a model system because S. salivarius ATCC 25975 is refractory to stable transformation by electroporation in our hands (20). The phagemids and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial phagemids and plasmids
| Plasmid | Descriptiona | Host organism(s)b | Source or reference |
|---|---|---|---|
| pIBI31 | Apr (E) | E. coli | 1B1 Corp. |
| pVA838 | Emr (E,S); Cmr (E) | E. coli, S. gordonii | 5 |
| pKRK1969 | Apr (E); Ftf+ | E. coli | 20 |
| pKRK1001 | Apr (E); pKRK1969 with a site-directed mutation, coding for S880STOP | E. coli | This study |
| pKRK1002 | Apr (E); pKRK1969 with site-directed mutations, coding for K636P, S875R, and S877P | E. coli | This study |
| pKRK1003 | Apr (E); pKRK1969 with site-directed mutations, coding for G696P, S875R, and S877P | E. coli | This study |
| pKRK1004 | Apr (E); pKRK1969 with site-directed mutations, coding for K739P, S875R, and S877P | E. coli | This study |
| pKRK1005 | Apr (E); pKRK1002 with the ftf region coding for Asp-635 to Ser-875 deleted | E. coli | This study |
| pKRK1006 | Apr (E); pKRK1003 with the ftf region coding for Asp-695 to Ser-875 deleted | E. coli | This study |
| pKRK1007 | Apr (E); pKRK1004 with the ftf region coding for Asp-738 to Ser-875 deleted | E. coli | This study |
| pKRK2969 | Emr (E,S); 3.29 kbp EcoRI/SphI of pKRK1969 in 7.0 kbp EcoRI/SphI of pVA838 | E. coli, S. gordonii | 20 |
| pKRK2914 | Emr (E,S); Ftf(Δ863–917) in pVA838 | E. coli, S. gordonii | 20 |
| pKRK2816 | Emr (E,S); Ftf(Δ766–917) in pVA838 | E. coli, S. gordonii | 20 |
| pKRK2001 | Emr (E,S); 3.29 kbp EcoRI/SphI of pKRK1001 in 7.0 kbp EcoRI/SphI of pVA838 | E. coli, S. gordonii | This study |
| pKRK2002 | Emr (E,S); 3.29 kbp EcoRI/SphI of pKRK1005 in 7.0 kbp EcoRI/SphI of pVA838 | E. coli, S. gordonii | This study |
| pKRK2003 | Emr (E,S); 3.29 kbp EcoRI/SphI of pKRK1006 in 7.0 kbp EcoRI/SphI of pVA838 | E. coli, S. gordonii | This study |
| pKRK2004 | Emr (E,S); 3.29 kbp EcoRI/SphI of pKRK1007 in 7.0 kbp EcoRI/SphI of pVA838 | E. coli, S. gordonii | This study |
Apr, ampicillin resistance; Emr, erythromycin resistance; Cmr, chloramphenicol resistance. Letters in parentheses indicate whether the resistance genes function in E. coli (E) or S. gordonii (S).
Plasmids and phagemids were expressed in E. coli NM522 (9) or S. gordonii LGR2 (24) as previously described (20). E. coli was grown in Luria-Bertani medium at 37°C, while cultures of S. gordonii were grown at 37°C without aeration in Todd-Hewitt broth supplemented with 1% inactivated horse serum and 0.6% yeast extract. For growth on solid medium, S. gordonii was cultured at 37°C on brain heart infusion medium containing 2% raffinose for 1 to 3 days. Erythromycin (40 μg ml−1) was added where appropriate.
Effect of proteinase inhibitors on the release of Ftf from S. salivarius.
Surface protein-releasing enzyme activity has been characterized in some pathogenic streptococci (16). Other studies have shown that oral streptococci release surface proteins, particularly antigens such as antigen A and protein P1, by the action of endogenous proteinases (7, 17). The implication of these studies is that these streptococci shed antibody-antigen complexes as a means of evading the host immune system (15).
When washed cells of S. salivarius were incubated for 5 min at 37°C with 10 mM sucrose in the presence of 10 mM NaF and 50 μg of chloramphenicol ml−1 to inhibit glycolysis and de novo protein synthesis, respectively (18), phenylmethylsulfonyl fluoride (35 μg ml−1) in conjunction with either aprotinin (2 μg ml−1), chymostatin (100 μg ml−1), leupeptin (500 ng ml−1), trypsin inhibitor (100 μg ml−1), EDTA (500 ng ml−1), benzamide (100 μg ml−1), or iodoacetamide (100 μg ml−1) failed to prevent the release of cell-associated Ftf (data not shown). These results suggested that either the release of the enzyme from the surface of S. salivarius was independent of a proteolytic event or a novel proteinase not affected by the inhibitors was responsible for the secretion process.
Cellular location of mutated Ftf in S. gordonii.
Site-directed mutagenesis of the ftf gene made use of the previously described mutagenic oligosaccharides in conjunction with the T7 modification of the Transformer Site-Directed Mutagenesis Kit supplied by Clontech Laboratories (21). This modified procedure allowed the construction of an ftf allele that expressed a truncated Ftf, Ftf with amino acids 880 through 917 deleted [Ftf(Δ880–917)], which was devoid of its hydrophobic domain and its positively charged C terminus (Fig. 1). Site-directed mutagenesis was also used to introduce BamHI sites into ftf genes such that in-frame deletions could be constructed (21). These in-frame deletions expressed Ftf devoid of portions of the proline-glycine-threonine-serine-rich wall-associated domain while maintaining their hydrophobic tails and positively charged C termini intact (Fig. 1). All mutated forms of the enzyme expressed in Escherichia coli retained the ability to hydrolyze sucrose and to form fructan except for Ftf(Δ635–875), expressed by pKRK1005. This inactive form of the enzyme was devoid of its entire wall-associated domain as well as 96% of the 73-amino-acid C-terminal spacer region linking it to the catalytic domain but retained its C-terminal hydrophobic domain with its positively charged C terminus (Fig. 1). The direct juxtaposition of the hydrophobic domain and the catalytic domain of the Ftf may have destabilized the tertiary structure of the enzyme. This hypothesis was supported by a previous observation that an altered Ftf truncated at Ser-609, and thus having 22 amino acids deleted from the C-terminal region of the ‘catalytic’ domain, was also inactive (20). These two observations suggest that the tertiary structure formed by the C-terminal amino acids of the catalytic domain of the Ftf is critical for the maintenance of catalytic function.
To study the effect of deletions of various C-terminal domains on the localization of the Ftf in a gram-positive streptococcus, the mutated ftf genes were cloned into the E. coli-Streptococcus shuttle vector pVA838 and transformed into S. gordonii. Cells harvested at various stages throughout the growth cycle were used to determine the amount of Ftf bound to the cell, as well as the amount secreted into the culture medium. Where deemed appropriate, the amount of Ftf activity released following incubation of washed cells with sucrose was also measured (18). Irrespective of the stage of growth, the relative percentage of each mutated Ftf present on the surface of the cell, as well as that subsequently released from washed cells in the presence of sucrose, was not significantly different from that measured in late-exponential phase (data not shown).
Deletion of the C-terminal hydrophobic domain resulted in the secretion of Ftf(Δ880–917) by the heterologous host, S. gordonii (Fig. 1). This result was in keeping with the data previously obtained with truncated Ftf’s expressed by plasmids pKRK2914 and pKRK2816 in S. gordonii that carried 3′ deletions of the ftf gene constructed by random exonuclease III digestion (20). However, in these two instances, portions of the C-terminal wall-associated domain of the Ftf had also been removed along with the hydrophobic domain (Fig. 1). In order to determine whether there was a specific role for the wall-associated domain in surface retention of Ftf, Ftf(Δ695–875), devoid of its wall-associated domain but retaining its hydrophobic C terminus intact, was expressed by pKRK2003 in S. gordonii. This mutated Ftf was secreted, indicating that the wall-associated domain was essential for the stable binding of Ftf to the surface of S. gordonii irrespective of the presence of the hydrophobic region (Fig. 1). Inclusion of 22% of the wall-associated domain together with the C-terminal hydrophobic domain resulted in 54% of the Ftf remaining attached to the surface of S. gordonii (Fig. 1). This result further supported the hypothesis that the wall-associated domain was as important as the hydrophobic domain in stabilizing the Ftf on the surface of the cell. The cell-associated Ftf, Ftf(Δ738–875), possessing only 22% of the wall-associated domain, was released from the surface of S. gordonii by its substrate, sucrose, by an increased (33%) amount compared with the intact parental enzyme (Fig. 1).
Conclusions.
The release of Ftf from the surface of S. salivarius was not inhibited by any of the proteinase inhibitors tested. It is possible that the release of the Ftf from the cell surface is not a proteolytic event but is caused by “tearing” of the enzyme from the surface by large fructan complexes. Alternatively, the enzyme may be autoproteolytic in the presence of its substrate, sucrose, and may be capable of self-cleavage from the surface of the cell.
The absence of the carboxy-terminal pentapeptide anchoring signal (LPXTG) found in other gram-positive surface proteins implicated the wall-spanning region and the hydrophobic domain in the attachment of the Ftf of S. salivarius to the surface of the cell (20). The results of this study confirmed the well-documented role of the hydrophobic domain in anchoring proteins to the plasma membranes of gram-positive bacteria (10). This study also supported the hypothesis that the wall-associated domain was just as essential as the hydrophobic C-terminal domain in stabilizing surface attachment (1, 20). The presence of an extended wall-associated domain possessing turn-promoting proline-glycine residues could theoretically allow the C-terminal wall-associated region of the Ftf to span the cell wall by randomly intercalating throughout the peptidoglycan-carbohydrate-teichoic acid matrix. The high serine-threonine content could further stabilize this association by allowing hydrogen bonding to these constituents of the cell wall. Whether this is the case or not, it is clear from the results of this study that both the C-terminal hydrophobic domain and the extended wall-associated domain of the Ftf of S. salivarius are required for stable attachment of the enzyme to a gram-positive cell surface.
Acknowledgments
This work was supported by a project grant awarded by the Australian National Health and Medical Research Council.
REFERENCES
- 1.Burne R A, Penders J E C. Characterization of the Streptococcus mutans GS5 fruA gene encoding exo-beta-d-fructosidase. Infect Immun. 1992;60:4621–4632. doi: 10.1128/iai.60.11.4621-4632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlsson J. A levansucrase from Streptococcus mutans. Caries Res. 1970;4:97–113. doi: 10.1159/000259632. [DOI] [PubMed] [Google Scholar]
- 3.Chambert R, Petit-Galtron M-F. Hyperproduction of exocellular levansucrase by Bacillus subtilis: examination of the phenotype of a sac-Uh strain. J Gen Microbiol. 1984;130:3143–3152. doi: 10.1099/00221287-130-12-3143. [DOI] [PubMed] [Google Scholar]
- 4.Chambert R, Petit-Galtron M-F. Polymerase and hydrolase activities of Bacillus subtilis levansucrase can be modulated by site-directed mutagenesis. Biochem J. 1991;279:35–41. doi: 10.1042/bj2790035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell D B, Macrina F L, Tobian J A, Jones K R, Evans R P. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982;19:345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 6.Ebusi S, Kato K, Kotani S, Misaki A. Structural differences in fructans elaborated by Streptococcus mutans and Streptococcus salivarius. J Biochem (Tokyo) 1975;78:879–887. doi: 10.1093/oxfordjournals.jbchem.a130993. [DOI] [PubMed] [Google Scholar]
- 7.Ferretti J J, Russell R R B, Dao M L. Sequence analysis of the wall-associated protein precursor of Streptococcus mutans antigen A. Mol Microbiol. 1989;3:469–478. doi: 10.1111/j.1365-2958.1989.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 8.Fischetti V A, Pancholi V, Schneewind O. Common characteristics of the surface proteins from gram-positive cocci. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1991. pp. 290–294. [Google Scholar]
- 9.Gough J A, Murray N E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- 10.Hansson M, Ståhl S, Nguyen T N, Bächi T, Robert A, Binz H, Sjölander A, Uhlén M. Expression of recombinant proteins on the surface of the coagulase-negative bacterium Staphylococcus xylosus. J Bacteriol. 1992;174:4239–4245. doi: 10.1128/jb.174.13.4239-4245.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingshead S K, Fischetti V A, Scott J R. Complete nucleotide sequence of type 6 M protein of the group A streptococci: repetitive structure and membrane anchor. J Biol Chem. 1986;261:1677–1686. [PubMed] [Google Scholar]
- 12.Jacques N A. Inhibition of the expression of cell-associated fructosyltransferase in Streptococcus salivarius by octyl β-d-glucopyranoside. J Gen Microbiol. 1985;131:3243–3250. doi: 10.1099/00221287-131-12-3243. [DOI] [PubMed] [Google Scholar]
- 13.Jacques N A. The fructosyltransferase of Streptococcus salivarius ATCC 25975. New Phytol. 1993;123:429–435. doi: 10.1111/j.1469-8137.1993.tb03754.x. [DOI] [PubMed] [Google Scholar]
- 14.Jacques N A, Wittenberger C L. Inactivation of cell-associated fructosyltransferase in Streptococcus salivarius. J Bacteriol. 1981;148:912–918. doi: 10.1128/jb.148.3.912-918.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkinson H F. Anchorage and release of Gram-positive bacterial cell-surface polypeptides. Trends Microbiol. 1995;3:333–335. doi: 10.1016/s0966-842x(00)88969-6. [DOI] [PubMed] [Google Scholar]
- 16.Lee S F. Identification and characterization of a surface protein-releasing activity in Streptococcus mutans and other pathogenic streptococci. Infect Immun. 1992;60:4032–4039. doi: 10.1128/iai.60.10.4032-4039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S F. Active release of bound antibody by Streptococcus mutans. Infect Immun. 1995;63:1940–1946. doi: 10.1128/iai.63.5.1940-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milward C P, Jacques N A. Secretion of fructosyltransferase by Streptococcus salivarius involves the sucrose-dependent release of the cell-bound form. J Gen Microbiol. 1990;136:165–170. doi: 10.1099/00221287-136-1-165. [DOI] [PubMed] [Google Scholar]
- 19.Mouw A R, Beachy E H, Burdett V. Molecular evolution of streptococcal M protein: cloning and nucleotide sequence of type 24 M protein gene and relation to other genes of Streptococcus pyogenes. J Bacteriol. 1988;170:676–684. doi: 10.1128/jb.170.2.676-684.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathsam C, Giffard G M, Jacques N A. The cell-bound fructosyltransferase of Streptococcus salivarius: the carboxyl terminus specifies attachment in a Streptococcus gordonii model system. J Bacteriol. 1993;175:4520–4527. doi: 10.1128/jb.175.14.4520-4527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathsam C, Jacques N A. Development of a technique for multiple site-directed mutagenesis of the ftf gene of Streptococcus salivarius containing palindromic sequences. FEMS Microbiol Lett. 1997;153:447–453. doi: 10.1111/j.1574-6968.1997.tb12609.x. [DOI] [PubMed] [Google Scholar]
- 22.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–105. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 23.Shiroza T, Kuramitsu H K. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J Bacteriol. 1988;170:810–816. doi: 10.1128/jb.170.2.810-816.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyatt J E, Willcox M D P, Russell R R B, Handley P S. Fibrillar strains of Streptococcus sanguis biotype 1 carry a surface protein which cross-reacts with antigen B from Streptococcus mutans Ingbritt. Oral Microbiol Immunol. 1988;3:162–168. doi: 10.1111/j.1399-302x.1988.tb00003.x. [DOI] [PubMed] [Google Scholar]