Abstract
The chromosomal acetylornithine deacetylase (argE) gene of Myxococcus xanthus was identified via homology to acetylornithine deacetylases from other bacterial species. A mutant carrying a disruption in argE was unable to grow on minimal media lacking supplemental arginine and formed fruiting bodies and spores in response to arginine starvation at high cell density.
Myxococcus xanthus is a gram-negative soil bacterium that, upon starvation for essential nutrients, initiates a complex developmental process culminating in the formation of multicellular fruiting bodies (for reviews, see references 3 and 15). Within completed fruiting bodies reside several hundred thousand differentiated and metabolically quiescent myxospores, which are able to withstand environmental stresses and remain dormant for at least 50 years (13). When nutrients are restored, myxospores germinate and form viable colonies. M. xanthus cells are bacteriolytic and feed on amino acids and peptides from prey bacteria, but they are unable to use sugars as an energy source due to the absence of key glycolytic enzymes such as pyruvate kinase (22).
Starvation for amino acids alone results in fruiting body formation and sporulation. Cells sense amino acid starvation as an indicator of overall starvation, at least in part, through a stringent response via the ribosome-associated (p)ppGpp synthetase enzyme RelA. This sensory mechanism is proposed to be one of the earliest developmental events, and a block in this mechanism results in an early developmental arrest and in sporulation deficiency (6, 18). Previous work has shown that starving cells for any essential amino acids (wild-type M. xanthus cannot synthesize leucine, isoleucine, or valine) or depriving an auxotroph of its newly essential amino acid (9–11) will elicit a developmental response. In this work we describe the identification and characterization of an M. xanthus open reading frame (ORF) with strong homology to the Escherichia coli N-acetylornithine deacetylase gene (argE), which catalyzes the conversion of acetylornithine to ornithine (5, 12).
Identification of a putative argE homolog.
An ORF with strong sequence similarity and identity to the E. coli argE was identified 300 bp upstream of the putative trcF gene in M. xanthus (4) and is transcriptionally oriented in the opposite direction with respect to trcF. A second divergently transcribed ORF was identified just upstream of this putative argE homolog. A diagram of this region, including a putative translational start site and Shine-Dalgarno sequence, is outlined in Fig. 1. Sequence determination and analysis of this region were performed as previously described (6). DNA sequence analysis identified two ORFs with >90% third-position GC codon usage, which is typical for genes in M. xanthus and other high-G+C-content organisms (1, 16). These ORFs were designated argE and orf2 (Fig. 1). Analysis of both ORFs by using BLAST (National Center for Biotechnology Information [NCBI], National Library of Medicine) revealed that one of these ORFs bears a high degree of sequence identity to argE in E. coli. An alignment comparison between the E. coli and M. xanthus ArgE homologs showed 33% identity and 46% similarity (Fig. 2; comparison was performed with Macaw version 2.0.5, NCBI). No significant identity or similarity was found between ORF 2 and anything in the NCBI database.
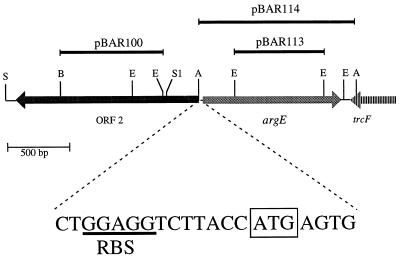
FIG. 1.
Partial restriction map of the 2.8-kb M. xanthus DK101 argE chromosomal region. Arrows indicate putative directions of transcription. The region 3′ to argE including the 3′ end of M. xanthus trcF is shown (for reference, see Garza et al. [4]). Restriction enzymes are abbreviated as follows: A, AccI; B, BamHI; E, EagI; S, SmaI (not all sites shown); S1, SacI. Solid lines indicate DNA regions used for insertion mutagenesis in orf2 (pBAR100) or argE (pBAR113) and expression of argE on the pBluescript lac promoter (pBAR114). A possible ribosome binding site (RBS) and initiator codon (ATG) for argE are shown.
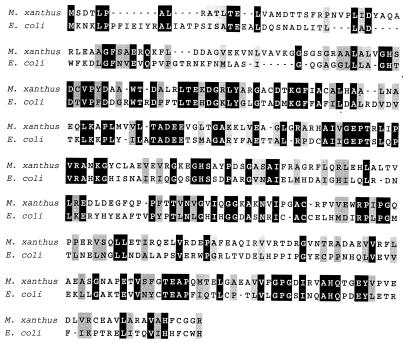
FIG. 2.
Best-fit alignments of the deduced acetylornithine deacetylase (argE) amino acid sequences from M. xanthus and E. coli. Black boxes with white lettering indicate amino acid identities, and gray boxes indicate similarities within groups by side chain.
Complementation of an E. coli argE mutant.
To demonstrate that our putative argE homolog encodes an acetylornithine deacetylase, we assayed its ability to complement a known E. coli argE mutant. To test this hypothesis, we constructed a plasmid containing the entire M. xanthus argE ORF and putative ribosome binding site under the control of the Plac promoter from pBluescript KS(+) (Stratagene, La Jolla, Calif.). A 1,282-bp AccI fragment from pBELF1R (4) was blunted with DNA polymerase Klenow fragment and ligated into pBluescript KS(+) linearized with SmaI (14). The orientation of the insert was verified by sequence analysis, and the resulting plasmid was named pBAR114 (Fig. 1). This plasmid, along with the vector, was transformed into E. coli strain BJW72 (17), which carries a point mutation in argE. Transformants were then screened for their ability to grow in the presence and absence of arginine (final concentration, 0.2 mg/ml). As shown in Fig. 3, BJW72 cells can grow in the absence of arginine only if they carry pBAR114 or are supplemented with arginine; however, they are unable to grow in the absence of arginine with the vector pBluescript.
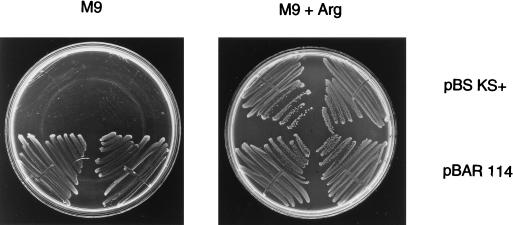
FIG. 3.
Complementation of an E. coli argE mutation by the M. xanthus argE gene. E. coli cells carrying a point mutation in argE were transformed with either pBluescript KS(+) (pBS KS+) or pBAR114 and assayed for growth on M9 minimal agar plates (14) with or without supplemental arginine (Arg). (Each quadrant of a single plate contains a separate isolate, and corresponding quadrants of the two plates contain the same isolate.) Both strains show growth on plates with supplemental arginine, but only the strain with pBAR114 grows in the absence of arginine.
M. xanthus argE is essential for arginine biosynthesis.
To test whether argE is essential for arginine biosynthesis in M. xanthus, we constructed an argE null mutant and tested whether it required arginine for growth on A1 minimal agarose plates (2). The argE null mutation was constructed by plasmid-insertion mutagenesis procedures, as previously described (6). This procedure leaves a tandem duplication of two truncated, nonfunctional genes. For the argE insertion, an EagI fragment internal to the argE ORF was cloned into pBGS18 (19), to give rise to the deletion plasmid pBAR113. This plasmid was then transformed into M. xanthus DK101 via electroporation (7). Because M. xanthus does not maintain pBGS18 as an extrachromosomal plasmid, kanamycin-resistant colonies result from plasmid insertions into the M. xanthus chromosome (20). The presence of a single copy of pBAR113 at the chromosomal locus was confirmed by Southern blot (data not shown). The argE null strain was designated MS2014. A similar approach was taken to construct an orf2 null mutant by using an internal BamHI to SacI fragment within orf2. The resulting null mutant was designated pBAR100.
Growth of the argE null mutant was tested on A1 agarose plates with or without 0.1 mg of arginine supplement/ml. Strain MS2014 was able to grow on A1 agarose only when supplemented with arginine, demonstrating that this strain is an arginine auxotroph. When the strain was grown in rich medium, such as CTT medium (6), no detectable growth phenotype was observed. In addition, the orf2 null strain MS2008 was examined for its ability to grow in the presence and absence of arginine. No detectable growth phenotype was observed, demonstrating that this gene is not involved in arginine biosynthesis.
Developmental phenotypes of the argE and orf2 null mutants.
To determine whether either the argE null mutant or orf2 null mutant was defective in development under standard laboratory conditions (8), both strains, along with the parent strain, DK101, were assayed for their ability to form fruiting bodies on TPM agar plates (6, 21). No defect in fruiting body formation was observed for either mutant strain as compared to the parental strain. Viable spore assays were also performed after 3 and 5 days, and the numbers of heat- and sonication-resistant CFU were determined (21). The argE null mutant formed viable spores at a level two- to threefold greater than that by the wild type; the mutant showed 229% (± 56%) wild-type developing cells after 3 days and 297% (± 57%) wild-type developing cells after 5 days, while the orf2 null mutant showed wild-type sporulation at 87% (± 22%) after 3 days and 91% (± 25%) after 5 days.
Previous work with M. xanthus auxotrophs showed that development could be initiated by simply removing the required amino acid (9–11). This work led to the hypothesis that starvation for any amino acid could lead to initiation of development. To further test this hypothesis, we examined whether the argE null mutant strain MS2014 could be induced to form fruiting bodies if placed at high cell density on A1 agarose plates in the absence of supplemental arginine. Under such conditions, MS2014 forms multicellular fruiting bodies while DK101 remains as a normal vegetative colony (Fig. 4). Inspection of the MS2014 cells after heating and sonication shows that these developing cells also form viable spores, approximately 4 × 106 spores/ml after 5 days. This is approximately 20% of the number of spores observed under complete starvation condition on TPM agar (21) as described above. Though the number of viable spores produced by MS2014 is lower under arginine-starvation conditions than that produced by complete starvation, it is significantly higher than the number of spores produced by wild-type cells under identical conditions, which is 100 spores/ml. These studies support the hypothesis that starvation of M. xanthus for any required amino acid at high cell density can initiate the developmental process.
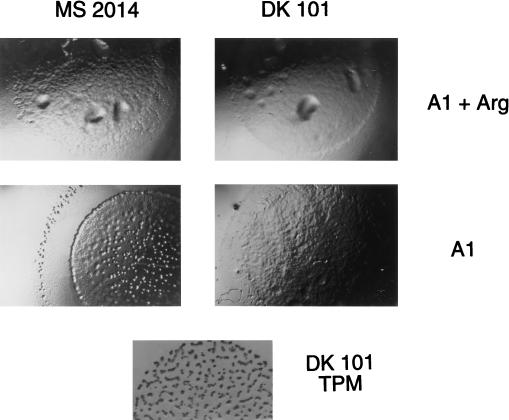
FIG. 4.
Fruiting body formation in the absence of supplemental arginine (Arg). Cells were spotted at high density (5 × 109 cells/ml) on 0.8% agarose A1 plates with or without 0.1 mg of supplemental arginine/ml. M. xanthus MS2014 (sglA1 argE::pBAR113) forms fruiting bodies on A1 minimal media in response to arginine starvation, while strain DK101 (sglA1) does not. Magnification, ×8. Both strains show normal growth in the presence of supplemental arginine. Development of strain DK101 on TPM starvation agar is shown for reference. Magnification, ×6.
Nucleotide sequence accession number.
The DNA sequence of the argE region has been submitted to GenBank under accession no. AF055904.
Acknowledgments
We thank Margaret Baer, Anthony Garza, and Jeff Pollack for helpful discussions and readings of this work. In addition, we thank Stacia Hoover and Dean Lavell for technical assistance with sequencing and loaning of computer software.
This research was supported in part by National Institutes of Health grant GM54592 to M.S.
REFERENCES
- 1.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 2.Bretscher A, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garza A, Pollack J, Harris B, Lee A, Keseler I, Licking E, Singer M. SdeK is required for early fruiting body development in Myxococcus xanthus. J Bacteriol. 1998;180:4628–4637. doi: 10.1128/jb.180.17.4628-4637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glansdorff N. Biosynthesis of arginine and polyamines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 408–433. [Google Scholar]
- 6.Harris B, Singer M, Kaiser D. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashefi K, Hartzell P. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 8.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 9.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manoil C, Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J Bacteriol. 1980;141:1062–1065. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manoil C. Initiation of the development of Myxococcus xanthus. Ph.D. thesis. Stanford, Calif: Stanford University; 1982. [Google Scholar]
- 12.Meinnel T, Schmitt E, Mechulam Y, Blanquett S. Structural and biochemical characterization of the Escherichia coli argE gene product. J Bacteriol. 1992;174:2323–2331. doi: 10.1128/jb.174.7.2323-2331.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reichenbach H. Myxobacteria: a most peculiar group of social prokaryotes. In: Rosenberg E, editor. Myxobacteria, development and cell interactions. New York, N.Y: Springer-Verlag; 1984. pp. 1–50. [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 15.Shimkets L J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimkets L J. The myxobacterial genome. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 85–108. [Google Scholar]
- 17.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- 19.Spratt B G, Hedge P J, Heesen S T, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 20.Stephens K, Hartzell P, Kaiser D. Gliding motility in Myxococcus xanthus: mgl locus, RNA, and predicted protein products. J Bacteriol. 1989;171:819–830. doi: 10.1128/jb.171.2.819-830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thony-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson B, Dworkin M. Comparative intermediary metabolism of vegetative cells and microcysts of Myxococcus xanthus. J Bacteriol. 1968;96:1456–1473. doi: 10.1128/jb.96.5.1465-1473.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]