Abstract
The chromosomal insertion sites of Tn10-containing Escherichia coli strains were amplified by inverse PCR, and the nucleotide sequences of the junctions were determined. In 95 strains analyzed, 88 unique Tn10 positions were determined and matched to the E. coli chromosome sequence. Two gaps in insertion site positions were noted, one including the terminus of DNA replication and another bounded by recombination hot spots RhsA and RhsB.
A collection of Escherichia coli strains with Tn10 insertions located at approximately 1-min intervals around the chromosome was reported in 1989 (12) and has been used in many laboratories for strain construction and genetic mapping. The versatility of this collection of strains is based on its regularity of map positions around the chromosome and its combination of Tn10 and positionally equivalent Tn10kan members. To clarify an occasional inconsistency in map position in certain members of the collection in our laboratory, we developed an inverse PCR scheme to allow determination of the precise positions of the Tn10 insertion sites by DNA sequence analysis. We have determined the nucleotide positions of nearly all of the Tn10 insertion sites in the collection of strains originally reported by Singer et al. (12) and subsequently catalogued by Berlyn et al. (2).
Strains used in this study were obtained either from the Carol Gross laboratory or from the E. coli Genetic Stock Center. DNA preparations were done by a modification of standard methods (10, 13). Cells from 5 ml of an overnight culture grown in Luria broth-tetracycline (10 μg/ml) were harvested by centrifugation and resuspended in 2.5 ml of lysis solution (25 mM Tris-HCl [pH 7.4], 50 mM glucose, 10 mM EDTA, 2-μg/ml lysozyme). Cells were lysed by the addition of 0.25 ml of 10% sodium dodecyl sulfate, and DNA was extracted once with an equal volume of phenol saturated with 0.3 M sodium acetate (NaOAc). The aqueous phase was retained and made 0.3 M in NaOAc by addition of 0.1 volume of 3 M NaOAc, and DNA was precipitated by addition of 2.5 volumes of ethanol. The precipitate was transferred to 0.5 ml of 70% ethanol by using a Pasteur pipette. Following a brief centrifugation (30 s at 13,000 × g), the supernatant was removed and the DNA was resuspended in 0.25 ml of 10 mM Tris-HCl (pH 7.4)–0.1 mM EDTA. Chromosomal DNA was digested with HpaII and circularized with DNA ligase preparatory to inverse PCR (8).
Inverse PCR was performed as described by Ochman et al. (8) by using Platinum Taq DNA Polymerase (Life Technologies). The PCR primers (Integrated DNA Technologies) were designed from the Tn10 sequence (5, 7, 9, 11) and are illustrated in Fig. 1. The product of the first PCR using primers 1 and 2 was diluted 1/10, and 1 μl was used as the template for a second round of PCR using primers 3 and 4. The sequences of the primers were as follows: primer 1, ACATGAAGGTCATCGATAGCAGGA; primer 2, GGCTGTTGAGTTGAGGTTGACGAA; primer 3, AACAGTAATGGGCCAATAACACCG; primer 4, CGAGTTCGCACATCTTGTTGTCTG.
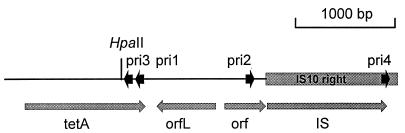
FIG. 1.
Map of a portion of Tn10 including tetA and IS10R. Open reading frames (orfs) are indicated, as are the positions of the PCR primers (pris) used in this study. The single HpaII site in this region is also shown.
PCR products were sequenced by using a PCR sequencing kit (Amersham). The sequencing primer was a 19-mer situated at positions −62 to −53 relative to the end of IS10R. Twenty- to 40-nucleotide-long sequences at the junction of IS10R were determined, and the positions were identified by a BLAST search (1) of the E. coli genome sequence (3).
Of 95 strains analyzed, 88 yielded sufficient sequences for confident definition of positions on the E. coli chromosome (Table 1 and Fig. 2). Two strains (CAG18463 and CAG12099) yielded short sequences that occurred twice in the genome sequence, once within 0.5 min of the reported map position and once distant. The site near the reported map position was taken as the site of insertion for these strains. The sequence derived from the junction of CAG18491 was very short, but inspection of the sequence near metE showed only two identities, one in the metR-metE intergenic region and one downstream from the metE coding region. We presumed that the intergenic insertion was more likely to yield a metE phenotype and placed the insertion site at that position. One additional sequence (from CAG18429 zjh-6::Tn10) was too short for unambiguous assignment, and no phenotype was available to assist in placement. The remaining five strains yielded sequences identical to others in the collection. These duplicate strains were CAG12074 = CAG18465, CAG12159 = CAG18459 = CAG12151, CAG18483 = CAG12080, CAG18498 = CAG18499, and CAG18709 = CAG18456. Each duplicate sequence was confirmed by analysis of strains obtained from the E. coli Genetic Stock Center.
TABLE 1.
Locations of Tn10 insertion sites
| Strain | Genotypea | Map position (min)
|
Nucleotide position | Dd | Gene | Accession no. | Position (bp) | Flanking sequence | |
|---|---|---|---|---|---|---|---|---|---|
| Reportedb | Calculatedc | ||||||||
| CAG18442 | thr-34::Tn10 | 0.0 | 0.0 | 425 | < | thrA | AE000111 | 425 | TGCCTGGCATTGCTTTCCAG |
| CAG12093 | car-96::Tn10 | 0.7 | 0.7 | 33,963 | < | carB | AE000113 | 13,304 | GGCAAAGCCGCCGTTGAGGG |
| CAG12095 | zab-3051::Tn10 | 1.9 | 1.8 | 84,350 | < | leuO | AE000118 | 805 | CACTTAACTCCCTTTCCCTT |
| CAG12025 | zad-220::Tn10 | 3.2 | 3.2 | 150,100 | > | yadC | AE000123 | 472 | GGTGAAGCGCCATTTTTAAT |
| CAG18436 | zae-502::Tn10 | 4.4 | 5.0 | 230,099 | > | yafC | AE000129 | 10,075 | CACTTTATCTGCCATTAACT |
| CAG18447 | proAB81::Tn10 | 5.7 | 5.6 | 261,286 | > | proA | AE000132 | 8,002 | TGCTGCGTATGGATAAATAC |
| CAG18439 | lacI42::Tn10 | 7.9 | 7.9 | 365,945 | > | lacI | AE000141 | 9,129 | CGCGCAGCCCGG |
| CAG12080 | zah-281::Tn10 | 7.5 | 8.0 | 371,180 | > | b0349 | AE000142 | 3,414 | CGCTTTGTGCCGATGGATGC |
| CAG18091 | zaj-3053::Tn10 | 9.0 | 8.5 | 393,633 | > | b0374 | AE000144 | 4,231 | TACCTGGTAATATTCTTCAC |
| CAG12148 | tsx-247::Tn10 | 9.3 | 9.3 | 430,667 | < | tsx | AE000147 | 9,036 | TACTTTGTGCCGATTACCGA |
| CAG12017 | zba-3054::Tn10 | 10.1 | 9.9 | 461,486 | > | b0441 | AE000150 | 7,331 | GGTTAAACAGGCGATTTTCG |
| CAG12154 | zba-3055::Tn10 | 10.9 | 10.8 | 500,025 | > | gsk | AE000154 | 782 | CGCTGGGCACCAAGTTTGTC |
| CAG12171 | purE79::Tn10 | 11.9 | 11.9 | 551,279 | < | purK | AE000158 | 5,564 | CGCATAACCTGCATCAGGAC |
| CAG12149 | zbe-601::Tn10 | 14.3 | 13.7 | 637,765 | > | b0604 | AE000166 | 1,930 | AGCAGAGCCAGTAAAAGTAT |
| CAG12077 | zbe-280::Tn10 | 14.7 | 14.1 | 655,952 | < | ybeG | AE000167 | 4,670 | TGCCAGGTGATGGCAGGAAT |
| CAG18433 | zbf-3057::Tn10 | 15.9 | 15.1 | 698,404 | < | asnB | AE000171 | 1,888 | GGTTATGTGTTCAATTTTTG |
| CAG12147 | nadA57::Tn10 | 16.9 | 16.8 | 781,532 | > | nadA | AE000177 | 8,094 | TGCAAAGCATCCCGCTTCTA |
| CAG18493 | zbi-29::Tn10 | 18.0 | 17.7 | 819,871 | > | ig,e b0786-b0787 | AE000181 | 3,833 | CACTTAACAAGTACCAGGTA |
| CAG12034 | zbi-3058::Tn10 | 18.7 | 19.3 | 897,129 | < | potI | AE000187 | 9,890 | TGTCGCGTATCCGCTGTTTT |
| CAG18478 | zbj-1230::Tn10 | 19.8 | 20.4 | 946,252 | > | ig, ycaD-b0899 | AE000192 | 1,345 | AGCAGAGTGTGAACTTACTG |
| CAG12094 | zcb-3059::Tn10 | 21.1 | 21.6 | 1,000,952 | < | b0940 | AE000196 | 4,074 | AGCTTCGCCCCAGCGCACGC |
| CAG18466 | zcc-282::Tn10 | 22.6 | 22.9 | 1,064,111 | < | yccE | AE000202 | 2,444 | TACTCTGCCCCTGAATTTGG |
| CAG12078 | zce-726::Tn10 | 24.6 | 24.9 | 1,154,216 | > | yceG | AE000210 | 6,298 | TGCTGCGCATCCGG |
| CAG18463 | zcf-117::Tn10 | 25.5 | 26.1 | 1,211,084 | > | b1160 | AE000215 | 1,599 | GACTTAACCGG |
| CAG18497 | fadR13::Tn10 | 26.6 | 26.6 | 1,234,734 | < | fadR | AE000217 | 2,438 | AGCCCAGCGCCAGACTGCGC |
| CAG12016 | zch-3060::Tn10 | 28.0 | 27.4 | 1,269,233 | < | ig, kdsA-cha | AE000220 | 156 | GACGTAGTATCCACACCAAG |
| CAG12169 | zci-506::Tn10 | 28.0 | 28.1 | 1,302,475 | > | oppC | AE000223 | 1,625 | ATCGGGCATTGTTATTCGCC |
| CAG18455f | trpB83::Tn10 | 28.3 | 28.4 | 1,317,596 | < | trpC | AE000224 | 5,761 | CGCATTGCCGCCATTTATAA |
| CAG12028f | zcj-233::Tn10 | 29.0 | 29.5 | 1,368,905 | > | b1309 | AE000229 | 1,260 | TGCTGAGTATGTCCGCCTGG |
| CAG12179f | mg1-500::Tn10 | 48.2 | 30.2 | 1,399,003 | > | ydaH | AE000231 | 9,081 | TGCTATGCGTAAACCAAAAC |
| CAG12081 | zda-3061::Tn10 | 30.4 | 30.4 | 1,412,228 | < | recT | AE000232 | 9,574 | TGCGCAGCCTGAGTAAAGCT |
| CAG18640f | zhj-3076::Tn10 | 79.7 | 30.9 | 1,434,058 | > | b1377 | AE000234 | 8,385 | TACTGTGCAGTGACTTCAAA |
| CAG12026 | trg-2::Tn10 | 32.1 | 32.2 | 1,491,930 | < | trg | AE000239 | 6,837 | CGTTATGCCTCTACTTTGTT |
| CAG18461 | zdd-235::Tn10 | 33.3 | 33.1 | 1,533,652 | < | yddE | AE000243 | 5,163 | AGCTGCGCACTATGTACGTG |
| CAG12151 | zdi-925::Tn10 | 38.6 | 38.3 | 1,776,386 | < | ig, b1695-b1696 | AE000264 | 9,608 | TGCGCCGCCTGCAGATTATT |
| CAG18464 | zdj-276::Tn10 | 39.4 | 39.5 | 1,834,488 | < | b1754 | AE000270 | 4,289 | AGCTGTGTGTCGACATAGCG |
| CAG18465 | zea-225::Tn10 | 40.3 | 40.3 | 1,869,535 | > | b1785 | AE000273 | 8,941 | GACATCGTGTGGGTGATAAA |
| CAG12068 | zeb-3190::Tn10 | 41.3 | 41.0 | 1,903,614 | < | ig, b1820-b1821 | AE000276 | 7,274 | GGCATAGCGATTGATGTGCA |
| CAG18486 | eda-51::Tn10 | 41.6 | 41.6 | 1,930,416 | < | eda | AE000279 | 1,608 | TGCTGGGTATGGACTACGGT |
| CAG12156 | uvrC279::Tn10 | 43.0 | 42.9 | 1,991,244 | > | uvrC | AE000284 | 5,249 | CGCCAAGTAGCAGCGGATGA |
| CAG18451 | zed-3069::Tn10 | 43.9 | 43.9 | 2,038,762 | < | b1972 | AE000288 | 8,420 | GGCAAAGCACCATAATCCTA |
| CAG12099 | zef-3129::Tn10 | 45.4 | 44.9 | 2,085,192 | > | ig, yeeF-b2015 | AE000293 | 1,586 | TACGAAGCCCGG |
| CAG12163f | zib-207::Tn10 | 81.8 | 46.9 | 2,173,967 | > | gatZ | AE000298 | 10,901 | CGCATCAAGATGAATTTTAC |
| CAG12021f | zbd-3105::Tn10 | 13.1 | 46.9 | 2,175,213 | > | gatY | AE000298 | 12,147 | TACCACGTACATTTTCATAT |
| CAG12098 | zei-722::Tn10 | 48.3 | 49.6 | 2,299,610 | < | napA | AE000309 | 11,314 | CAACCTGCACCTGCTGACCG |
| CAG12177 | zej-298::Tn10 | 49.6 | 50.0 | 2,318,294 | > | atoS | AE000311 | 3,377 | ATCCGCGCATTAAATGCAGA |
| CAG12178 | zfa-723::Tn10 | 50.4 | 50.5 | 2,341,772 | < | yfaL | AE000313 | 4,289 | GGCGCAATCTATTCTTCTGG |
| CAG18484 | zfb-223::Tn10 | 51.5 | 51.5 | 2,387,022 | < | b2274 | AE000317 | 5,073 | TGCTCCGTATCTTCTGACCA |
| CAG18467 | zfd-1::Tn10 | 53.4 | 53.7 | 2,491,451 | < | ig, b2374-b2375 | AE000325 | 12,877 | TACTTACCATGCAGAAAGGA |
| CAG18468 | nupC510::Tn10 | 54.1 | 54.1 | 2,511,317 | > | nupC | AE000327 | 4,976 | GGCCTGGCATTCTTCTTCCT |
| CAG18470 | purC80::Tn10 | 55.7 | 55.9 | 2,595,619 | > | purC | AE000334 | 10,116 | AACTCAGCTTGCTTTTGCAT |
| CAG18481 | zfh-208::Tn10 | 57.5 | 57.4 | 2,665,159 | > | b2536 | AE000340 | 2,865 | GGCCGAGTAGCCAGCTGCCT |
| CAG18480 | nadB51::Tn10 | 58.2 | 58.4 | 2,708,518 | < | nadB | AE000344 | 312 | GGCGTAGCGCCAGTGAAAGT |
| CAG12158 | pheA18::Tn10 | 58.9 | 59.0 | 2,736,076 | < | pheA | AE000346 | 3,848 | TGCTGAGTGCGGATTAATTT |
| CAG18642 | zfi-3131::Tn10 | 59.4 | 60.9 | 2,826,375 | < | srlD | AE000354 | 6,308 | TACCCAGCTTGGTCGCGTAT |
| CAG12173 | cysC95::Tn10 | 61.9 | 62.2 | 2,886,785 | < | cysI | AE000360 | 1,394 | AGCAGAGCGTTTCCTGCCGT |
| CAG12079 | fuc-3072::Tn10 | 63.2 | 63.2 | 2,932,885 | > | fucP | AE000364 | 932 | CACCTTATATGATCATCGTG |
| CAG12135 | recD1901::Tn10 | 63.5 | 63.6 | 2,948,961 | < | recD | AE000365 | 5,008 | CGCGCGTCTGGTTTGCGATG |
| CAG12168 | zgf-210::Tn10 | 66.0 | 66.5 | 3,082,773 | > | speA | AE000377 | 3,072 | CACCGTGTATTCGTTACGTT |
| CAG18472 | nupG511::Tn10 | 66.9 | 66.9 | 3,104,395 | < | nupG | AE000379 | 2,008 | CGCCCAGCAGCATTGAGAAG |
| CAG18475 | metC162::Tn10 | 67.9 | 67.9 | 3,150,485 | > | metC | AE000383 | 2,934 | GGCTGCGTGCTATTTCCCTG |
| CAG12184 | tolC210::Tn10 | 68.4 | 68.5 | 3,176,389 | > | tolC | AE000385 | 6,033 | ACCAGTGCGTCCTTGCAGTT |
| CAG12152 | zgj-3075::Tn10 | 69.5 | 69.3 | 3,215,934 | < | air | AE000389 | 1,688 | AGCCAGGTGTCCAGTGTCAG |
| CAG12072 | zha-203::Tn10 | 70.9 | 71.8 | 3,332,800 | > | nlp | AE000399 | 6,432 | TGCGTAGCTACACTAAACCG |
| CAG12153 | zhc-6::Tn10 | 72.1 | 72.5 | 3,364,914 | > | b3219 | AE000401 | 6,566 | GACAACGCGCTTATTCGGCT |
| CAG12075f | zhe-3083::Tn10 | 74.7 | 73.6 | 3,412,834 | > | acrF | AE000405 | 4,993 | GACCGTGCAGGATACGGTGA |
| CAG12071f | zhd-3082::Tn10 | 73.3 | 73.9 | 3,429,813 | < | smg | AE000407 | 246 | TGCTCAACCTTGAAACTCGT |
| CAG18456 | zhe-3084::Tn10 | 74.0 | 75.5 | 3,503,687 | > | yhfT | AE000413 | 6,756 | GGCCAAGCCAGTAAAGAACA |
| CAG18638f | zhh-21::Tn10 | 77.8 | 76.8 | 3,564,175 | > | glgP | AE000419 | 2,951 | CGCTAAGCGTGGGCGATGAA |
| CAG18450f | zhf-5::Tn10 | 75.8 | 77.4 | 3,590,125 | > | ig, ugpB-livF | AE000421 | 8,047 | GATGGGGCACGGATAAGCGG |
| CAG18492f | zic-4901::Tn10 | 82.7 | 82.5 | 3,826,553 | < | ig, glts-yicE | AE000443 | 259 | AGCAAAGCGGGCATTTTAGC |
| CAG18452f | zhf-3085::Tn10 | 75.8 | 82.7 | 3,837,058 | > | nlpA | AE000443 | 10,764 | AGCTGCGCCCCCTCGAGTTC |
| CAG18499 | zid-501::Tn10 | 83.9 | 83.5 | 3,873,001 | < | ig, yidW-yidX | AE000446 | 8,448 | CGCCTGATATCCCTTTTCAG |
| CAG18501 | zie-296::Tn10 | 84.5 | 84.7 | 3,931,475 | < | rbsD | AE000452 | 854 | CGCTGCGCCCGAGAGGGCTT |
| CAG18431 | ilv-500::Tn10 | 85.2 | 85.2 | 3,952,161 | < | ilvD | AE000453 | 10,898 | GGCCAAGTACGTTTTTCACA |
| CAG18491 | metE3079::Tn10 | 86.4 | 86.4 | 4,010,549 | < | ig, metR-metE | AE000458 | 8,157 | GACCGG |
| CAG18496 | fadAB101::Tn10 | 86.7 | 86.8 | 4,025,408 | > | fadA | AE000460 | 559 | TACATGGCAGGATCTGCGCG |
| CAG18495 | zih-35::Tn10 | 87.4 | 87.6 | 4,063,253 | > | b3872 | AE000463 | 5,333 | TGCAGAGCATTAAATTCGAA |
| CAG18477 | zij-501::Tn10 | 89.1 | 89.3 | 4,143,393 | > | pflD | AE000469 | 3,582 | CGCGCGGCGGCTACTTCACG |
| CAG12185 | argE86::Tn10 | 89.4 | 89.5 | 4,151,734 | > | argE | AE000470 | 1,041 | CGCTTCGTAGTGATAACGTT |
| CAG18500 | thi-39::Tn10 | 90.3 | 90.4 | 4,192,143 | > | thiC | AE000473 | 4,927 | TGCTTAACATCTTCTTTATT |
| CAG12164f | malF3089::Tn10 | 91.4 | 91.4 | 4,241,898 | > | malF | AE000476 | 11,030 | GGCTTAGCTTTTCATCACCC |
| CAG18427 | zje-2241::Tn10 | 94.1 | 92.8 | 4,303,986 | > | yjcS | AE000482 | 2,055 | CGCTGGGCATCGTCAAAATC |
| CAG18488 | zjd-2231::Tn10 | 93.7 | 93.9 | 4,356,936 | < | cadB | AE000486 | 731 | TGCTGGGTACTGGTTTAGCA |
| CAG12073 | cycA30::Tn10 | 95.5 | 95.4 | 4,427,714 | < | cycA | AE000492 | 2,708 | AGCGAAGTCACTAAAAGATT |
| CAG12019 | zjg-920::Tn10 | 96.2 | 95.8 | 4,442,377 | > | ytfN | AE000493 | 7,248 | CGCTTAACCTGAACATTGAA |
| CAG18462f | zdh-603::Tn10 | 37.1 | 95.8 | 4,446,109 | > | chpS | AE000494 | 219 | GGCAGAGCGTGGAGGCGCGA |
| CAG18430 | zji-202::Tn10 | 98.6 | 99.0 | 4,595,002 | > | mdoB | AE000507 | 382 | TGCTTGTACTCGATTTTTAC |
Only the position of Tn10 is given.
Map position as reported in reference 2.
Map position calculated from the DNA sequence position.
D, direction. The flanking sequence was read clockwise (>) or counterclockwise (<) on the genetic map.
ig, intergenic. Flanking genes or open reading frames are listed.
Strains from this laboratory and the E. coli Genetic Stock Center were analyzed.
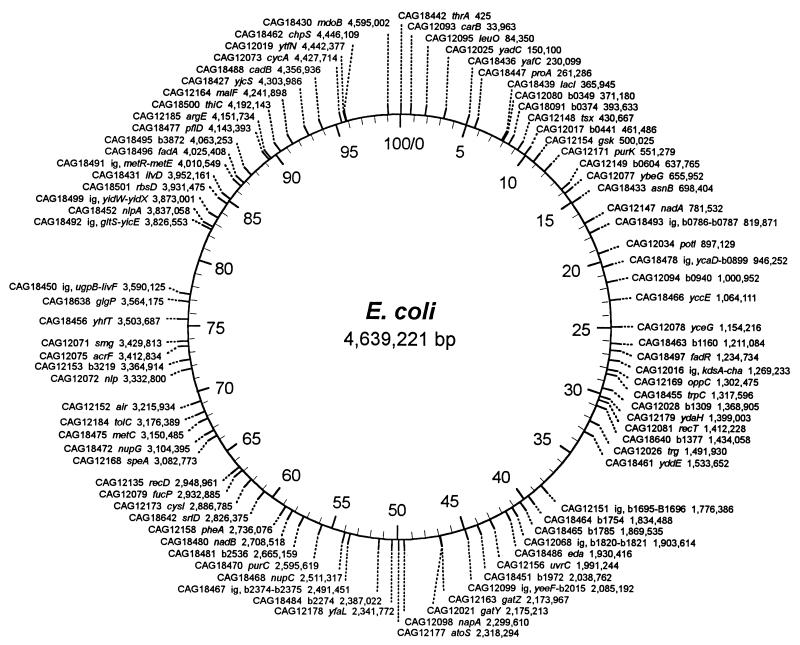
FIG. 2.
Positions of Tn10 insertions on the E. coli map. Shown for each strain are its designation, the gene or open reading frame disrupted by the insertion, and the base pair position. Numbering on the inside of the circle is in minutes. ig, intergenic.
Eleven of the Tn10 insertion sites were in intergenic regions, 51 were in coding regions of known genes, and 26 were in potential open reading frames identified by sequence analysis (3). For the majority of strains, the nucleotide positions of the Tn10 insertion site fell within 1 min of the position determined by genetic mapping. Six of the sequences differed in map position by greater than 2 min. All strains whose Tn10 positions differed from the mapped position by greater than 1.5 min were obtained from the E. coli Genetic Stock Center and reanalyzed. In most cases, there was agreement between the strains in our laboratory collection and those obtained from the E. coli Genetic Stock Center. In several cases, cross-contamination of cultures was evident. In most cases, the mixture was resolved by isolation of single colonies from the cultures. It is not clear whether the positional differences we noted were caused by culture contamination that occurred prior to the distribution of strains to this laboratory and the E. coli Genetic Stock Center or some other artifact of the original genetic analysis.
There are two noticeable gaps in this particular collection of transposon-containing strains, each about 5 min long. The gap at 33 to 38 min contains the DNA replication terminus and recombination hot spot sites dif and RhsE. The gap at 77 to 82 min is bounded by recombination hot spot sites RhsB and RhsA. Three of the strains with transposons originally mapped to these two gaps (CAG18640, CAG12163, and CAG18462) now contain the transposon at a grossly different location on the chromosome. It seems likely that the failure of the transposons to be maintained at these locations is due to the same features that have led to the characterization of these regions as “recombinationally unstable” (4, 6).
Acknowledgments
We thank Jonathan Narita, Mitchell Singer, and Mary Berlyn for advice during the course of this work.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Berlyn M K B, Low K B, Rudd K E, Singer M. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schoechter M, Umbarger H E, editors. Escherichia coli and Salmonella. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Corre J, Cornet F, Patte J, Louarn J M. Unraveling a region-specific hyper-recombination phenomenon: genetic control and modalities of terminal recombination in Escherichia coli. Genetics. 1997;147:979–989. doi: 10.1093/genetics/147.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halling S M, Simons R W, Way J C, Walsh R B, Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci USA. 1982;79:2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin R J, Capage M, Hill C W. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J Mol Biol. 1984;177:1–18. doi: 10.1016/0022-2836(84)90054-8. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T T, Postle K, Bertrand K P. Sequence homology between the tetracycline-resistance determinants of Tn10 and pBR322. Gene. 1983;25:83–92. doi: 10.1016/0378-1119(83)90170-1. [DOI] [PubMed] [Google Scholar]
- 8.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 219–227. [Google Scholar]
- 9.Postle K, Nguyen T T, Bertrand K P. Nucleotide sequence of the repressor gene of the Tn10 tetracycline resistance determinant. Nucleic Acids Res. 1984;12:4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 11.Schollmeier K, Hillen W. Transposon Tn10 contains two structural genes with opposite polarity between tetA and IS10R. J Bacteriol. 1984;160:499–503. doi: 10.1128/jb.160.2.499-503.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1987. pp. 2.4.1–2.4.5. [Google Scholar]