Abstract
In Saccharomyces cerevisiae, an open reading frame, YOL061w, encodes a polypeptide with sequence similarity to the four known 5-phosphoribosyl-1(α)-pyrophosphate synthetase (PRS) genes since it contains a divalent cation binding site and a phosphoribosyl pyrophosphate binding site. We regard YOL061w as the fifth member of the PRS gene family, PRS5. Loss of Prs5p has a significant impact on PRS enzyme activity, causing it to be reduced by 84%. On the other hand, Δprs5 strains are not affected in growth or in the size of their nucleotide pools. However, simultaneous deletion of PRS1 and PRS5 or PRS3 and PRS5 rendered the strains inviable, which implies that PRS5 plays an important role in the maintenance of PRS function in S. cerevisiae.
The enzyme 5-phosphoribosyl-1(α)-pyrophosphate synthetase (PRS; ATP:d-ribose-5-pyrophosphotransferase; EC 2.7.6.1) catalyzes the biosynthesis of phosphoribosyl pyrophosphate (PRPP) from ribose-5-phosphate and ATP (9). PRPP is required for the production of purine, pyrimidine, and pyridine nucleotides and the amino acids histidine and tryptophan (13, 15). In Saccharomyces cerevisiae, there are at least four genes capable of encoding PRS (3). The PRS2-, PRS3-, and PRS4-predicted polypeptides are 318 to 320 amino acids long, whereas the PRS1-predicted polypeptide sequence is longer and more divergent since it contains an in-frame insertion of 105 amino acids bearing no similarity to any PRS product or any other known gene product. This insertion, which is neither an intron nor processed by protein splicing, has been named nonhomologous region 1-1 (NHR1-1) (3, 4). The contributions of the PRS gene products to the cell’s well-being do not appear to be equal, but none of the genes per se is essential. Measurements of growth rates and enzyme activity suggested that Prs1p might well be the key member encoded by the PRS gene family (4).
The predicted polypeptide of YOL061w discovered on chromosome XV in the course of the European Yeast Genome Sequencing Project (10) is 496 amino acids long and contains the characteristic motifs of PRS enzymes, the divalent cation binding site (DCbs) and the PRPP binding site (PRPPbs) (2, 7), suggesting that it may be encoded by the fifth member of the yeast PRS gene family. This open reading frame (ORF) encodes two potential NHR regions, 116 and 70 amino acids long, which in analogy to Prs1p have been designated NHR5-1 and NHR5-2. NHR5-1 lies N-terminal to the DCbs, and NHR5-2 is located between the DCbs and the PRPPbs (Fig. 1A), the same relative position as that of NHR1-1. PRS5 is transcribed to give an mRNA of approximately 1.6 kb; this is in accordance with neither NHR5-1 nor NHR5-2 being spliced, which suggests, as is the case for NHR1-1, that they are not introns. Furthermore, Northern analysis showed that deletion of PRS5 had no effect on the transcriptional levels of the other four PRS genes and that PRS5 transcription was not affected by deletion of any of the other PRS genes.
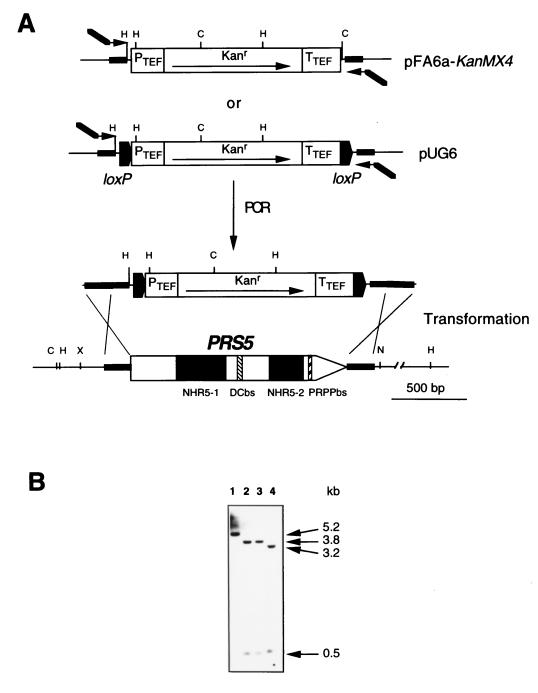
FIG. 1.
PRS5 disruption. (A) Schematic representation of the disruption cassettes and the PRS5 locus. Arrows at either end of the modules represent the oligonucleotides used for PCR, arrowheads correspond to the cassette amplification regions (black bars), and thick lines of the arrows represent the 40- to 41-bp extension used to target the PCR fragment to the PRS5 flanking sequences. These oligonucleotides were used to amplify the KanMX4 plasmid pFA6a-KanMX4 (16) or the loxP-KanMX-loxP plasmid pUG6 cassette (6, 12). Yeast wild-type strains were transformed with these DNA products (5). The Δprs5 strains were obtained by recombination (indicated by ×) between the DNA regions upstream and downstream of the PRS5 locus and their homologous sequences flanking the corresponding cassette. The positions of NHR5-1 and NHR5-2 as well as those of the DCbs and PRPPbs are indicated. (B) Southern blot of HindIII/ClaI-digested genomic DNA of the wild-type strain, YN94-1 (lane 1), the Δprs5::loxP-KanMX-loxP MATa (YN96-54) and MATα (YN96-55) strains (lanes 2 and 3, respectively), and the Δprs5::KanMX4 MATa strain (YN96-1) (lane 4) hybridized with an XbaI/NsiI fragment containing the PRS5 gene as shown in panel A. The wild-type strain gives a 5.2-kb signal corresponding to the HindIII genomic fragment containing the PRS5 gene. When hybridized with the same probe, YN96-54 (lane 2) and YN96-55 (lane 3) give rise to two signals: a 0.5-kb fragment corresponding to the region between the HindIII site upstream of PRS5 and the HindIII site in the promoter PTEF of the loxP-KanMX-loxP module and a 3.8-kb fragment corresponding to the DNA contained between this HindIII site and the HindIII site downstream of PRS5. DNA from YN96-1 (lane 4) gives the same 0.5-kb upstream fragment, but the downstream fragment is smaller, 3.2 kb, since this cassette has an additional ClaI site present in the 3′ end of the KanMX4. Restriction enzyme abbreviations: C, ClaI; H, HindIII; N, NsiI; X, XbaI. PTEF and TTEF are the promoter and terminator sequences, respectively, of the Ashbya gossypii TEF gene (16).
To investigate the role of Prs5p in vivo, we created a null mutant by targeted gene disruption. The entire PRS5 ORF (1,490 bp) was replaced by either the KanMX4 module (16) or its derivative, the loxP-KanMX-loxP cassette (6). Plasmid pUG6, containing the loxP-KanMX-loxP cassette and pFA6-KanMX4, shares the sequence of the multiple cloning site, allowing the same oligonucleotides to be used for PCR amplification of both cassettes. Two DNA fragments of 1,542 and 1,691 bp comprising the KanMX4 module or the loxP-KanMX-loxP cassette flanked by 41 and 40 bp of DNA homologous to the regions immediately upstream and downstream of the PRS5 coding sequence were obtained by PCR by using the corresponding plasmids as templates with the primers PRS5-SFH1 (5′-CTTTGTTGGAGGTTGCTACGAGGCTAGGAACGCAG TC TGGCAGC TGAAGC T TCG TACGC TG - 3′) and PRS5-SFH2 (5′-CCCTATTTTTATCAATAAAAAAATGAACACATCAATGCCAATAGGCCACTAGTGGATCTG-3′) (12) (Fig. 1A). The PCR amplification products were used to transform (5) the yeast strains YN94-1 (MATa ade2-1 his3-11 leu2-3 trp1-1 ura3-1 can1-100) and YN94-2 (MATα; isogenic to YN94-1), and the resulting transformants were selected on complete medium containing 200 mg of Geneticin G418 (Boehringer, Mannheim, Germany) per liter. The correct integration of the Kanr cassette was verified by PCR and Southern hybridization (Fig. 1B). Thus, we obtained four S. cerevisiae strains: two Δprs5::KanMX4 strains (YN96-1 MATa and YN96-2 MATα) and two Δprs5::loxP-KanMX-loxP strains (YN96-54 MATa and YN96-55 MATα). YN96-1 and YN96-2 were used for matings as described below, while YN96-54 and YN96-55 were transformed with plasmid pSH47, which carries on a URA3-based plasmid the Cre recombinase gene from bacteriophage P1 under the control of the inducible GAL1 promoter (6). When grown in galactose-containing medium, the Cre recombinase is induced and the KanMX module is excised by a recombination event between the two loxP sites, leaving behind a single loxP site at the PRS5 locus. The correct excision of the KanMX module was checked by PCR and Southern blotting. In this way, strains YN96-68 and YN96-69 (Δprs5::loxP) in both mating types were obtained. The pSH47 plasmid (URA3+) was removed from these strains by streaking the cells onto plates containing 5-fluoroorotic acid (5-FOA; Sigma-Aldrich, Poole, Dorset, United Kingdom), which counterselects URA3+ plasmids (1).
The creation of the PRS5 null mutant strains shows that disruption of PRS5 is not a lethal event. Furthermore, there is little or no effect on growth since the Δprs5 strains have doubling times of 1.9 to 2.0 h, virtually identical to that of the wild type, which has a doubling time of 1.7 to 1.9 h. PRS enzyme activity was measured in crude cell extracts prepared from mid-log-phase cultures of the Δprs5 strain (YN96-69) and assayed by thin-layer chromatography (4). The Δprs5 strain retained only 16% of the wild-type activity since it synthesizes 4.6 ± 0.3 nmol of PRPP min−1 mg of protein−1 (mean ± standard deviation), in contrast to the wild type, which produced 28 ± 4 nmol of PRPP min−1 mg−1.
By using high-performance liquid chromatography, we analyzed the effect of deleting each of the PRS genes on the nucleotide profile of the yeast cell. Total nucleotides were extracted from wild-type and Δprs::loxP strains after growth of the strains in complete medium to approximately mid-log phase (11). The extracts were resuspended in 150 μl of 7 mM KH2PO4 (pH 4.0). Fifty microliters of the resuspended extract was used to determine the nucleotide pools as described by Strauch et al. (14). Nucleotide standards (98% pure) were from Sigma-Aldrich.
The nucleotide content of YN96-66 (Δprs1::loxP) was drastically reduced in comparison to that of the wild type, and a deletion in PRS3 (YN96-67) also had a dramatic effect. Δprs2, Δprs4, and Δprs5 strains (YN97-7, YN97-6, and YN96-69, respectively) had profiles differing only slightly from that of the wild type (Table 1). It is unlikely that the reduction in the nucleotide content observed for the Δprs1 and Δprs3 strains (YN96-66 and YN96-67) was caused by degradation of nucleotides, since all peaks were reduced and there were no abnormally high nucleoside monophosphate peaks, as would be expected if nucleoside di- and triphosphates were degraded. This finding was confirmed by adding known amounts of ATP and GTP to extracts of the wild-type strain and observing that no significant degradation of them occurred as the result of the extraction procedure. These results are in agreement with previous observations indicating that Prs1p and Prs3p apparently make a more important contribution to the yeast metabolism than the other members of the family (4).
TABLE 1.
Nucleotide content of wild-type and Δprs strains
| Strain | % of nucleotides produceda,b
|
|||
|---|---|---|---|---|
| UXP | CXP | AXP | GXP | |
| YN94-1 (wild type) | 99 ± 0.8 | 100 ± 0.6 | 101 ± 1.5 | 99 ± 1.1 |
| YN96-66 (Δprs1::loxP)c | 25 ± 7.0 | 12 ± 0.5 | 45 ± 9.0 | 37 ± 6.0 |
| YN96-67 (Δprs3::loxP)c | 26 ± 4.0 | 31 ± 9.8 | 55 ± 4.0 | 38 ± 3.0 |
| YN97-7 (Δprs2::loxP)c | 72 ± 13 | 97 ± 1.5 | 91 ± 9.5 | 80 ± 8.2 |
| YN97-6 (Δprs4::loxP)c | 75 ± 15 | 102 ± 2.0 | 70 ± 15 | 86 ± 19 |
| YN96-69 (Δprs5::loxP) | 101 ± 15 | 80 ± 13 | 89 ± 12 | 96 ± 10 |
UXP, UMP + UDP + UTP; CXP, CDP + CTP (CMP is not detectable under the experimental conditions used); AXP, AMP + ADP + ATP; GXP, GMP + GDP + GTP.
Values are the percentages of nucleotides produced by each strain with respect to that of the wild type and represent the average ± standard deviation of at least three independent determinations.
These deletion mutant strains were created as described for Δprs5::loxP in the text by using appropriate primers for each gene.
To further characterize the possible role of Prs5p in the production of PRPP in yeast and to establish its relationship with products of the other members of the PRS gene family, we constructed strains disrupted in PRS5 and one of each of the other four PRS genes. Double-disrupted Δprs2 Δprs5 (YN97-89) and Δprs4 Δprs5 (YN97-90) strains were constructed by transforming Δprs5::loxP mutants with the appropriate disruption cassette in the same manner as described above. In spite of repeated attempts, it was not possible to obtain the double disruptants Δprs1 Δprs5 and Δprs3 Δprs5. To determine the reason for this failure, we constructed heterozygous diploids (PRS1/Δprs1::HIS3 Δprs5::KanMX4/PRS5 and PRS3/Δprs3::TRP1 Δprs5::KanMX4/PRS5) by crossing the corresponding haploid strains (YN94-5 [YN94-2 Δprs1::HIS3] × YN96-1 and YN94-9 [YN94-2 Δprs3::TRP1] × YN96-1). More than 130 tetrads were analyzed after sporulation of the diploids, but no viable HIS+G418r or TRP+G418r haploids were recovered. Microscopic examination of the spores corresponding to the double disruptants indicated that they had undergone germination but had not progressed beyond two or three cell divisions. This suggested that the double-mutant combinations Δprs1 Δprs5 and Δprs3 Δprs5 were lethal.
To confirm this result, a heterozygous diploid containing wild-type and deleted versions of PRS1 and PRS5 but with a copy of PRS1 in a URA3-based plasmid (pVT1) was constructed. A similar experiment was performed for PRS3 by using plasmid pVT3. As a result, viable Δprs1::HIS3 Δprs5::KanMX4 and Δprs3::TRP1 Δprs5::KanMX4 isolates were recovered, but they always contained the corresponding plasmid, pVT1 or pVT3. These strains were sensitive to media containing 5-FOA, indicating that any cells losing the plasmid were inviable (Fig. 2). Wild-type and single-deletion mutants carrying pVT1 and pVT3 gave rise to colonies on 5-FOA-containing media. These data confirm that the PRS5 null mutant is synthetically lethal in combination with either Δprs1 or Δprs3. Therefore, while PRS5 is not an essential gene, the loss of Prs5p cannot be tolerated together with the loss of either Prs1p or Prs3p, indicating that the maintenance of the cell’s requirement for PRPP is a complicated issue that could involve interaction between Prs5p and Prs1p or Prs3p. Two-hybrid experiments to investigate this further are under way.
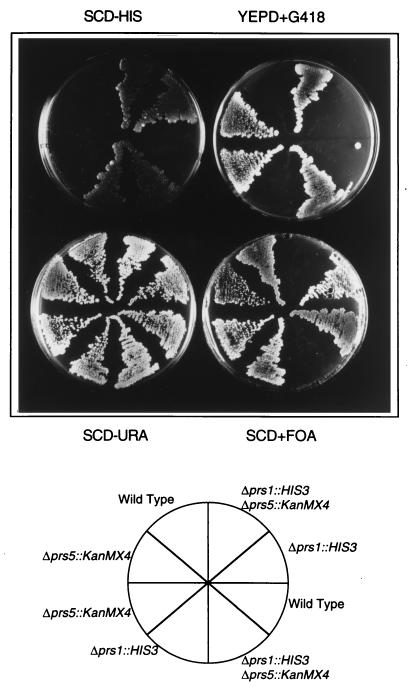
FIG. 2.
Δprs5 is lethal in combination with Δprs1. PRS1/Δprs1::HIS3 Δprs5::KanMX4/PRS5 diploids carrying pVT1 were sporulated, and tetrads were dissected. Two tetrads were streaked on media for selection of single disruptants (synthetic complete dextrose minus histidine [SCD-HIS] and yeast extract-peptone-dextrose plus Geneticin G418 [YEPD+G418]) and for the maintenance (SCD-URA) or loss (SCD+FOA) of plasmid pVT1. Yeast media were prepared as described by Kaiser et al. (8). Similar results were obtained with the combination of Δprs3 and Δprs5.
Our analysis of the PRS gene family in S. cerevisiae has shown that Prs1p and Prs3p may play a more important role in PRPP biosynthesis than the other members of the family (4). In this study, we have shown that either Prs1p or Prs3p is essential in the absence of Prs5p. To be able to understand the level of functional interaction among the PRS gene products, it will be necessary to analyze the phenotypes associated with strains bearing combinations of multiple disruptions of the PRS genes and to determine to what extent PRS activity is influenced by each member of this gene family.
Acknowledgments
The work was supported by BBSRC and a fellowship from the Spanish Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) to Y.H.
We thank B. M. Pearson for performing the tetrad dissection, B. Hove-Jensen (University of Copenhagen) for helping with the enzyme assay, and A. T. Carter and M. Cleaton-Roberts for initial help with the nucleotide extraction procedure. We thank J. H. Hegemann, University of Düsseldorf, for providing us with the plasmids for the Cre-loxP system. We are grateful to Mervyn Bibb (John Innes Centre) for valuable discussions.
REFERENCES
- 1.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 2.Bower S G, Harlow K W, Switzer R L, Hove-Jensen B. Characterization of the Escherichia coli prsA1-encoded mutant phosphoribosylpyrophosphate synthetase identifies a divalent cation-nucleotide binding site. J Biol Chem. 1989;264:10287–10291. [PubMed] [Google Scholar]
- 3.Carter A T, Narbad A, Pearson B M, Beck K F, Logghe M, Contreras R, Schweizer M. Phosphosribosylpyrophosphate synthetase: a new gene family in Saccharomyces cerevisiae. Yeast. 1994;10:1031–1044. doi: 10.1002/yea.320100805. [DOI] [PubMed] [Google Scholar]
- 4.Carter A T, Beiche F, Hove-Jensen B, Narbad A, Barker P J, Schweizer L M, Schweizer M. PRS1 is a key member of the gene family encoding phosphoribosylpyrophosphate synthetase in Saccharomyces cerevisiae. Mol Gen Genet. 1997;254:148–156. doi: 10.1007/s004380050402. [DOI] [PubMed] [Google Scholar]
- 5.Gietz R D, Woods R A. High efficiency transformation with lithium acetate. In: Johnson J R, editor. Molecular genetics of yeast: a practical approach. Oxford, United Kingdom: IRL Press; 1994. pp. 121–131. [Google Scholar]
- 6.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hove-Jensen B, Harlow K W, King C J, Switzer R L. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem. 1986;261:6765–6771. [PubMed] [Google Scholar]
- 8.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- 9.Khorana H G, Fernandes J F, Kornberg A. Pyrophosphorylation of ribose-5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J Biol Chem. 1958;230:941–948. [PubMed] [Google Scholar]
- 10.Mannhaupt G, Vetter I, Schwarzlose C, Mitzel S, Feldmann H. Analysis of a 26 kb region of the left arm of yeast chromosome XV. Yeast. 1996;12:67–76. doi: 10.1002/(sici)1097-0061(199601)12:1<67::aid-yea884>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Ochi K. Changes in nucleotide pools during sporulation in Streptomyces griseus in submerged culture. J Gen Microbiol. 1987;133:2787–2795. [Google Scholar]
- 12.Pearson B M, Hernando Y, Schweizer M. Construction of PCR-ligated long flanking homology cassettes for use in the functional analysis of six unknown open reading frames from the left and right arms of Saccharomyces cerevisiae chromosome XV. Yeast. 1998;14:391–399. doi: 10.1002/(SICI)1097-0061(19980315)14:4<391::AID-YEA235>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 13.Sakakibara Y. dnaR function of the prs gene of Escherichia coli in initiation of chromosome replication. J Mol Biol. 1992;226:989–996. doi: 10.1016/0022-2836(92)91047-s. [DOI] [PubMed] [Google Scholar]
- 14.Strauch E, Takano E, Baylis H A, Bibb M J. The stringent response in Streptomyces coelicolor A3(2) Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 15.Switzer R L. Regulation and mechanism of phosphoribosylpyrophosphate synthetase. J Biol Chem. 1969;244:2854–2863. [PubMed] [Google Scholar]
- 16.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]