Abstract
Epidermal growth factor receptor (EGFR) is expressed in various types of cancer and is associated with the malignant biological behavior of cancer cells. In the present study, the expression of EGFR in hepatocellular carcinoma (HCC) tissues and liver cancer cells was detected by immunohistochemical staining, western blotting and immunofluorescence. Furthermore, a lentivirus was transduced into HepG2 liver cancer cells to knock down EGFR expression. Cell proliferation and migration, and the expression levels of epithelial-mesenchymal transition (EMT) markers were assessed by EdU staining, Cell Counting Kit-8, colony formation, wound healing and Transwell assays, and western blotting. The results revealed that EGF/EGFR can mediate EMT through the Akt/glycogen synthase kinase-3β (GSK-3β)/Snail signaling pathway to promote HepG2 cell proliferation and migration. Inhibition of the activation of the EGFR signaling pathway can help to partially reverse the EMT phenotype, and inhibit the proliferation and migration of HepG2 cells. In conclusion, the EGFR/Akt/GSK-3β/Snail signaling pathway serves an important role in HCC progression, and inhibition of the activation of the EGFR signaling pathway may be a valuable strategy in liver cancer treatment.
Keywords: hepatocellular carcinoma, epidermal growth factor receptor, epithelial-mesenchymal transition, proliferation, migration
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer worldwide (1–3), which is prone to metastasis, recurrence and poor prognosis (4–6). Since HCC is an insidious tumor, most patients are diagnosed with liver cancer at an advanced stage and only receive systemic therapies (7). A variety of targeted drugs have been studied and used in clinical trials, such as sorafenib, lenvatinib and regorafenib (8–10). However, resistance to these drugs can emerge after several months of treatment (11). Therefore, it is crucial to investigate the molecular mechanisms underlying HCC occurrence and development for the treatment of HCC. Overexpression and abnormal activation of epidermal growth factor receptor (EGFR) is considered an important factor leading to tumorigenesis and development of cancer, such as non-small cell lung cancer, breast cancer and HCC (12–14). Downstream signaling pathways of EGFR can promote biological effects, such as proliferation, migration, angiogenesis and inhibition of apoptosis in tumor cells (15–17). Epithelial-mesenchymal transition (EMT) is an important step in the process of tumorigenesis and development (18,19), which involves multiple signaling pathways that regulate gene expression by modulating major transcription factors, thereby promoting cell invasion and metastasis (20–23). However, the molecular mechanism underlying EGFR-mediated EMT in HCC is currently unclear.
Glycogen synthase kinase-3β (GSK-3β) is a ubiquitously expressed serine/threonine protein kinase, that is involved in the regulation of various key cellular processes, including cell proliferation, cell survival and cell signaling (24). GSK-3β is a key downstream component of the PI3K/Akt pathway, and its activity can be inhibited by the Akt-mediated phosphorylation of GSK-3β at Ser9 (25). In addition, GSK-3β can be regulated by Wnt to participate in the EMT process (26). Snail is a zinc finger transcription factor that regulates cellular EMT process by inhibiting E-cadherin transcription (27). However, whether EGFR is involved in cell EMT via the Akt/GSK-3β/Snail pathway in HCC is currently unknown.
The present study aimed to explore the expression and function of EGFR in HCC and to analyze the molecular mechanism of EGFR-mediated EMT in HepG2 cells.
Materials and methods
Patients and HCC tissue specimens
A total of 40 patients who had received curative resection or biopsies for HCC between January 2021 and January 2023 at The First Affiliated Hospital of Bengbu Medical College (Bengbu, China) were enrolled in the present study. The First Affiliated Hospital of Bengbu Medical College is the teaching hospital of Anhui University of Science & Technology. The morphology of tissue samples from 40 patients was observed and analyzed by light microscopy, some of which did not meet the research criteria. For example, the adjacent tissue was not paracancerous, too little tissue was obtained or the tissue cells were not obvious in immunohistochemical staining. Therefore, 20 pairs of cancer tissues and adjacent tissues (2 cm away from cancerous tissues) were selected, and were stained by immunohistochemistry and analyzed. All clinical specimens were collected from patients after they gave written informed consent in accordance with a protocol approved by the Ethics Committee of Anhui University of Science & Technology (Huainan, China). Detailed clinicopathological characteristics of the patients are provided in Table SI. Tumors were staged according to the AJCC Cancer Staging Manual (8th edition) (28).
Immunohistochemistry
Clinical specimens were fixed with 4% paraformaldehyde (cat. no. BL539A; Biosharp Life Sciences) at room temperature for 24 h, embedded in paraffin and sectioned into 4-µm slices. Sections were deparaffinized in xylene three times (5 min each time) and were rehydrated in a descending ethanol series. After deparaffinization and hydrated, tissue sections were incubated in sodium citrate antigen retrieval solution (pH 6.0; cat. no. KGIHC001; Nanjing KeyGen Biotech Co., Ltd.) at 100°C for 30 min. After natural cooling, the slides were washed three times with PBS. Subsequently, 3% H2O2 was added to the sections and they were blocked with 10% goat serum (cat. no. SP-9000; Universal SP Kit; OriGene Technologies, Inc.) at room temperature for 10 min. The slides were then incubated with anti-EGFR (1:50; cat. no. 4267; Cell Signaling Technology, Inc.) and anti-phosphorylated (p)-EGFR (1:200; cat. no. 3777; Cell Signaling Technology, Inc.) at 4°C overnight to detect specific. Biotinylated goat anti-rabbit IgG (cat. no. SP-9000; 1:100; Universal SP Kit; OriGene Technologies, Inc.) was then added and incubated at room temperature for 10 min, followed by incubation with HRP-conjugated streptavidin at room temperature for 10 min. Diaminobenzidine chromogenic solution was added and incubated for 5 min at room temperature. The stained tissue sections were counterstained with hematoxylin, dehydrated in a graded ethanol series and permeabilized in xylene for 5 min. Finally, the slices were sealed and examined under a light microscope.
Cell culture
The HepG2 liver cancer cell line (HB-8065) was purchased from American Type Culture Collection, and was cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Biological Industries; Sartorius AG) at 37°C with 5% CO2 to maintain a constant cell growth environment. The Huh7 liver cancer cell line (BFN60800691) and HHL-5 normal liver cell line (BFN6072012687) were purchased from BLUEFBIO, and were cultured in DMEM (Biosharp Life Sciences) containing 10% fetal bovine serum at 37°C with 5% CO2 to maintain a constant cell growth environment. To ensure their identity and purity, the cells were identified using the short tandem repeat method.
Lentivirus transduction
To knockdown the target gene EGFR (NM_005228.3), a short hairpin (sh)EGFR lentivirus vector, which was constructed and validated by sequencing performed by Sangon Biotech Co., Ltd., was used. The shRNA sequences used in the present study are listed as follows: pLVE3753, 5′-CCGGCCTCCAGAGGATGTTCAATAACTCGAGTTATTGAACATCCTCTGGAGGTTTTTTG-3′; pLVE3754, 5′-CCGGGCTGGATGATAGACGCAGATACTCGAGTATCTGCGTCTATCATCCAGCTTTTTTG-3′; pLVE3755, 5′-CCGGGCCACAAAGCAGTGAATTTATCTCGAGATAAATTCACTGCTTTGTGGCTTTTTTG-3′; and negative control (NC) pLVT1, scrambled sequence: 5′-CCGGGTTCTCCGAACGTGTCACGTACTCGAGTACGTGACACGTTCGGAGAACTTTTTTG-3′. The shEGFR was synthesized and cloned into the pMAGIC1.1 vector (Addgene, Inc.) to construct the pMAGIC1.1-shEGFR plasmid. Subsequently, pMAGIC1.1-shEGFR (100 µg), pCMV-dR8.9 (65 µg) and pCMV–VSV-G (35 µg) (both Addgene, Inc.) were transfected into 293T cells (The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences) using CalPhos™ Mammalian Transfection Kit (cat. no. 631312; Clontech Laboratories, Inc.) for 4 h at 37°C, after which, the medium was replaced with fresh medium. After 72 h, the 293T cell supernatant was collected. Lentivirus particles were obtained by purification, and the virus titers were determined. HepG2 cells (5×104 cells/well) were seeded in 6-well plates. Based on a multiplicity of infection value of 20, the appropriate volume of the virus was added to the cell culture medium for infection after culturing for 24 h at 37°C. After 24 h infection at 37°C, the medium was replaced with fresh medium. The selection of stable cell lines was performed with puromycin (cat. no. HY-B1743A; MedChemExpress) at 2 µg/ml 48 h and 1 µg/ml puromycin was used for maintenance. Western blotting was used to screen the most effective cells for subsequent experiments.
Western blotting
Adherent HepG2 cells treated with or without 100 ng/ml EGF (cat. no. GMP-10605-HNAE; Sino Biological, Inc.) or 100 ng/ml EGF + 3 µM MK-2206 (cat. no. HY-108232; MedChemExpress) at 37°C for 24 h were washed with PBS and lysed with RIPA lysis buffer (Nanjing KeyGen Biotech Co., Ltd.) containing protease inhibitors. Subsequently, proteins (25 µg/lane), quantified using the BCA method, were separated by SDS-PAGE on 10% gels and were then transferred to PVDF membranes, before being blocked with 5% skim milk for 1 h at room temperature. The membranes were then incubated with the following primary antibodies at 4°C overnight to detect specific proteins: EGFR (cat. no. 4267), p-EGFR (Tyr1068) (cat. no. 3777), Akt (cat. no. 9272), GSK-3β (cat. no. 9315), p-GSK-3β (Ser9) (cat. no. 5558), E-cadherin (cat. no. 3195), Vimentin (cat. no. 5741), Snail (cat. no. 3879), β-actin (cat. no. 4970) (all 1:1,000; Cell Signaling Technology, Inc.), p-Akt (Ser473) (cat. no. 4060; 1:2,000; Cell Signaling Technology, Inc.), MMP2 (cat. no. ab92536), MMP9 (cat. no. ab76003) (both 1:500; Abcam). An HRP-conjugated anti-rabbit IgG secondary antibody (cat. no. 7074; 1:3,000; Cell Signaling Technology, Inc.) was then applied at room temperature for 1 h. An automatic gel imaging analysis system (JS-M6P; Shanghai Peiqing Science & Technology Co., Ltd.) was used to analyze the target proteins and the gray values of the bands were semi-quantified with ImageJ V1.8.0 software (National Institutes of Health).
Immunocytochemistry
HHL-5, HepG2 and Huh7 cells (2.5×104 cells/well) were seeded in 24-well plates containing cover slips and the cells were fixed with 4% paraformaldehyde for 10 min at room temperature after adherence. Endogenous peroxidase was blocked with 3% H2O2 for 15 min at room temperature and 10% goat serum (cat. no. SP-9000; Universal SP Kit; OriGene Technologies, Inc.) was applied for 10–15 min at room temperature. Subsequently, anti-EGFR (cat. no. 4267; 1:50; Cell Signaling Technology, Inc.) was added and incubated overnight at 4°C, after which, a biotinylated goat anti-rabbit IgG secondary antibody polymer (cat. no. SP-9000; 1:100; Universal SP Kit; OriGene Technologies, Inc.) was added and incubated at room temperature for 10–15 min. HRP-conjugated streptavidin working solution was then added and incubated at room temperature for 10–15 min. Finally, diaminobenzidine chromogenic solution was added and incubated at room temperature for 5 min, cells were counterstained with hematoxylin and were observed under a light microscope.
Indirect immunofluorescence
HHL-5, HepG2 and Huh7 cells (2.5×104 cells/well) were seeded in 24-well plates containing cover slips, and the cells were fixed with 4% paraformaldehyde for 10 min at room temperature after adherence. After cells were blocked with 10% goat serum (cat. no. SP-9000; Universal SP Kit; OriGene Technologies, Inc.) for 10–15 min at room temperature, the following primary antibodies were added and incubated overnight at 4°C: EGFR (cat. no. 4267; 1:50; Cell Signaling Technology, Inc.), E-cadherin (cat. no. 3195; 1:1,600; Cell Signaling Technology, Inc.) and Vimentin (cat. no. 5741; 1:200; Cell Signaling Technology, Inc.). Subsequently, an Alexa Fluor® 488-labeled secondary anti-rabbit IgG antibody (1:250; cat. no. 4412; Cell Signaling Technology, Inc.) was added and incubated at 37°C for 30 min. After DAPI counterstaining, the slides were sealed and observed under a fluorescence microscope, and images were captured.
EdU proliferation assay
HepG2 cells (5×104 cells/well) were seeded in 24-well plates and treated with or without 100 ng/ml EGF for 24 h at 37°C or 100 ng/ml EGF + 3 µM MK-2206 for 24 h at 37°C before being replaced with medium containing EdU (10 µM; EdU-594 cell proliferation detection kit; Beyotime Institute of Biotechnology). The medium was removed after continued incubation for 2 h, and the cells were fixed with 4% paraformaldehyde for 15 min at room temperature and washed three times with PBS. Triton X-100 was used for permeabilization for 15 min at room temperature, and the slides were washed three times with PBS. Click reaction solution was then added and incubated for 30 min at room temperature in the dark. Hoechst-33342 staining was performed for 10 min at room temperature. The cells were then observed under an inverted fluorescence microscope, images were captured and the cell proliferation rate was calculated according to the red fluorescence ratio.
Colony formation assay
HepG2 cells were seeded in 6-well plates (1,000 cells/well), treated with 100 ng/ml EGF for 2 weeks at 37°C and the medium was discarded after 2 weeks. After washing, the samples were fixed with 4% paraformaldehyde for 15 min at room temperature and further stained with 0.1% crystal violet for 10 min at room temperature. Cell proliferation was determined by visually counting the number of cell colonies in the sample. Each colony was 0.3–1 mm in size.
CCK-8 assay
HepG2 cells (4×103 cells/well) were inoculated into 96-well plates, and after treatment with 100 ng/ml EGF for 1, 2, 3 and 4 days at 37°C, 10 µl CCK-8 solution (Beyotime Institute of Biotechnology) was added and incubated at 37°C for 2 h. Absorbance was measured at 450 nm and cell viability was calculated using GraphPad Prism 8 software (Dotmatics).
Wound healing assay
HepG2 cells (2.5×105 cells/well) were seeded in a 12-well plate. After 24 h, cells grew to ~100% confluence and a 10-µl pipette tip was used to generate a scratch. After washing three times with PBS, the medium was replaced with serum-free medium containing 100 ng/ml EGF for 48 h at 37°C or 100 ng/ml EGF + 3 µM MK-2206 for 48 h at 37°C. Images were captured under a light microscope at 0 and 48 h after scratching. Wound healing rate (%) was calculated as follows: (0 h wound area −48 h wound area)/0 h wound area ×100.
Transwell migration assay
HepG2 cells were pretreated with 100 ng/ml EGF for 6 h at 37°C. A total of 3×105 cells/ml resuspended in 100 µl serum-free RPMI-1640 medium were added to the upper chamber of a Transwell system (pore size, 8 µm), and 600 µl RPMI-1640 medium containing 10% fetal bovine serum was added to the lower chamber. After 48 h at 37°C, non-migrating cells in the upper chamber were wiped away and the cells were fixed with 4% paraformaldehyde for 10 min at room temperature, stained with 0.5% crystal violet for 10 min at room temperature and observed under a light microscope, and images were captured. The number of migratory cells was recorded.
Statistical analysis
All experiments were repeated at least three times and measured in three independent experiments. Data are presented as the mean ± SD and GraphPad Prism 8 was used for statistical analysis. Differences between two groups were compared using a paired Student's t-test, and differences between three or more groups were compared using one-way ANOVA followed by Bonferroni's test. P<0.05 was considered to indicate a statistically significant difference.
Results
EGFR is highly expressed in HCC tissues and cells
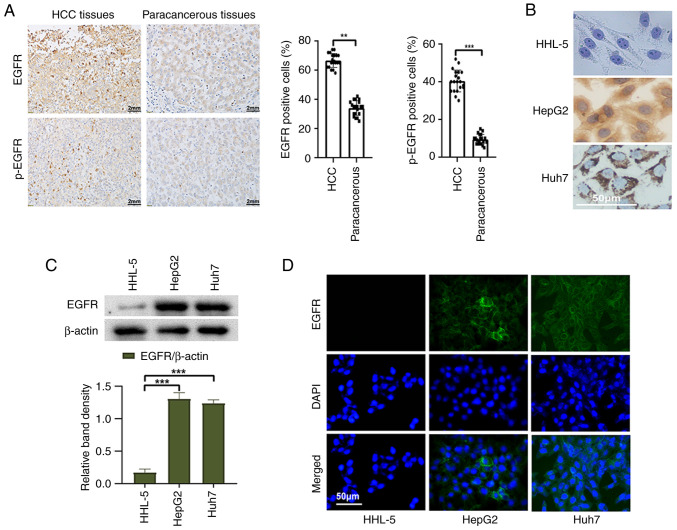
To detect the expression of EGFR in HCC tissues and cells, a series of experiments were conducted. First, 20 HCC tissues and corresponding paracancerous tissues were selected for immunohistochemical staining. The positive rates of EGFR and p-EGFR in HCC tissues were significantly higher than those in paracancerous tissues (Fig. 1A). Furthermore, the expression of EGFR in liver cancer cells and normal liver cells was examined using western blot analysis. The results revealed a significantly higher expression of EGFR in liver cancer cells compared with those in normal liver cells (Fig. 1C). Immunocytochemistry and immunofluorescence analyses provided additional confirmation of these results (Fig. 1B and D). These results serve as evidence for the high expression of EGFR in liver cancer cells and tissues.
Figure 1.
Expression of EGFR in HCC tissues and liver cancer cells. (A) Representative images of immunohistochemical staining (magnification, ×400), and statistical analysis of the positive rates of EGFR and p-EGFR expression in HCC tissues and paracancerous tissues. (B) Immunocytochemistry staining, (C) western blotting and (D) indirect immunofluorescence was used to detect the expression of EGFR in normal liver cells and liver cancer cells. **P<0.01 and ***P<0.001. EGFR, epidermal growth factor receptor; HCC, hepatocellular carcinoma; p-, phosphorylated.
Knockdown of EGFR reduces HepG2 cell proliferation
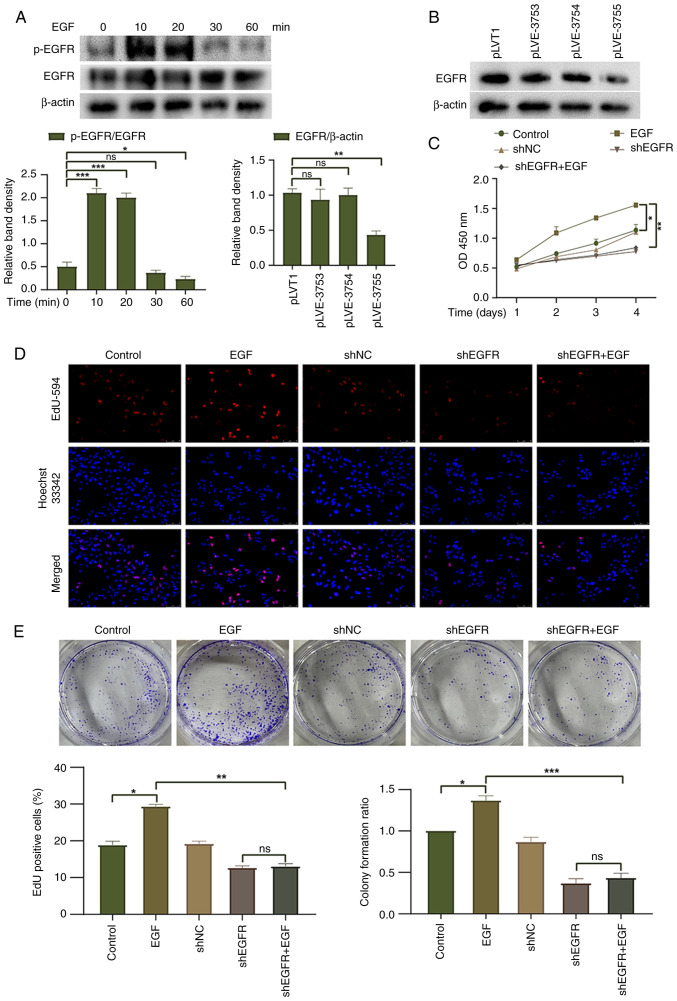
To investigate the biological effects of EGFR on HepG2 liver cancer cells, EGFR activation was induced in HepG2 cells following treatment with EGF (100 ng/ml). The peak level of EGFR phosphorylation was reached 10 min after EGF treatment and gradually decreased thereafter (Fig. 2A). Furthermore, an EGFR knockdown lentivirus was transduced into HepG2 cells. Western blotting showed that pLVE-3755 had the best knockdown effect on EGFR and was thus used for subsequent experiments (Fig. 2B). CCK-8, EdU and colony formation assays were used to evaluate the proliferation and colony-forming ability of HepG2 cells, respectively. The EGF group exhibited increased cell proliferation and clonogenic ability compared with the control group, suggesting that EGFR activation promotes cell proliferation (Fig. 2C-E). EGF treatment did not enhance the cell proliferation or clonogenic ability of HepG2 cells with EGFR knockdown, and there was no significant difference observed between the shEGFR group and the shEGFR + EGF group. To further validate the role of EGFR activation in HepG2 cells, the EGF group was compared with the shEGFR + EGF group. The findings revealed that EGFR knockdown had a significant inhibitory effect on the proliferation and clonogenic ability of HepG2 cells. These results indicated that EGFR activation may promote the proliferation of HepG2 cells, whereas downregulation of EGFR expression exerts an inhibitory effect on the proliferation of HepG2 cells.
Figure 2.
Effect of EGFR on cell proliferation. (A) Western blotting was used to detect the differences in EGFR activation induced by EGF at different times. (B) Western blotting was used to detect the knockdown efficiency of three EGFR knockdown lentiviruses in HepG2 cells. (C) Cell Counting Kit-8 assay and (D) EdU-594 proliferation assay were used to detect the proliferation of HepG2 cells (magnification, ×200). (E) Colony formation assay was used to detect the colony-forming ability of HepG2 cells. *P<0.05, **P<0.01 and ***P<0.001. EGFR, epidermal growth factor receptor; NC, negative control; ns, not significant; p-, phosphorylated; sh, short hairpin.
Knockdown of EGFR reduces HepG2 cell migration
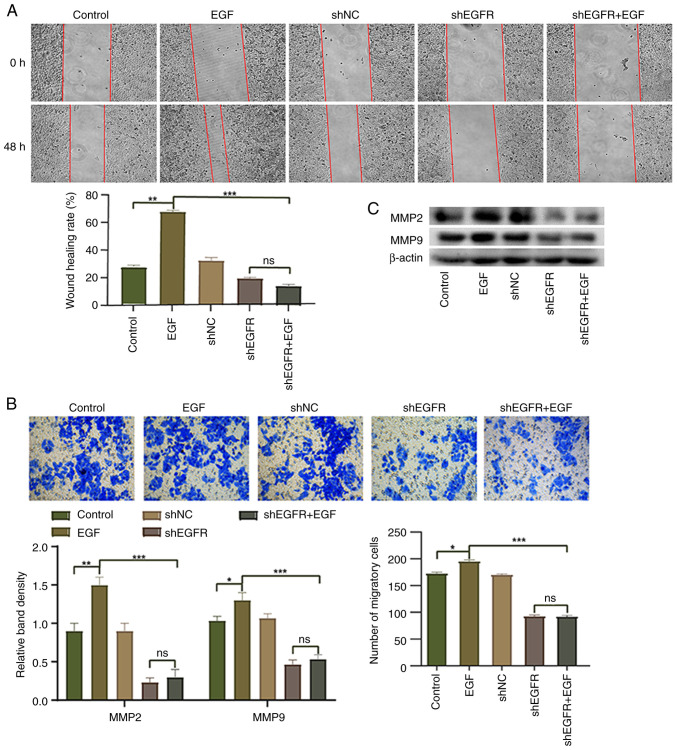
EGFR has been reported to serve an important role in cell migration and invasion (29). Therefore, the present study conducted wound healing and Transwell assays to investigate the effect of EGFR knockdown on the migration of HepG2 cells. Compared with in the control group, the migration of HepG2 cells in the EGF group was significantly upregulated, whereas the migration of HepG2 cells in the shEGFR + EGF group was significantly inhibited (Fig. 3A and B). Western blotting was performed to detect the expression levels of MMP9 and MMP2, and the expression levels of MMP9 and MMP2 were increased in the EGF group compared with the control group, whereas the expression levels of these proteins were significantly decreased in the shEGFR + EGF group (Fig. 3C). These results suggested that EGFR activation induced by EGF may increase cell migration, whereas downregulation of EGFR expression can decrease the migration of HepG2 cells.
Figure 3.
Effect of EGFR on HepG2 cell migration. (A) Wound healing assay (magnification, ×100) and (B) Transwell assay (magnification, ×200) were used to evaluate the migration of HepG2 cells. (C) Western blotting was used to detect the expression levels of MMP2 and MMP9 in HepG2 cells. *P<0.05, **P<0.01 and ***P<0.001. EGFR, epidermal growth factor receptor; NC, negative control; ns, not significant; sh, short hairpin.
Knockdown of EGFR partially reverses HepG2 cell EMT
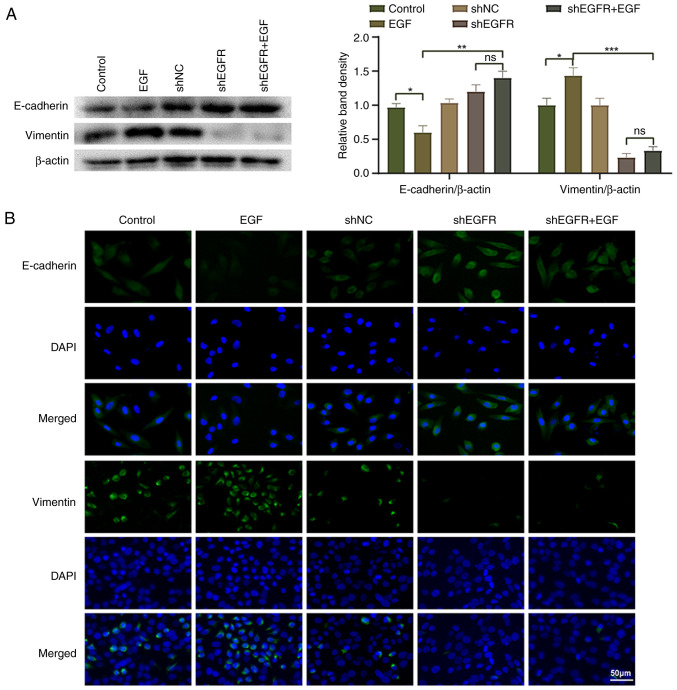
To investigate whether EGFR regulates the EMT process of HepG2 cells, the present study analyzed the expression of EMT markers in HepG2 cells. The results of western blotting and indirect immunofluorescence showed that compared with the control group, the expression levels of E-cadherin were significantly decreased, whereas those of vimentin were significantly increased in the EGF group, which is a typical feature of the EMT process (Fig. 4). Compared with those in the EGF group, the expression levels of E-cadherin were significantly increased and those of vimentin were significantly decreased in the shEGFR + EGF group. These results indicated that EGFR may promote the EMT process of HepG2 cells, thereby promoting HepG2 cell migration and invasion.
Figure 4.
Regulation of HepG2 cell epithelial-mesenchymal transition by EGFR. (A) Western blotting and (B) indirect immunofluorescence was used to detect the expression levels of E-cadherin and vimentin in HepG2 cells. *P<0.05, **P<0.01 and ***P<0.001. EGFR, epidermal growth factor receptor; NC, negative control; ns, not significant; sh, short hairpin.
EGFR mediates HepG2 cell EMT through the Akt/GSK-3β/Snail signaling pathway
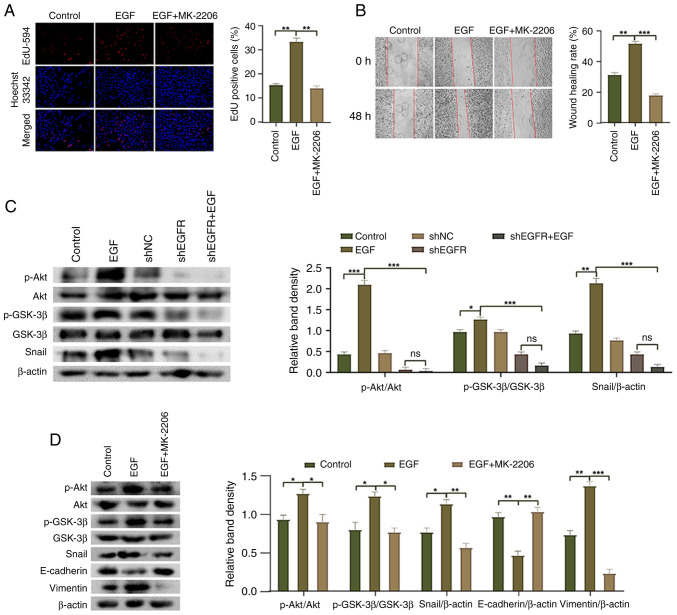
The Akt/GSK-3β/Snail pathway has been reported to be involved in the EMT process (30); however, it is unclear whether EGFR mediates the EMT of HepG2 cells through this pathway. The present study conducted western blot analysis and revealed that compared with the control group, the phosphorylation of Akt and GSK-3β, and the expression levels of Snail were significantly increased in the EGF group, whereas the expression levels of AKT and GSK-3β were not significantly altered (Fig. 5C). By contrast, the phosphorylation of Akt and GSK-3β, and the expression levels of Snail were significantly decreased in the shEGFR + EGF group compared with the EGF group. Additionally, following treatment with the Akt inhibitor MK-2206, compared with the EGF group, the phosphorylation of Akt and GSK-3β, and the expression levels of Snail and vimentin were decreased, whereas the expression levels of E-cadherin were increased (Fig. 5D). Moreover, the proliferation and migration of HepG2 cells were significantly decreased in response to MK-2206 treatment compared with those in the EGF group (Fig. 5A and B). These results indicated that EGFR regulates the EMT process of HepG2 cells by modulating the Akt/GSK-3β/Snail pathway, affecting cell proliferation, migration and EMT marker expression.
Figure 5.
Molecular mechanism underlying EGFR-mediated epithelial-mesenchymal transition in HepG2 cells. (A) EdU-594 assay was used to detect the proliferation of HepG2 cells (magnification, ×200). (B) Wound healing assay was used to detect the migration of HepG2 cells (magnification, ×100). (C) Western blotting was used to detect the phosphorylation of Akt and GSK-3β, and the expression of Snail in HepG2 cells. (D) Western blotting was used to detect the phosphorylation of Akt and GSK-3β, and the expression levels of Snail, E-cadherin and Vimentin in HepG2 cells after MK-2206 treatment. *P<0.05, **P<0.01 and ***P<0.001. EGFR, epidermal growth factor receptor; NC, negative control; ns, not significant; p-, phosphorylated; sh, short hairpin.
Discussion
The present study aimed to determine the molecular mechanism by which EGFR mediates EMT to promote the progression of liver cancer. The results revealed that EGFR mediated EMT through the Akt/GSK-3β/Snail signaling pathway, thereby promoting cell proliferation and migration. EGFR and p-EGFR were highly expressed in HCC tissues, and knockdown of EGFR significantly inhibited the proliferation and migration of HepG2 cells.
EMT is the process by which epithelial cells acquire mesenchymal cell phenotypes, which is an important step in tumor development and metastasis (31). One of the markers of EMT is the loss of E-cadherin function (32); notably, Snail can strongly inhibit the expression of the E-cadherin protein (33). EMT is regulated by various signaling pathways, including receptor tyrosine kinases, transforming growth factor-β and STAT3 (34). In the present study, treatment with EGF to activate EGFR led to a decrease in E-cadherin expression and an increase in vimentin expression in HepG2 cells. The opposite results were obtained in response to EGFR knockdown, thus indicating that EGFR can promote EMT. Investigations into the molecular mechanism underlying EGFR-mediated EMT indicated that activation of EGFR promoted the phosphorylation of Akt and GSK-3β, whereas knocking down EGFR inhibited the phosphorylation of Akt and GSK-3β. Notably, the present study revealed that activation of EGFR also promoted the expression of the downstream cytoplasmic transcription factor Snail. The effects of EGF treatment were reversed following EGFR knockdown. Moreover, the Akt inhibitor (MK-2206) inhibited the phosphorylation of Akt and GSK-3β, decreased Snail expression, and partially reversed the cell EMT phenotype, thus reducing cell migration and proliferation. These findings indicated that EGF/EGFR may be involved in regulating the EMT of HCC through the Akt/GSK-3β/Snail signaling pathway, promoting proliferation and migration.
Although previous studies have reported the effect of EGFR on EMT in HCC (35,36), to the best of our knowledge, the mechanism by which EGFR mediates EMT through the Akt/GSK-3β/Snail signaling pathway in HCC has not been reported. The present study revealed that the HepG2 cell EMT phenotype could be reversed, and the proliferation and migration of HepG2 cells could be reduced, by inhibiting the Akt/GSK-3β/Snail signaling pathway. However, a number of factors are involved in the progression of HCC; therefore, further research is required to better define the mechanisms underlying the occurrence of HCC and to identify possible treatment methods.
In conclusion, the present study revealed that EGF/EGFR can mediate EMT through the Akt/GSK-3β/Snail signaling pathway, thus promoting the proliferation and migration of HepG2 cells. Inhibiting EGFR activation may partially reverse the EMT phenotype, thus inhibiting the proliferation and migration of HepG2 cells. This knowledge may lead to innovative approaches for the treatment of liver cancer.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Huaiyong Gan (Department of Pathology, The First Affiliated Hospital of Bengbu Medical College) for his work in clinical tumor tissue sample collection.
Funding Statement
This work was supported by grants from the 2022 Graduate Innovation Fund Project of Anhui University of Science & Technology (grant no. 2022CX2144), the National Natural Science Fund of China (grant nos. 82071862 and 81872017), and the Medical Special Cultivation Project of Anhui University of Science & Technology (grant no. YZ2023H1A005).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JG, ZH, XS, QS, WR and XH designed and performed the research. SZ and XT analyzed data and drafted the manuscript. JG and XT confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Anhui University of Science & Technology (approval no. 2021005). The study obtained the written informed consent of all participants.
Patient consent for publication
Patients provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 2.Dat VHX, Nhung BTH, Chau NNB, Cuong PH, Hieu VD, Linh NTM, Quoc NB. Identification of potential microRNA groups for the diagnosis of hepatocellular carcinoma (HCC) using microarray datasets and bioinformatics tools. Heliyon. 2022;8:e08987. doi: 10.1016/j.heliyon.2022.e08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y, Ren Y, Dong X, Kan X, Zheng C. An overview of hepatocellular carcinoma after insufficient radiofrequency ablation. J Hepatocell Carcinoma. 2022;9:343–355. doi: 10.2147/JHC.S358539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M, Gupta R, Qiu M, Deczkowska A, Weiner A, et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature. 2021;592:450–456. doi: 10.1038/s41586-021-03362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steeg PS, Theodorescu D. Metastasis: A therapeutic target for cancer. Nat Clin Pract Oncol. 2008;5:206–219. doi: 10.1038/ncponc1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Liang J, Cao N, Gao J, Xie Y, Zhou S, Tang X. ASIC1α up-regulates MMP-2/9 expression to enhance mobility and proliferation of liver cancer cells via the PI3K/AKT/mTOR pathway. BMC Cancer. 2022;22:778. doi: 10.1186/s12885-022-09874-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C, Zhang H, Zhang L, Zhu AX, Bernards R, Qin W, Wang C. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20:203–222. doi: 10.1038/s41575-022-00704-9. [DOI] [PubMed] [Google Scholar]
- 8.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Lin Y, Zhang J, Zhang Y, Li Y, Liu Z, Li Q, Luo M, Liang R, Ye J. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:447. doi: 10.1186/s13046-019-1412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faivre S, Rimassa L, Finn RS. Molecular therapies for HCC: Looking outside the box. J Hepatol. 2020;72:342–352. doi: 10.1016/j.jhep.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Huang A, Yang XR, Chung WY, Dennison AR, Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:146. doi: 10.1038/s41392-020-00264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanaya H, Natarajan A, Komposch K, Li L, Amberg N, Chen L, Wculek SK, Hammer M, Zenz R, Peck-Radosavljevic M, et al. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. 2014;16:972–977. doi: 10.1038/ncb3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z. ErbB Receptors and Cancer. Methods Mol Biol. 2017;1652:3–35. doi: 10.1007/978-1-4939-7219-7_1. [DOI] [PubMed] [Google Scholar]
- 14.Thompson SM, Jondal DE, Butters KA, Knudsen BE, Anderson JL, Stokes MP, Jia X, Grande JP, Roberts LR, Callstrom MR, Woodrum DA. Heat stress induced, ligand-independent MET and EGFR signalling in hepatocellular carcinoma. Int J Hyperthermia. 2018;34:812–823. doi: 10.1080/02656736.2017.1385859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds AR, Tischer C, Verveer PJ, Rocks O, Bastiaens PI. EGFR activation coupled to inhibition of tyrosine phosphatases causes lateral signal propagation. Nat Cell Biol. 2003;5:447–453. doi: 10.1038/ncb981. [DOI] [PubMed] [Google Scholar]
- 16.Niu J, Li W, Liang C, Wang X, Yao X, Yang RH, Zhang ZS, Liu HF, Liu FY, Pei SH, et al. EGF promotes DKK1 transcription in hepatocellular carcinoma by enhancing the phosphorylation and acetylation of histone H3. Sci Signal. 2020;13:eabb5727. doi: 10.1126/scisignal.abb5727. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y, Xu R, Liu X, Zhang Y, Song L, Cai S, Zhou S, Xie Y, Li A, Cao W, Tang X. LY3214996 relieves acquired resistance to sorafenib in hepatocellular carcinoma cells. Int J Med Sci. 2021;18:1456–1464. doi: 10.7150/ijms.51256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 19.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Yang WH, Lan HY, Huang CH, Tai SK, Tzeng CH, Kao SY, Wu KJ, Hung MC, Yang MH. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol. 2012;14:366–374. doi: 10.1038/ncb2455. [DOI] [PubMed] [Google Scholar]
- 21.Yeh HW, Hsu EC, Lee SS, Lang YD, Lin YC, Chang CY, Lee SY, Gu DL, Shih JH, Ho CM, et al. PSPC1 mediates TGF-β1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat Cell Biol. 2018;20:479–491. doi: 10.1038/s41556-018-0062-y. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 24.Lin J, Song T, Li C, Mao W. GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim Biophys Acta Mol Cell Res. 2020;1867:118659. doi: 10.1016/j.bbamcr.2020.118659. [DOI] [PubMed] [Google Scholar]
- 25.Pecoraro C, Faggion B, Balboni B, Carbone D, Peters GJ, Diana P, Assaraf YG, Giovannetti E. GSK3β as a novel promising target to overcome chemoresistance in pancreatic cancer. Drug Resist Updat. 2021;58:100779. doi: 10.1016/j.drup.2021.100779. [DOI] [PubMed] [Google Scholar]
- 26.Lv Q, Wang J, Xu C, Huang X, Ruan Z, Dai Y. Pirfenidone alleviates pulmonary fibrosis in vitro and in vivo through regulating Wnt/GSK-3β/β-catenin and TGF-β1/Smad2/3 signaling pathways. Mol Med. 2020;26:49. doi: 10.1186/s10020-020-00173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 28.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, Sullivan DC, et al. 8th edition. Springer International Publishing; Cham, Switzerland: 2017. AJCC Cancer Staging Manual; p. 291. [Google Scholar]
- 29.Liu X, Adorno-Cruz V, Chang YF, Jia Y, Kawaguchi M, Dashzeveg NK, Taftaf R, Ramos EK, Schuster EJ, El-Shennawy L, et al. EGFR inhibition blocks cancer stem cell clustering and lung metastasis of triple negative breast cancer. Theranostics. 2021;11:6632–6643. doi: 10.7150/thno.57706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Cao N, Gao J, Liang J, Liang Y, Xie Y, Zhou S, Tang X. ASIC1a stimulates the resistance of human hepatocellular carcinoma by promoting EMT via the AKT/GSK3β/Snail pathway driven by TGFβ/Smad signals. J Cell Mol Med. 2022;26:2777–2792. doi: 10.1111/jcmm.17288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed Pharmacother. 2021;133:110909. doi: 10.1016/j.biopha.2020.110909. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 34.Montanari M, Rossetti S, Cavaliere C, D'Aniello C, Malzone MG, Vanacore D, Di Franco R, La Mantia E, Iovane G, Piscitelli R, et al. Epithelial-mesenchymal transition in prostate cancer: An overview. Oncotarget. 2017;8:35376–35389. doi: 10.18632/oncotarget.15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65:798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Wang B, Liu T, Wu JC, Luo SZ, Chen R, Lu LG, Xu MY. STAT3 aggravates TGF-β1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed Pharmacother. 2018;98:214–221. doi: 10.1016/j.biopha.2017.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.