Abstract
A novel segregational stability system was identified on plasmid R485, which originates from Morganella morganii. The system is composed of two overlapping genes, stbD and stbE, which potentially encode proteins of 83 and 93 amino acids, respectively. Homologs of the stbDE genes were identified on the enterotoxigenic plasmid P307 from Escherichia coli and on the chromosomes of Vibrio cholerae and Haemophilus influenzae biogroup aegyptius. The former two homologs also promote plasmid stability in E. coli. Furthermore, the stbDE genes share homology with components of the relBEF operon and with the dnaT gene of E. coli. The organization of the stbDE cassette is reminiscent of toxin-antitoxin stability cassettes.
Bacterial chromosomes and the low-copy-number plasmids which bacteria often harbor have developed a variety of mechanisms which promote their segregational stability (9, 24, 25). First, active partition systems ensure that each daughter cell receives a copy of the newly replicated plasmid at cell division (7, 20, 25). There is increasing evidence that active partitioning of the chromosome may also occur (7, 13, 18). Second, the resolution by site-specific recombination of dimers and higher-order multimers which arise by homologous recombination optimizes the number of chromosomes and plasmids available for segregation at cell division (23). Third, toxin-antitoxin systems specified by some plasmids compromise the survival of those plasmid-free segregants which do arise (6, 10, 11). In this study, a novel two-gene stability system reminiscent of toxin-antitoxin systems was identified on plasmid R485, which originates from the nosocomial pathogen Morganella morganii. This system is also present in a number of other pathogenic bacteria.
Isolation of a stability region from plasmid R485.
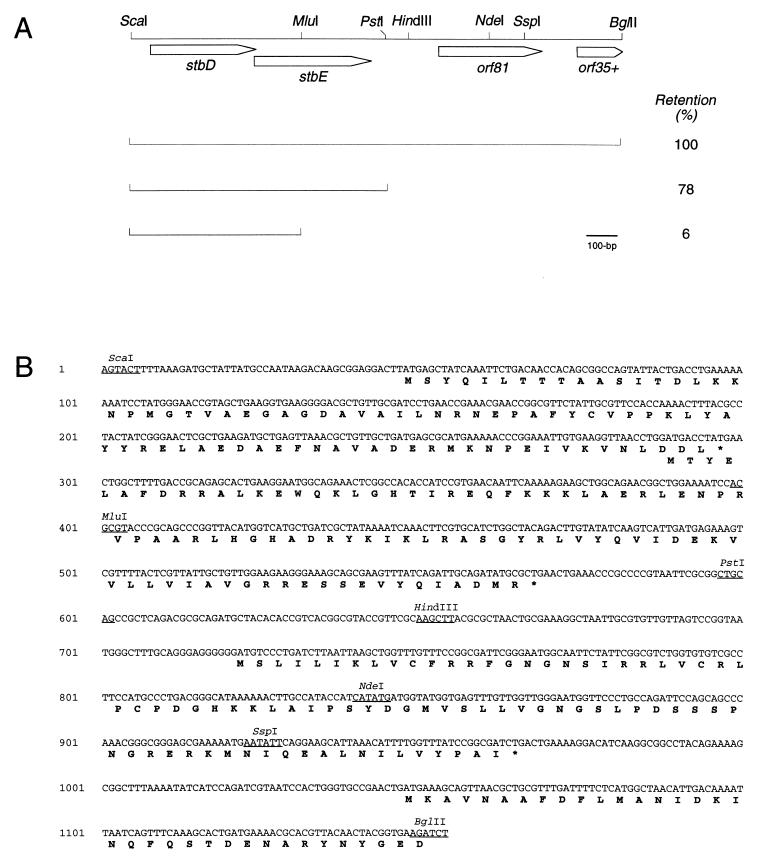
Plasmid R485, which specifies resistance to sulfonamides, originates from Morganella morganii and can conjugate to and replicate in Escherichia coli (8). R485 is a member of plasmid incompatibility group X (3, 8, 12). To isolate segregational stability determinants from R485, a library of 4- to 6-kb fragments generated by partial Sau3AI digestion of R485 was constructed by insertion into the BamHI site of the stability probe vector pALA136. The latter plasmid harbors both the moderate-copy-number ColE1 replicon and the unit copy number P1 replicon (16). In a wild-type host, pALA136 replicates by the ColE1 origin and can be isolated and manipulated with ease. However, in a polA host, the ColE1 origin is nonfunctional, and replication switches to a low copy number under the control of the P1 replicon. As the plasmid does not possess accessory stability genes, it is unstable in this host in the absence of selective pressure. Insertion of a stability locus will restabilize the plasmid (16, 21). The library of R485 fragments in pALA136 was transformed with selection for pALA136-encoded chloramphenicol resistance into the polA strain, BR825 (15). Colonies from this transformation were replica plated once on solid medium containing chloramphenicol and then successively 10 times on medium without antibiotic selection. At the end of this procedure, approximately 10% of colonies retained chloramphenicol resistance. Control experiments with pALA136 resulted in <1% chloramphenicol resistance. The apparent increase in stability of plasmids isolated from chloramphenicol-resistant colonies was retested by retransforming candidate plasmids into the polA strain. Following growth for approximately 25 generations in the absence of selective pressure as described elsewhere (16), the test plasmids were found to be maintained at levels of 40 to 100% compared to pALA136, which was maintained at a frequency of <2%. One plasmid with a high level of segregational stability which was chosen for further study contained a 4.8-kb insert. Subcloning refined the region within this insert required for stability to a 1,154-bp fragment (Fig. 1).
FIG. 1.
(A) Genetic organization of the stbDE genes and flanking regions from plasmid R485. Arrows indicate the length and orientation of open reading frames. Lines beneath the map indicate regions cloned in the stability probe vector, pALA136. The levels of retention conferred by these fragments after approximately 25 generations in the absence of selective pressure in a polA strain are shown. (B) Nucleotide sequence of the 1,154-bp ScaI-BglII fragment from plasmid R485. The inferred amino acid sequences of the four open reading frames are shown by the single-letter code.
Organization of the R485-derived stability region.
The 1,154-bp fragment contained four open reading frames which would be transcribed in the same direction (Fig. 1). One of these open reading frames extends beyond the region which was sequenced. Subcloning further delineated the stability locus to a 597-bp region which harbors two overlapping open reading frames (stbD and stbE) which potentially encode proteins of 83 and 93 amino acids, respectively. The stbD gene alone, as present on a ScaI-MluI fragment, was insufficient to confer stability (Fig. 1). Attempts to clone the stbE gene alone in pALA136 were unsuccessful. Plasmid constructs which were stabilized as a result of the presence of stbDE contained less than 50 bp of R485-derived sequences upstream of stbD (Fig. 1B). These sequences contained no close match to the E. coli ς70 consensus promoter sequence. It is likely that, in these constructs, the stbDE genes are expressed from a vector-derived promoter.
Identification of stbDE homologs.
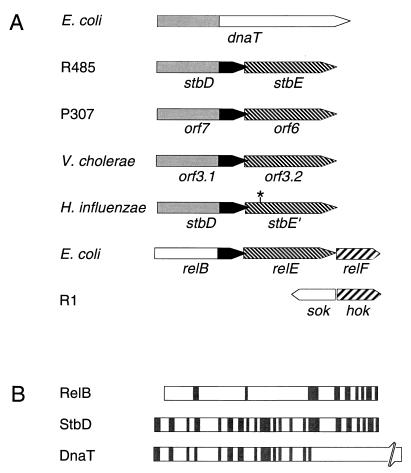
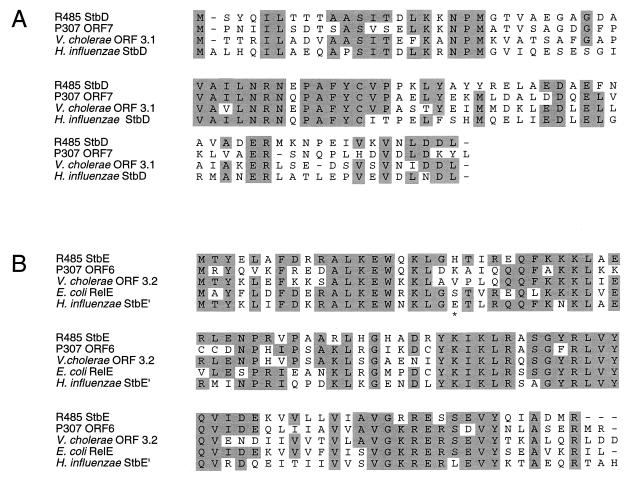
Database searching identified stbDE homologs of hitherto unknown function on the enterotoxigenic plasmid P307 (22) (GenBank accession no. M26308) and on the chromosomes of Vibrio cholerae (1) (GenBank accession no. X64097), the causative agent of cholera, and Haemophilus influenzae biogroup aegyptius (GenBank accession no. AF018635), the causative agent of Brazilian purpuric fever, which is a highly fatal pediatric septicemia (Fig. 2A). The stbE nucleotide sequence of the latter requires a +1 frameshift to maintain its integrity. In each case the stbD and stbE genes overlap, albeit by different numbers of nucleotides. Pairs of StbD proteins are collinear and share 37 to 54% identity; pairs of StbE proteins are collinear and share 55 to 60% identity (Fig. 3). Furthermore, StbE is collinear with and ∼60% identical to the RelE protein encoded by the E. coli chromosome whereas the 26 C-terminal amino acids of StbD and RelB are 46% identical (Fig. 2 and 3). The relB gene is positioned upstream of and overlaps with relE (Fig. 2A). Curiously, the third gene, in this operon is homologous to the hok gene, which encodes the toxin component of the Hok-Sok toxin-antitoxin stability system of plasmid R1 (5). The relBEF operon is involved in the stringent response to amino acid starvation (2). Components of this operon may have been recruited to form parts of chromosome stabilization mechanisms. Alternatively, as the relBEF operon appears not to be highly conserved, at least among those bacteria whose genomes have been sequenced, the chromosomal rel genes in E. coli may have been captured from plasmid-borne sequences.
FIG. 2.
(A) Comparative organizations of the stbDE genes from plasmid R485 and homologous genes from plasmid P307 and from the chromosomes of V. cholerae, E. coli, and H. influenzae biogroup aegyptius. Homologous regions are denoted by similar shadings. The asterisk indicates the position of the −1 frameshift in the stbE gene of H. influenzae biogroup aegyptius. (B) Homology between the RelB, DnaT, and putative StbD proteins. The DnaT and StbD proteins are aligned from their N termini, whereas the RelB and StbD proteins are aligned from their C termini. No gaps were introduced into the alignments. Shaded lines represent amino acids which are identical between proteins.
FIG. 3.
Homology in the StbD (A) and StbE (B) families of proteins. Residues which are identical in a majority of family members are shaded. Dashes indicate gaps introduced to optimize the alignments. Protein sequences were aligned with the PILEUP program (4) with subsequent manual modification. Note that for the StbE′ protein from H. influenzae biogroup aegyptius, a hypothetical +1 frameshift, indicated by an asterisk in the protein sequence, was introduced in the nucleotide sequence to maintain the integrity of the gene.
Whereas the C-terminal one-third of StbD and RelB share significant identity, the 58 N-terminal amino acids of StbD are 36% identical to the 58 N-terminal amino acids of DnaT, also encoded by the E. coli chromosome (Fig. 2). The homology between orf3.1 of V. cholerae and dnaT has previously been noted (1). StbD appears to be a natural hybrid between DnaT and RelB.
The stbDE homologs are functional stability cassettes.
The stbDE homologs from plasmid P307 and V. cholerae were subcloned as a 1,101-bp BglII-XmnI fragment in BamHI-EcoRV-cleaved pALA136 and as a 1,133-bp NsiI-BamHI fragment in PstI-BamHI-cleaved pFH450, respectively. Plasmid pFH450 is a derivative of pALA136 with a multiple cloning site. The P307- and V. cholerae-derived fragments conferred approximately 90 and 70% retention, respectively, following growth under nonselective conditions for approximately 25 generations in the polA strain, BR825. In contrast, the stbDE homolog from H. influenzae biogroup aegyptius, which was cloned as an end-filled 981-bp PmlI-BstXI fragment in HpaI-cleaved pFH450, conferred no detectable stability, which probably reflects the −1 frameshift in the N-terminal segment of the stbE gene.
Conclusions.
Plasmid R485 from the nosocomial pathogen M. morganii contains a pair of overlapping genes (stbDE) which confers a high level of segregational stability on a low-copy-number heterologous replicon in E. coli. Closely related genes, two of which were shown to be functional plasmid stability determinants, are present in a number of other gram-negative pathogens. The organization of the stbDE cassette is highly reminiscent of toxin-antitoxin stability cassettes which have been identified on a number of plasmids, viz., two overlapping genes encoding approximately 10-kDa proteins (6, 10, 11). The inability to clone stbE in the absence of stbD further suggests that the StbE protein may be toxic to its host. In toxin-antitoxin stability systems, the toxic activity of one protein is normally repressed by the partner antitoxin, which may be either a protein or an antisense RNA. When a plasmid-free variant arises, the antitoxin decays more rapidly than the toxin. This releases the latter to act on its intracellular target, which results in cell death or stasis. A number of different intracellular targets have been identified for different toxin-antitoxin systems (10). If the stbDE system is indeed proven to be a toxin-antitoxin system, the lack of homology between its components and components of other toxin-antitoxin systems suggests that the intracellular target of StbD-StbE differs from heretofore characterized targets. The homology between StbD and the essential host protein DnaT (17) fortuitously may provide a clue as to the target in this case: StbD may interact with a protein with which DnaT normally interacts, thereby poisoning the primosome of which DnaT is a component throughout replication (14, 19). Alternatively, the homology between StbD and DnaT may indicate that StbE can interact with both of these proteins through homologous residues or regions on these proteins. StbD and StbE may normally be physically associated, but in the absence of StbD, for example, following plasmid loss and StbD decay, StbE may substitute for a component of the primosome with which DnaT interacts, thereby disrupting primosome assembly or maturation. These hypotheses remain to be tested.
StbD appears to be a hybrid between DnaT and RelB, whereas StbE is homologous to RelE (Fig. 2). The potential significance of the homology between StbD and DnaT has been alluded to in the preceding section. The questions of the importance of the homology which the stbDE genes share with the relBEF operon, which itself does not function as a plasmid stability cassette (5), and why components of the relBEF operon have been hijacked as parts of both the stbDE and R1 hok-sok (5) stability systems remain to be elucidated. However, the homology which StbE and RelE share suggests a mechanism different from that proposed above by which the stbDE cassette might promote stability: StbE may interact with a factor with which RelE usually interacts, thereby disrupting a critical cellular function.
The stbDE homologs identified to date have been found only in pathogenic bacteria. This suggests that these genes may function in virulence or its control. In the case of the plasmid-located genes, the stbDE genes simply may contribute to virulence plasmid segregational stability. This hypothesis could be tested by inactivating the stbDE cassette on plasmids R485 and P307 and determining the effect on plasmid stability either in laboratory strains or in their natural hosts. In the instance of chromosomal stbDE genes, it is less clear how a toxin-antitoxin system can promote chromosome stability, although putative toxin-antitoxin systems also have been identified on the E. coli chromosome (10). The construction of chromosomal stbDE mutants in V. cholerae and the characteristics and virulence of the resulting mutants will assist in determining the biological significance of the chromosomal stbDE genes.
Nucleotide sequence accession number.
The nucleotide sequence of the 1,154-bp ScaI-BglII fragment from plasmid R485 has been deposited in the GenBank database under accession no. AF072126.
Acknowledgments
This work was funded by Wellcome Trust Research Career Development Fellowship 040822/Z/94/Z.
I thank Stuart Austin, Martine Couturier, Paul Manning, Leonard Mayer, Lyndsay Radnedge, and Andrew Spiers for generous gifts of strains and plasmids.
REFERENCES
- 1.Barker A, Clark C A, Manning P A. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J Bacteriol. 1994;176:5450–5458. doi: 10.1128/jb.176.17.5450-5458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bech F W, Jorgensen S T, Diderichsen B, Karlstrom O H. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couturier M, Bex F, Bergquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genetics Computer Group. Program manual for the GCG package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 5.Gerdes K, Bech F W, Jorgensen S T, Lobner-Olesen A, Rasmussen P B, Atlung T, Boe L, Karlstrom O, Molin S, von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdes K, Gultyaev A P, Franch T, Pedersen K, Mikkelsen N D. Antisense RNA-regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 8.Hedges R W, Datta N, Coetzee J N, Dennison S. R factors from Proteus morganii. J Gen Microbiol. 1973;77:249–259. doi: 10.1099/00221287-77-2-249. [DOI] [PubMed] [Google Scholar]
- 9.Hiraga S. Chromosome partition in Escherichia coli. Curr Opin Genet Dev. 1993;5:789–801. doi: 10.1016/s0959-437x(05)80100-5. [DOI] [PubMed] [Google Scholar]
- 10.Holcik M, Iyer V N. Conditionally lethal genes associated with bacterial plasmids. Microbiology. 1997;143:3403–3416. doi: 10.1099/00221287-143-11-3403. [DOI] [PubMed] [Google Scholar]
- 11.Jensen R B, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 12.Jones C S, Osborne D J, Stanley J. Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J Gen Microbiol. 1993;139:735–741. doi: 10.1099/00221287-139-4-735. [DOI] [PubMed] [Google Scholar]
- 13.Lin D C-H, Grossman A D. Identification and characterization of a bacterial chromosome partition site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Nurse P, Marians K J. The ordered assembly of the φX174 primosome. III. PriB facilitates complex formation between PriA and DnaT. J Biol Chem. 1996;271:15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]
- 15.Ludtke D N, Eichorn B G, Austin S J. Plasmid-partition functions of the P7 prophage. J Mol Biol. 1989;209:393–406. doi: 10.1016/0022-2836(89)90005-3. [DOI] [PubMed] [Google Scholar]
- 16.Martin K A, Friedman S A, Austin S J. Partition site of the P1 plasmid. Proc Natl Acad Sci USA. 1987;84:8544–8547. doi: 10.1073/pnas.84.23.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masai H, Arai K-I. Escherichia coli dnaT gene function is required for pBR322 plasmid replication but not for R1 plasmid replication. J Bacteriol. 1989;171:2975–2980. doi: 10.1128/jb.171.6.2975-2980.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohl D, Gober J. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 19.Ng J Y, Marians K J. The ordered assembly of the φX174 primosome. II. Preservation of primosome composition from assembly through replication. J Biol Chem. 1996;271:15649–15655. doi: 10.1074/jbc.271.26.15649. [DOI] [PubMed] [Google Scholar]
- 20.Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 21.Radnedge L, Davis M A, Youngren B, Austin S J. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J Bacteriol. 1997;179:3670–3675. doi: 10.1128/jb.179.11.3670-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saul D, Spiers A J, McAnulty J, Gibbs M G, Bergquist P L. Nucleotide sequence and replication characteristics of RepFIB, a basic replication region of IncF plasmids. J Bacteriol. 1989;171:2697–2707. doi: 10.1128/jb.171.5.2697-2707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherratt D J, Arciszewska L K, Blakely G, Colloms S, Grant K, Leslie N, McCulloch R. Site-specific recombination and circular chromosome segregation. Philos Trans R Soc Lond Biol Sci. 1995;347:37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- 24.Wake R G, Errington J. Chromosome partitioning in bacteria. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- 25.Williams D R, Thomas C M. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]