SUMMARY
The genomic architecture and molecular mechanisms controlling variation in quantitative disease resistance loci are not well understood in plant species and have been barely studied in long-generation trees. Quantitative trait loci mapping and genome-wide association studies were combined to test a large single nucleotide polymorphism (SNP) set for association with quantitative and qualitative white pine blister rust resistance in sugar pine. In the absence of a chromosome-scale reference genome, a high-density consensus linkage map was generated to obtain locations for associated SNPs. Newly discovered associations for white pine blister rust quantitative disease resistance included 453 SNPs involved in wide biological functions, including genes associated with disease resistance and others involved in morphological and developmental processes. In addition, NBS-LRR pathogen recognition genes were found to be involved in quantitative disease resistance, suggesting these newly reported genes are qualitative genes with partial resistance, they are the result of defeated qualitative resistance due to avirulent races, or they have epistatic effects on qualitative disease resistance genes. This study is a step forward in our understanding of the complex genomic architecture of quantitative disease resistance in long-generation trees, and constitutes the first step towards marker-assisted disease resistance breeding in white pine species.
Keywords: white pine blister rust, GWAS, QTL mapping, linkage mapping, sugar pine
INTRODUCTION
Plants have evolved sophisticated molecular responses to defend themselves from a variety of pathogens. Although pathogens may employ a variety of infection strategies, common molecular processes have been observed in immune responses including pathogen recognition, signal transduction, and defense responses (Corwin and Kliebenstein, 2017). Recent genomic and molecular methods employed in model and crop species have allowed a good understanding of the genes, gene families, and pathways involved in these processes (Nelson et al., 2018). Most of this knowledge, however, comes from the study of large-effect qualitative disease resistance loci involved in pathogen recognition, while our understanding of the molecular mechanisms controlling variation in small-effect quantitative disease resistance loci is still limited in plant species and almost non-existent in long-generation trees (Poland et al., 2009; Neale and Kremer, 2011; Kovalchuk et al., 2013; Corwin and Kliebenstein, 2017; Elfstrand et al., 2020). Greater attention to the study of disease responses is warranted in long-generation tree species as theoretical work suggests long-lived plants may (i) have higher levels of polymorphism and rates of evolution of disease resistance than short-lived plants (Bruns et al., 2015), (ii) be more reliant on systemic-induced resistance to respond to pathogens (Bonello et al., 2006), and (iii) have experienced expansions in important gene families related to defense (Hamberger et al., 2011; Porth et al., 2011; Warren et al., 2015; De La Torre et al., 2020).
Qualitative disease resistance is controlled by a single major gene (often referred to as resistance gene [R-gene]), which confers complete or near-complete resistance (often referred to as major gene resistance [MGR]), segregates as simple Mendelian loci producing discrete classes of susceptible and resistant individuals, and typically encodes proteins involved in pathogen recognition (Jones and Dangl, 2006). These large-effect genes are easier to detect in genome-wide association studies (GWAS) or quantitative trait locus (QTL) mapping; therefore, a body of literature has accumulated in different, mainly commercial, plant species (Nelson et al., 2018). In contrast, quantitative disease resistance is controlled by multiple genes of small effect which confer partial resistance and produce individuals with continuously varying (quantitative) resistance (Young, 1996; Quesada et al., 2010). As for most complex traits, dissecting the genomic basis of quantitative disease resistance has proven to be challenging, and the molecular mechanisms influencing phenotypic variation are not well understood (Nelson et al., 2018). Despite these challenges, a greater knowledge of this type of resistance is valuable for breeding purposes due to its more stable and durable nature (McDonald and Linde, 2002; Ayliffe et al., 2008). Although qualitative and quantitative disease resistance often have been studied as a dichotomy, some studies suggested they should be considered as extremes in a continuum. R-genes with partial quantitative resistance have been identified as QTLs in some species, suggesting overlap and interplay between MGR and quantitative resistance (Dowkiw and Bastien, 2007; Poland et al., 2009).
Cronartium ribicola, an exotic fungal pathogen causing white pine blister rust (WPBR), is currently a major threat to North American five-needle pines (subgenus Strobus) (Kinloch et al.,1970; Kinloch, 2003; Nesmith et al., 2019). Individuals impacted by WPBR have shown levels of qualitative (MGR), and/or quantitative resistance to the pathogen (Sniezko et al., 2008; King et al., 2010; Schoettle et al., 2014; Sniezko et al., 2014, 2020). MGR produces an hypersensitive response triggering rapid cell death in tissues surrounding the infection (Kinloch and Littlefield, 1977; St. Clair, 2010). Four MGR genes have been identified: Cr1 in sugar pine (Pinus lambertiana), Cr2 in western white pine (Pinus monticola; Kinloch et al., 1999), Cr3 in southwestern white pine (Pinus strobiformis; Kinloch and Dupper, 2002), and Cr4 in limber pine (Pinus flexilis; Schoettle et al., 2014). However, two avirulent strains of C. ribicola, capable of overcoming MGR, have been documented in western white pine and sugar pine (Kinloch et al., 2004). As a result, breeding programs have focused on assessing quantitative resistance after inoculating trees with avirulent strains (Sniezko et al., 2014). Despite the importance of WPBR quantitative resistance, the genetic basis is largely unknown. A pathogenesis-related gene in western white pine, PmCh4B, was found to be associated with quantitative resistance to WPBR through candidate gene-based association (Liu et al., 2011). However, large-scale genome-wide analyses are necessary to account for all segregating variation in quantitative traits and to reduce long breeding cycles through marker-assisted selection (Neale and Kremer, 2011).
Sugar pine is an economically and ecologically important species that is naturally distributed from Baja California (Mexico) to Oregon, with a latitudinal range of 30–43 degrees N, a longitudinal range of 115–124 degrees W, and an elevational range of 0–3.0 km. It is the only Strobus pine with a published reference genome (Stevens et al., 2016; Crepeau et al., 2017) and transcriptome (Gonzalez-Ibeas et al., 2016), and it also has multiple field site resources such as progeny trials and a two-generations full-sib cross designed for QTL mapping (Jermstad et al., 2011; Vázquez-Lobo et al., 2017). This paper aims to identify loci associated with WPBR quantitative resistance through the combination of genome-wide association studies (GWAS) and QTL and linkage mapping. Our main questions were the following. (i) What is the genomic architecture (number of genes, effect sizes, and their genomic locations) of WPBR quantitative resistance? (ii) Are genes conferring WPBR quantitative resistance mainly involved in defense or do they show wider biological functions? (iii) Are genomic responses to quantitative and qualitative disease resistance extremes along a continuum or do they represent a dichotomy?
RESULTS
Population structure
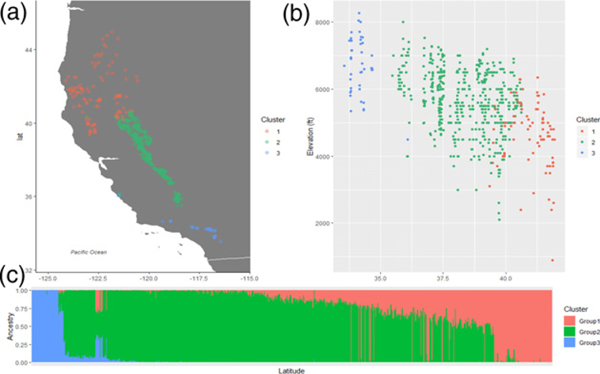
FastSTRUCTURE ancestry plots identified three distinctive genetic clusters distributed along latitude and elevation (Figure 1). Individuals from the Klamath Mountains and Northern Sierra (northern California and southern Oregon) clustered together in cluster 1, whereas individuals from the central Sierra and Transverse ranges (southern California) were separated in clusters 2 and 3, respectively. Similarly, the principal component analysis (PCA) based on the single nucleotide polymorphism (SNP) data also showed three potential clusters (Figure S1).
Figure 1.
Population structure based on fastSTRUCTURE results. (a) Geographic map showing genetic clusters in natural populations of sugar pine. (b) Genetic clusters along elevational gradients. (c) Barplot of ancestry levels per individual, as obtained by fastSTRUCTURE. Colors represent genetic clusters.
Genome-wide association study
The GWAS identified 30 SNPs that were significantly associated with quantitative disease resistance (percentage of progeny which had no symptoms or bark reactions after FDR multiple testing correction P < 0.05; Table S1). Significant SNPs were found on 22 different scaffolds with seven SNPs co-located on the same scaffold number 60 229 (linkage group 3 at 79.121 cM, valine-glutamine (VQ) gene PILA_28470 in the sugar pine reference genome v1.5). Minor allele frequencies for significant SNPs were between 0.051 and 0.493, with an average of 0.1109. Effect sizes for significant SNPs were between 6.2% and 14.5%, with an average of 9.3%, and no significant association was found between effect size and minor allele frequencies (Figure S2). Nineteen genes showed evidence of additive effects, and seven showed departures from additivity due to dominance effects. Heritability was estimated for quantitative resistance at 0.247. Traits not tested in the GWAS had heritability estimated at 9.97×10−6 (bark reactions), 2.59×10−4 (no recorded symptoms), 1.00×10−6 (survival), and 0.272 (normal cankers). Main functional categories included stress and defense response (VQ motif gene PILA_28470, no apical meristem [NAM] gene PILA_08207, microsomal glutathione S-transferase gene PILA_26414, and ubiquitin PPAR signaling pathway gene PILA_08019); carbohydrate metabolism (glucan endo-1–3-beta-glucosidase gene PILA_05171, UDP-glycosyltransferase gene PILA_05211, and phosphoglycerate gene PILA_30234); secondary metabolism (protochlorophyllide reductase gene PILA_29041 and serine carboxypeptidase-like gene PILA_20641), and cell wall organization (pectin biosynthesis ARAD1 exostosin gene PILA_09735).
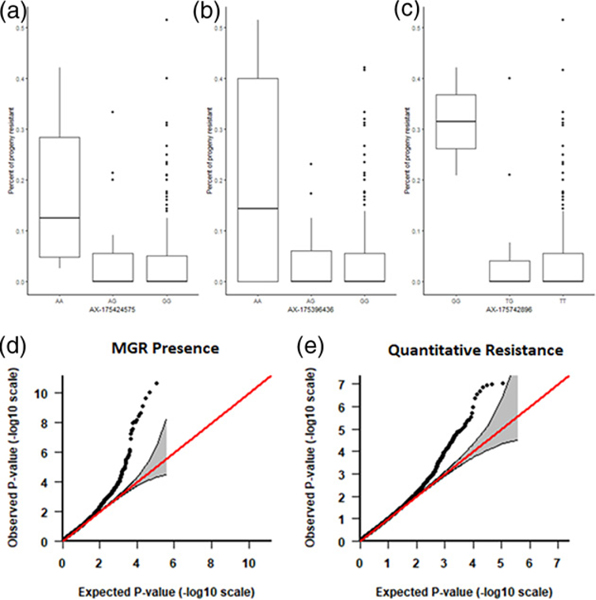
A total of 27 SNPs (from 17 genes) were identified as significantly associated with MGR status in parental trees (Table S2). Of these SNPs, three pairs of SNPs were co-located on the same scaffolds. Minor allele frequencies for these SNPs ranged between 0.051 and 0.284, with an average of 0.117. Effect sizes for significant SNPs ranged between 8.3% and 21.4%, with an average of 11.7%. There was evidence of additive effects for 13 of these SNPs, and dominance effects for four of them. Main functional categories of significant SNPs included biotic and abiotic stress in genes such as PILA_21059, leucine-rich repeat (LRR) genes PILA_07835 and PILA_16685; cytochrome P450 gene PILA_01359; glutathione peroxidase gene PILA_31072; MORC family CW-type zinc finger gene PILA_02308; WRKY transcription factor gene PILA_30972; and E3 ubiquitin-protein ligase gene PILA_18319. QQ-plots for both GWAS and boxplots for individual SNPs for quantitative resistance were generated (Figure 2).
Figure 2.
Boxplots for selected SNPs identified as associated with quantitative resistance in our GWAS. (a) AX-175424575 is associated with a gene involved in the stress response and protein binding. (b) AX-175396436 is associated with a gene with annotations relating to glycotransferase. (c) AX-175742896 is associated with a gene with peptidase activity. (d,e) QQ-plots for (d) parental major gene presence and (e) quantitative resistance.
Linkage map
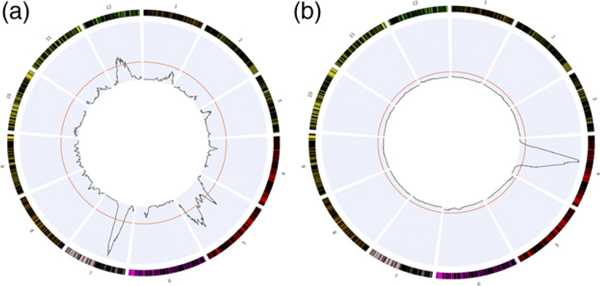
A consensus map containing 12 linkage groups (LGs) was generated through anchoring the SNPs of our two linkage maps (Figure 3, Table S3). The first of these maps contained 3949 SNPs that were co-located in 2012 unique locations, whereas the second map contained 4755 SNPs co-located in 2318 unique locations. We found 8159 SNPs that were heterozygous in both parents and retained 2075 after filtering for anchoring our linkage maps. The root mean squared error between maps had a maximum of 5.75 (LG 5) and a minimum of 1.16 (LG 1), with an average RMSE of 3.93 across all linkage groups. The consensus map contained 8702 SNPs in 5527 unique loci. SNPs were grouped into 12 linkage groups, with an average of 460.6 SNPs per linkage group, covering an average distance of 161.9 cM per linkage group. The total map length was 1943.1 cM (Table 1).
Figure 3.
Consensus linkage map for sugar pine showing 12 linkage groups and results of the QTL analysis. (a,b) LOD scores (black lines) for (a) backcross 1 and (b) backcross 2. The horizontal red line represents a 95% significance threshold for each test generated by randomly shuffling trait values relative to genetic information for 1000 permutations. The threshold is determined by the most extreme 5% of LOD scores generated by these permutations.
Table 1.
Summary of the consensus linkage map showing the length in centimorgans (cM), number of SNP markers, and root mean squared error (RMSE) for each of the 12 linkage groups (LGs) in sugar pine
| LG | Consensus map |
||
|---|---|---|---|
| Length | SNPs | RMSE | |
| 1 | 155.315 | 513 | 1.16 |
| 2 | 136.744 | 535 | 1.46 |
| 3 | 173.587 | 508 | 6.93 |
| 4 | 154.586 | 360 | 2.45 |
| 5 | 195.015 | 590 | 5.75 |
| 6 | 155.936 | 415 | 3.46 |
| 7 | 180.841 | 377 | 4.79 |
| 8 | 163.972 | 514 | 2.25 |
| 9 | 164.901 | 383 | 5.27 |
| 10 | 158.648 | 470 | 5.34 |
| 11 | 147.580 | 393 | 4.11 |
| 12 | 155.991 | 469 | 4.26 |
| Total | 1943.116 | 5527 | |
QTL mapping
QTLs associated with quantitative disease resistance (bark reactions or symptom-free) were found in five regions of four linkage groups (Figure 3, Table S4). Our first pseudobackcross identified two QTLs on LG 5 and a single QTL each on LGs 7 and 12. An additional QTL on LG 4 was identified by our second pseudobackcross. These regions of significant LOD contained 423 SNPs which lie within the identified QTL. Of these, 175 SNPs were located in coding regions of the sugar pine genome. Gene ontology analysis indicated biological processes involved in defense responses such as the response to oxidative stress, as well as cellular components involved in the cell wall. Ontologies also indicated processes which are not directly involved in defense, such as metabolic processes, signal transduction, and developmental processes. Defense response-related gene families identified in this analysis were zf-C3HC4 (PILA_08798), guanine nucleotide-binding protein (PILA_15748), mitogen-activated protein kinase (PILA_16422), and FA desaturase 2, which was also identified to be involved in the oxidative stress response (PILA_14905). Other gene families identified related to oxidative stress were the thioredoxin family (PILA_16337), the zf_PARP family (PILA_16990), and the SMART (COIL) family (PILA_26986). Gene families involved in cellular components of the cell wall were also observed.
QTL mapping analysis identified 12 SNPs from genes involved in NBS-LRR disease resistance distributed across three regions of the sugar pine genome. The first region contained two SNPs (PILA_30024 gene, super scaffold 5997) at position 59.81 cM of LG 7. Five SNPs (PILA_09968 gene, scaffold 47917) and one SNP (PILA_06586 gene, super scaffold 3789) were found on LG 12 at 66.08 and 68.89 cM, respectively. A group of three NBS-LRR-related genes (PILA_25059 scaffold 62717; PILA_23861 scaffold 100032; and PILA 28990 scaffold 60793) was found on LG 5 between 40.60 and 43.75 cM. In addition, PILA_24437 scaffold 420245 was found at 89.18 cM and PILA_04003 scaffold 72706 was found at 94.01 cM in LG 4 (Figure 3).
Gene enrichment analysis
The Biological Networks Gene Ontology (BiNGO) gene enrichment analysis of WPBR-associated sugar pine genes did not yield any significantly enriched ontologies after P-value correction for multiple testing.
Correlations between phenotype and environment
No significant correlation was observed between the presence/absence of MGR in parental trees and environmental variables. Significant correlations were found between parental tree mortality and latitude, longitude, mean annual precipitation, mean summer precipitation, annual heat moisture index, mean annual radiation, and extreme maximum temperature (Figure S3). The percentage of progeny with bark reactions and the percentage of progeny with no symptoms or bark reactions were both correlated to the mean annual radiation in the environment of parental trees (Figure S3). No significant correlation between latitude and quantitative disease resistance was found in the dataset (Figure S4).
DISCUSSION
Genomic architecture of WPBR quantitative disease resistance
This study found a largely polygenic basis of quantitative disease resistance, with hundreds of genes of mostly additive gene action and small effect sizes, conferring resistance to WPBR. This is coincident (i) with previous studies in other plant–fungus pathosystems in maize (Zea mays), soybean (Glycine max), and Arabidopsis (Arabidopsis thaliana) (reviewed in Corwin and Kliebenstein, 2017) and (ii) with expectations for highly polygenic complex traits in conifers (Neale and Wheeler, 2019). Both QTL mapping and GWAS analyses suggested a widespread genomic distribution of significant SNPs, with QTLs located in 9 of the 12 linkage groups. In the absence of a chromosome-scale reference genome for sugar pine, this is the best representation of the WPBR genomic architecture available to date. Clusters of significant SNPs were found in the QTL analysis but not in the GWAS analysis, as expected for highly outcrossing, long-generation species with large population sizes in which linkage disequilibrium decays rapidly in natural populations but slowly in mapping (controlled cross) populations. Recent genome-wide studies in long-generation trees have found that rare alleles play an important role in explaining phenotypic variance in complex traits (De La Torre et al., 2019; Piot et al., 2020). In our study, we also found evidence of a significant but low correlation between minor allele frequencies and effect sizes (Figure S2).
Quantitative disease resistance relies on a variety of mechanisms and gene families
Our study identified several gene families; some of them were MGR-related gene families previously identified in other white pines (such as LRRs), some were implicated in other forms of disease resistance, and others were not directly implicated in disease or defense responses. Collectively, these gene families indicated a breadth of mechanisms for the quantitative response of sugar pine to WPBR. These mechanisms include stress and defense responses such as pathogen detection, necrosis of infected cells, ubiquitin-dependent protein catalysis, the response to oxidative stress, and immune effector processes. Other related mechanisms involved cell wall and cell membrane processes; developmental processes involved in reproduction and regionalization; carbohydrate, lipid, phosphorus, DNA, and secondary metabolism; and signal transduction and regulation of translation (Tables S1 and S4).
Other significantly associated genes were not primarily involved in plant defense. NAM genes PILA_08207 and PILA_30970 were identified in both the QTL mapping and the GWAS. NAM genes are involved in floral development and are overexpressed in the boundaries between plant organs (Aida and Tasaka, 2006; Cheng et al., 2012), but have also been shown to be involved in biotic and abiotic stress responses (Tweneboah and Oh, 2017). This dual function may underlie previous findings indicating strong correlations between plant flowering time and disease resistance (Collins et al., 1999). Furthermore, expression data from Norway spruce (Picea abies), showed MA_10002g002 (a PILA_30970 ortholog) was overexpressed in both tissues related to apical growth (vegetative shoots and early season buds) as well as tissues responding to pests or pathogens (adelgid-infected needles) (Congenie.org; Nystedt et al., 2013). Genes regulating morphological and developmental traits have been hypothesized to confer quantitative disease resistance in species such as maize (Thompson and Bergquist, 1984; Bian et al., 2014), clover (Trifolium) (Bradley et al., 2003), and rice (Oryza sativa) (Albar et al., 1998). These findings further support this hypothesis and demonstrate the diverse mechanisms underlying quantitative resistance in sugar pine.
A large group of genes not directly involved in defense were identified as being involved in abiotic stress, with 15 of these genes identified by the QTL mapping and one identified in the GWAS. These genes were from a diverse group of families, including the PARP, NBS-LRR, thioredoxin, and heat shock protein (HSP)70 families. HSPs function by chaperoning other proteins to maintain their proper configuration (Lee et al., 2012). HSPs have been identified as important chaperones for NBS-LRR gene family members directly involved in disease resistance (Elmore et al., 2011), and an HSP70 family member in tobacco (Nicotiana tabacum) has been identified as necessary for the hypersensitive response (Kanzaki et al., 2003). Additionally, members of gene families which are primarily known for their involvement in disease resistance such as the NBS-LRR gene family and WRKY can also be involved in abiotic resistance. WRKY genes in Arabidopsis have been proposed as flexible transcription factors that play a role in both plant defense and abiotic stress. In rice, a single WRKY gene decreases resistance to rice blast while increasing cold tolerance when overexpressed. In addition to their functions in disease resistance, some NBS-LRR genes have been proposed to act as anti-freeze proteins, which confer additional cold tolerance (Muthukumaran et al., 2011). Overall, the numerous genes involved in both quantitative defense and abiotic stress indicate a potential for cross-talk in WPBR, although more research studies are needed to draw a conclusion.
Are genomic responses to quantitative and qualitative disease resistance extremes along a continuum or do they represent a dichotomy?
Although qualitative and quantitative disease resistance have mostly been studied as separate mechanisms, some studies suggested they might only be two ends of a continuum that vary from complete to partial resistance conferred by R-genes (Poland et al., 2009; Nelson et al., 2018). The reasoning behind this is that allelic variants of R-genes have been associated with quantitative resistance and have been found to be co-located with R-genes in species such as rice (Wang et al., 1994), maize (Xiao et al., 2007), and potato (Solanum tuberosum) (Gebhardt and Valkonen, 2001). In our study, we found a number of genes that belong to the same LRR gene family and are in close proximity (and likely in linkage disequilibrium) with previously discovered Cr1 alleles for MGR, but show partial quantitative resistance to WPBR. Potential explanations for this are the following: The newly reported genes are R-genes with partial resistance; they are the result of defeated MGR due to avirulent races; or they have epistatic effects on MGR.
The NBS-LRR gene family is widely implicated in qualitative disease response across a broad taxonomic scope for both hosts and pathogens, with evidence for conferring resistance to viral pathogens in potato (Boris et al., 2012), bacteria in both rice and Arabidopsis (Xu et al., 2018), and fungus in white pine species (Liu and Ekramoddoullah, 2007; Sniezko et al., 2014; Liu et al., 2017). The action of these genes is part of the first step of a plant immune system response that is triggered when a host perceives damage-associated molecular patterns as a consequence of a recent pathogen infection (Flor, 1971; Corwin and Kliebenstein, 2017).
This study identified 14 SNPs in genes involved in NBS-LRR quantitative disease resistance distributed across four regions of the sugar pine genome, including LGs 4, 5, 7, and 12 (Table 2). Although we could not map our LRR MGR-associated SNPs, our results suggest that previously identified scaffolds genetically or physically linked to Cr1 (Stevens et al., 2016) map to two of the same LGs 5 and 7 (Table S5). All of the associated LRR SNPs have low effect sizes, which is consistent with quantitative resistance but may also suggest partial MGR resistance.
Table 2.
GWAS and QTL mapping results showing LRR genes involved in both quantitative resistance and MGR responses in sugar pine
| Analysis | Resistance | Marker | Scaffold | Gene | LG | PFAM domain |
|---|---|---|---|---|---|---|
| GWAS | MGR | AX-175441014 | scaffold589011 | PILA_07835 | NA | NB-ARC, LRR_1, RPW8, NACHT |
| GWAS | MGR | AX-175545318 | super2806 | PILA_16685 | NA | LRR_1, Pkinase_Tyr, Pkinase, LRRNT_2 |
| QTL mapping | Quantitative | AX-175540379 | scaffold420245 | PILA_24437 | 4 | LRR_2 |
| QTL mapping | Quantitative | AX-175585823 | scaffold72706 | PILA_04003 | 4 | NB-ARC, TIR, LRR_1, NACHT, Arch_ATPase, LRR_3 |
| QTL mapping | Quantitative | seq-rs55884-SP | scaffold60793 | PILA_28990 | 5 | F-box, LRR_1 |
| QTL mapping | Quantitative | seq-rs56868-SP | scaffold62717 | PILA_25059 | 5 | LRR_1, Pkinase_Tyr, Pkinase, LRRNT_2 |
| QTL mapping | Quantitative | AX-175449157 | scaffold100032 | PILA_23861 | 5 | LRR_1, NB-ARC, TIR, LRR_3, NACHT |
| QTL mapping | Quantitative | AX-175681833 | super5997 | PILA_30024 | 7 | TIR, NB-ARC, LRR_1, DUF1863 |
| QTL mapping | Quantitative | AX-175719692 | super5997 | PILA_30024 | 7 | TIR, NB-ARC, LRR_1, DUF1863 |
| QTL mapping | Quantitative | AX-175472959 | super3789 | PILA_06586 | 12 | LRR_1, Pkinase_Tyr, LRRNT_2, Pkinase |
| QTL mapping | Quantitative | AX-175611159 | scaffold47917 | PILA_09968 | 12 | NB-ARC, TIR, LRR_1, NACHT, Arch_ATPase, LRR_3 |
| QTL mapping | Quantitative | AX-175668639 | scaffold47917 | PILA_09968 | 12 | NB-ARC, TIR, LRR_1, NACHT, Arch_ATPase, LRR_3 |
| QTL mapping | Quantitative | AX-175925756 | scaffold47917 | PILA_09968 | 12 | NB-ARC, TIR, LRR_1, NACHT, Arch_ATPase, LRR_3 |
| QTL mapping | Quantitative | AX-175948941 | scaffold47917 | PILA_09968 | 12 | NB-ARC, TIR, LRR_1, NACHT, Arch_ATPase, LRR_3 |
| QTL mapping | Quantitative | seq-rs37720-SP | scaffold47917 | PILA_09968 | 12 | NB-ARC, TIR, LRR_1, NACHT, Arch_ATPase, LRR_3 |
An alternative explanation is that the involvement of LRR genes in quantitative resistance is a result of defeated MGR due to avirulent races. Although R-genes are believed to be maintained by strong selection, pathogen evolution may reduce the strength and effectiveness of R-genes, converting them in quantitative resistance genes (Poland et al., 2009). This pattern of ‘defeated’ MGR has been observed in poplar (Populus) (Dowkiw and Bastien, 2007), rice (Li et al., 1999), and wheat (Triticum) (Nass et al., 1981; Brodny, 1986). In our study, our samples came from populations exposed to a vcr1 strain of C. ribicola which had overcome MGR in sugar pine; therefore, this hypothesis of ‘defeated’ MGR as a cause for LRR quantitative resistance cannot be excluded.
Finally, LRR quantitative resistance genes may have epistatic effects on the MGR response to WPBR. This is supported by previous mapping studies in other plant species in which major-effect qualitative disease resistance genes were found to be associated with small-effect quantitative resistance genes that epistatically affected the major-effect locus (Debener et al., 1991; Martin et al., 1993; Moscou et al., 2011). In the barley-stem rust pathosystem, quantitative disease resistance loci modulate the transcriptome to shape the pathogen recognition response of qualitative disease resistance loci (Druka et al., 2008; Moscou et al., 2011). Epistatic effects from other gene families are also expected on MGR since the strength of MGR in plants has previously been shown to be affected by other loci (Hu et al., 1997; Poland et al., 2009), and genes involved in molecular processes which are upstream and downstream of hypersensitive responses may modify disease resistance (Belkhadir et al., 2004).
Our study found other non-LRR genes with potential epistatic effects on the MGR response to WPBR in sugar pine. Associations with MGR were identified in loci scattered across 6 out of 12 linkage groups. They include a member of the MORC gene family located on LG 9. MORC genes are known to be involved in multiple mechanisms of plant defense, ranging from pathogen recognition to programmed cell death, in other plants (Lu et al., 2017). Both cell death and pathogen recognition are known to be important characteristics of the hypersensitive response induced by the Cr1 gene in sugar pine (Kinloch and Littlefield, 1977; Kinloch et al., 1999). Additionally, a MORC gene identified in Arabidopsis (AtMORC1) is a component of the hypersensitive response associated with LRR genes in the species. Another gene in this linkage group, encoding an E3 ubiquitin (PILA_18319), is also shown to be required for hypersensitive responses in Arabidopsis and rice (Zeng et al., 2004; Yang et al., 2006). Also of interest is a glutathione peroxidase gene (PILA_21059) identified in LG 6. These genes are mainly involved in reducing oxidative stress (Mittler et al., 2004). Hypersensitive responses in plants involve reactive oxidative bursts which signal the production of cellular protectants such as glutathione peroxidase in nearby cells (Tenhaken et al., 1995). Perhaps the most relevant gene detected by this analysis (PILA_30972) encodes a WRKY transcription factor. Chimeric proteins containing both NBS-LRR and WRKY domains have been shown to act as dual resistance genes in Arabidopsis, providing defense from multiple pathogens (Narusaka et al., 2009). Some NBS-LRR genes hypothesized to be involved in direct pathogen detection contain their own WRKY domains (DeYoung and Innes, 2006), further supporting this family’s role in regulating defense response of resistance genes.
Combining disease symptoms for GWAS leads to a higher heritability of quantitative resistance traits
Quantitative disease resistance is believed to involve multiple different mechanisms which may exhibit different symptoms as they respond to the disease (Kolpak et al., 2013). Furthermore, despite the continuous nature of inheritance of quantitative resistance, variation in quantitative resistance occurs in the presence or absence of various stem, bark, and needle symptoms (Liu et al., 2013). To account for this, past studies have used a 0–9 severity index based on combinations of these symptoms when measuring infection to WPBR. When measured using this severity index, trees from our common garden were highly bimodal. To address this bimodal distribution and ensure that symptoms were biologically related to resistance, we previously created a new scoring system in which trees with symptoms that were associated with higher survival (symptom-free and bark reactions) were classified as resistant, and trees with any other symptoms were classified as susceptible. Trees in this study were considered resistant if they had bark reactions or were entirely free of symptoms. Bark reactions have previously been associated with higher survival in Strobus pines exposed to C. ribicola in Pinus monticola and Pinus lambertiana (Sniezko et al., 2014). An unknown quantity of symptom-free trees may have escaped exposure. As such, their resistance would not be effectively determined by our measurements. Numerous trees recorded as clean in our 2016 QTL field trial were recorded as having disease symptoms in 2009, demonstrating that trees recorded as symptom-free may have simply recovered completely from previous symptoms. Combining these two traits was further supported by the higher heritability estimates for bark reactions and being symptom-free when measured together (0.247) than for either being symptom-free (2.59×10−4) or having bark reactions (9.97×10−6) when measured independently.
This study is a step forward in our understanding of the complex genomic architecture of quantitative disease resistance in the WPBR pathosystem. The new discovery of hundreds of genes of small effects involved in defense and other functions is a significant contribution towards marker-assisted breeding for disease resistance in sugar pine and other white pine species.
EXPERIMENTAL PROCEDURES
Sample collection and DNA extraction for whole-genome re-sequencing
Seeds from ten individuals spanning sugar pine’s natural distribution with the exception of Baja California were collected for genomic analysis. Prior to extraction, seeds were soaked in water at room temperature for 4 days, and haploid megagametophytes were dissected from each seed. DNA was extracted with a Qiagen DNeasy mini-prep Plant kit (Qiagen, Hilden, Germany) and DNA quality and concentration were evaluated using picogreen on a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Illumina’s TruSeq Nano DNA Library Prep Kit (Illumina Inc, San Diego, CA, USA) was used to construct libraries for sequencing. Steps prior to amplification included DNA fragmentation (200 ng starting material and 550 bp target insert size), followed by end repair and size selection of fragments, adenylation of 3′ ends, and ligation of adapters. PCR enrichment was performed in eight cycles. Barcoded libraries were combined into normalized pools and sequenced to greater than 10-fold coverage on an Illumina HiSeq 3000 using 150 bp paired-end reads at the University of California Davis Genome Center.
SNP calling
Raw reads from whole-genome re-sequencing data of 10 sugar pine individuals were aligned to the 34 GB sugar pine reference genome version 1.0 (Stevens et al., 2016) using Bowtie2 v2.2.9 (Langmead and Salzberg, 2012). SNP calling was done using SAMtools v1.3.1, followed by BEDtools v2.25.0 and BCFtools v1.3.1 (Li et al., 2009; Li and Barrett, 2011) using default parameters. A total of 715.9 million SNPs were called. Filtering criteria included the removal of SNPs with a quality <20, depth of coverage <8, mapping quality = 0, and indels. All SNPs were given a score based on the sum of 16mer frequency sums of the 30 bp forward or reverse adjacent to the SNP. When the score was higher than 300, SNPs were discarded. Only SNPs present in scaffolds of 1 kb or larger were called. Selected SNPs were later re-mapped to reference genome version 1.5 (Crepeau et al., 2017; https://treegenesdb.org/FTP/Genomes/Pila/v1.5).
Sample collection and DNA extraction for genotyping
Seeds from populations spanning the species’ natural geographic range were collected from the Placerville gene bank in California. In addition, needles were collected from a previously established two-generation full-sib cross in Happy Camp, northern California. Prior to extraction, seeds were soaked in water and 30% hydrogen peroxide (3%) overnight. Eight to ten megagametophyte haploid tissues for each family were pooled together to infer the maternal genotype. DNA was extracted from needles and megagametophytes using the Qiagen DNeasy mini-prep Plant kit and an Eppendorf automated pipetting workstation. The extraction protocol included 1 day of tissue lysis and incubation at 96°C, followed by several steps of precipitation and filtering. DNA quality and concentration were assessed using nanopore and picogreen on a Qubit Fluorometer, respectively.
SNP genotyping and filtering
Samples were genotyped using two SNP arrays, a 600 k Affymetrix (Thermo Fisher Scientific) SNP array and a custom-based multi-species Illumina Infinium SNP array comprising 80 k SNP markers from which 20 k were designed for Douglas fir and 60 k for sugar pine. Genome Studio 2.0.4 (Illumina, 2015) was used to call genotypes, filter, and generate genotyping statistics for all samples and SNPs from the Illumina SNP array. Genotype calling and SNP filtering for the Affymetrix SNP array was done with Axiom Analysis Suite (Version3.1.51.0 Applied Biosystems, Thermo Fisher Scientific). Individuals were discarded from the Affymetrix analysis which had Dish QC < 0.82 and QC call rate < 86.2. Individuals from the Illumina array were discarded if they had a GenCall threshold < 0.15 and a call rate < 0.7 for the GWAS analysis or < 0.8 for the QTL mapping. SNPs from the Affymetrix analysis that were of conversion types other than NoMinorHom (genotyping data above thresholds and only two clusters observed) and PolyHighRes (genotyping data above thresholds with polymorphic SNPs) were discarded. These SNPs were further filtered with call rates ≤ 0.01 removed from further analysis. Illumina data were filtered to discard SNPs with Cluster Separation ≤ 0.1 and call frequency ≤ 0.8 for the GWAS analysis or ≤ 0.7 for the QTL mapping. The combined SNP arrays from both platforms resulted in 1015 individuals with 125 236 SNPs for the GWAS analysis, and 616 individuals (614 progeny and two parents) with 88 200 SNPs for QTL mapping and linkage map construction. The distribution of minor allele frequencies for all SNPs can be found in Figure S5.
Population structure
Population structure was determined using PCA with the Adegenet R package (Jombart, 2008). Individuals in the PCA were clustered using a k-means clustering algorithm in the R package factoextra (1–10 clusters with 100 bootstraps). The optimal number of clusters was selected using the silhouette algorithm in the same R package. A Bayesian cluster analysis using fastSTRUCTURE (Raj et al., 2014) was also conducted using 10 independent runs of K = 2–10. Each run used 80–90 iterations, with an average of 88 iterations per run. The optimal value of K, representing the number of genetic lineages, was selected using the program chooseK.py (Raj et al., 2014). Ten replicates of each cluster analysis were aligned and visualized using CLUMPP (Jakobsson and Rosenberg, 2007). Input files for both Adegenet and fastSTRUCTURE were created using Plink v 1.07 (Purcell et al., 2007).
Phenotypic data from common gardens
Phenotypic data were obtained from two previously established common gardens maintained by the US Forest Service, and located at the Happy Camp Outplant Site, northern California. In both common gardens the secondary host for the pathogen, Ribes spp., was grown between rows to ensure inoculation of study trees. Since all trees in the study were exposed to a virulent strain of C. ribicola (vcr1) which overcomes MGR, any WPBR resistance seen in the trial can be assumed to be quantitative. Trees grown in the common gardens were assessed for disease phenotypes based on the presence of the following symptoms: normal active cankers, normal active blights, normal bark reaction, blights, bark reactions, no disease symptoms (clean). Survival and cause of death was also assessed, and were used to create a combined category for progeny that had died from rust.
In the first common garden, full-sib progeny from putatively resistant parent trees from throughout the species’ natural range were screened for the presence of MGR and later grown and evaluated for qualitative resistance, vigor, and survival. These progeny trees were planted in a randomized complete block design between years 1986 and 2003 with measurements taken between years 1994 and 2010. In the second common garden, a two-generation full-sib controlled cross between two individuals (5038 × 5500) whose offspring had previously exhibited a high level of quantitative resistance was established in June 2000 (Jermstad et al., 2011). Phenotypes and genotypes were obtained for 614 individuals in this common garden as well as genotype information for both parents. The maternal parent (5038) was the same individual used to generate the sugar pine reference genome (Stevens et al., 2016; Crepeau et al., 2017). Due to the Mendelian inheritance of Cr1, progeny testing gave accurate information about the genotype of the parent trees growing in natural populations. We used 955 of these parent trees’ genotypes (RR, Rr, rr) for association mapping (see below).
Genome-wide association study
In this study, two different GWAS were performed. In the first one, parental trees that had more than 10 progeny alive at the time of measurement or had progeny older than 5 years were selected for analyses resulting in a total of 280 trees with combined phenotypic and genotypic data. Parental trees were assessed for disease resistance based on the symptoms observed in their progeny grown in a common garden. Progeny trees that were symptom-free or had bark reactions were classified as resistant. Bark reactions, unlike other WPBR symptoms, are associated with increased survival in white pines. Principal components with highest eigenvalues (Figure S1) were used to account for population structure. A kinship matrix was used to account for relatedness. Both principal components and kinship were incorporated as co-variates in the following mixed linear model:
where genotype data, kinship, and population structure are fixed effects represented by , random additive effects are represented by , and residuals are represented bye.
In the second GWAS, parental trees’ presence/absence of MGR were treated as a binary trait. Homozygous or heterozygous individuals having MGR were recorded as resistant, and those that lacked any MGR alleles were recorded as susceptible. This included 955 trees with both MGR and genotypic information. Heritability was estimated for each trait using genetic and residual variance calculated in TASSEL. Additive effects, dominant effects, and effect sizes (proportion of phenotypic variance explained by the SNP marker) were also calculated in TASSEL.
Linkage map construction
Linkage maps were developed from SNPs sequenced in individuals from a two-generations controlled cross (see above). Before mapping, two pseudobackcrosses were generated by selecting SNPs segregating as homozygous for one parent and heterozygous for the other. SNPs were examined for phase changes and filtered in ASMap v.0.4 (Taylor and Butler, 2017) and R/qtl (Broman et al., 2003) R packages. Initial maps were constructed with ASMap (Taylor and Butler, 2017) using the MSTmap function with the Kosambi distance function and a P-value threshold of 1×10−6. Pairwise estimations of logarithm of the odds (LOD), obtained from a test of linkage disequilibrium, and pairwise recombination (r) were obtained for each SNP pair. Co-located SNPs (r = 0) were placed in the same bins with ASMap.
A consensus map (in this case, the same as the sex-averaged map) was created by merging the maps from the two parental backcrosses described above. Co-located SNPs from backcrosses were placed into bins containing 3047 and 3353 loci, respectively, in order to reduce loci for mapping to those representing unique locations. Additional SNPs segregating as heterozygous for both parents were filtered to exclude SNPs that deviated significantly from Hardy–Weinberg equilibrium (P < 0.05) or had more than 3% missing data. These SNPs were mapped together with SNPs from ASMap in Joinmap V.5 (Van Ooijen, 2018) to serve as anchors for an averaged sex map. Before mapping with Joinmap, SNPs were filtered to exclude SNPs with LOD scores higher than 1 and SNPs with a locus genotyping frequency > 0.01. The two maps for each of 12 linkage groups were merged to create averaged sex maps in LPmerge (Endelman and Plomion, 2014). The package was used to generate 10 consensus maps for each of the 12 linkage groups, each with a different maximum interval ranging from 1 to 10. Consensus maps for each linkage group were selected by choosing the maximum interval which resulted in the lowest average root mean squared error. The final consensus map was plotted with Circos v0.69–5 (Krzywinski et al., 2009). Linkage groups for SNPs which were found to be significant in the GWAS but which did not segregate in the linkage map were determined by matching scaffold information between GWAS SNPs and SNPs used in the consensus map.
QTL mapping
WPBR quantitative resistance was also evaluated using QTL mapping. As in the GWAS analysis, individuals that were symptom-free or only had bark reactions were classified as resistant. Resistance status was then used as a binary trait for interval mapping in R/qtl (Broman et al., 2003). The expectation-maximization algorithm was used for mapping. A permutation test (n = 1000) was run for each model to determine a 5% significance level for LOD scores. Functional annotations for each gene associated with a significant SNP were obtained from the PILA.1_5.functionalannotations.tsv file at the TreeGenes database (treegenesdb.org) (Baker et al., 2018; Falk et al., 2018; Wegrzyn et al., 2019). Annotations for SNPs represented by coding sequences were taken from the reference sugar pine genome V1.5, file CDS.FA from the TreeGenes database (Baker et al., 2018; Falk et al., 2018; Wegrzyn et al., 2019).
Gene enrichment analysis
A gene enrichment analysis was performed using a hypergeometric test in BiNGO v.3.0.3 (Maere et al., 2005). This analysis was performed separately for a set of genes identified by the GWAS for parental MGR status, and for a combined set of all genes identified from both the GWAS and QTL for quantitative disease resistance. Gene ontology terms were considered enriched if P-values were lower 0.05 after FDR correction.
Phenotype by environment correlations
Correlations between the natural environment of parental trees (included in the GWAS study) and disease traits for parents and offspring were examined. Latitude, longitude, and 22 environmental variables obtained from ClimateWNA (Wang et al., 2016) were examined for correlations with parental tree mortality, presence/absence of MGR in parental trees, the percentage of progeny that had bark reactions, and the percentage of progeny with bark reactions or no symptoms. Correlations between these variables and heatmaps were done in R (version 3.6.1).
Supplementary Material
Figure S1. Population structure based on PCA results. (a) PCA of SNP data with PC1 and PC2 represented by the X and Y axes. Clusters are differentiated by color. (b) Geographic location of genetic clusters across the species’ distribution range. (c) The number of clusters was selected using the silhouette algorithm in the R package factoextra testing 1–10 clusters with 100 bootstraps. The silhouette algorithm suggested three distinct clusters for our PCA. (d) The first five principal components explain most of the variation in the dataset.
Figure S2. Minor allele frequency versus effect size for all SNPs in this study.
Figure S3. Heatmap of correlations between phenotypes and environment. Color legend specifies whether correlation was positive or positive. Rows marked with “X” denote non-significant (P < 0.05) correlations.
Figure S4. Correlation between WPBR quantitative resistance and genetic clusters identified in Figure 1. Ancestry correlates with latitude and elevation; therefore no significant correlation with quantitative resistance was found for any of these variables (F(1, 273) = 1.5, P = 0.7).
Figure S5. Minor allele frequency distribution for all SNPs in this study.
Table S1. Results from the GWAS for WPBR quantitative disease resistance showing significant SNPs (FDR-corrected P < 0.05). Columns indicate the SNP marker, the scaffold position for each SNP, linkage group positions, and the linkage group ID for each SNP. Also included are additive and dominance effects (add_effect, dom_effect) for each SNP, F values from the F-test for additive and dominance effects (add_F, dom_F) and P-values for the F-test for additive and dominance effects (add_P, dom_P), minor allele frequencies (MAFs), structural and functional annotations based on version 1.5 of the sugar pine reference genome, and gene ontologies.
Table S2. Results from the GWAS for WPBR qualitative disease resistance (parental MGR) showing significant SNPs (FDR-corrected P < 0.05). Columns indicate the SNP marker, scaffold position for each SNP, and linkage group positions for each SNP. Also included are additive and dominance effects (add_effect, dom_effect) for each SNP, F values from the F-test for additive and dominance effects (add_F, dom_F) and P-values for the F-test for additive and dominance effects (add_P, dom_P), minor allele frequencies (MAFs), structural and functional annotations based on version 1.5 of the sugar pine reference genome, and gene ontologies.
Table S3. Consensus linkage map for 8703 SNPs distributed in 12 linkage groups in sugar pine. Variables include SNP marker, position (pos, in cM), linkage group (LG), and scaffold position in the sugar pine reference genome.
Table S4. Results of the QTL mapping for WPBR quantitative disease resistance showing SNPs from regions determined to be associated with QTLs. Variables include consensus linkage group (LG), position (pos, in cM), logarithm of the odds (LOD), SNP marker, scaffold ID, structural and functional annotations from version 1.5 of the sugar pine reference genome, and gene ontologies.
Table S5. Scaffolds associated with the major resistance gene Cr1 in sugar pine that contained SNPs mapped in our linkage map. Variables include scaffold ID, SNP ID, location of the SNP in the linkage group (cM), and linkage group number (LG).
ACKNOWLEDGMENTS
This project was supported by the US Department of Agriculture/National Institute of Food and Agriculture (award # 2017-67013-26214) awarded to ADLT and DBN at the University of California, Davis, and by the NAU School of Forestry new faculty start-up funds awarded to ADLT. The authors would like to thank Deems Burton and Dean Davis for establishing and maintaining the common gardens and for their WPBR assessments; staff at the Dorena Genetic Resource Center for the 2016 assessment of the QTL trial; Randi Famula for lab support; and students who helped with DNA extraction such as Annalisa Romero, Rosa Mateos, Mai-Khan Tran, and Justin Ndihokubwayo.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
DATA AVAILABILITY STATEMENT
Sequencing raw reads are deposited in the NCBI SRA (https://www.ncbi.nlm.nih.gov/sra) under bioproject PRJNA174450.
References
- Aida M. and Tasaka M. (2006) Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol 9, 72–77. [DOI] [PubMed] [Google Scholar]
- Albar L, Lorieux M, Ahmadi N, Rimbault I, Pinel A, Sy AA, Fargette D. and Ghesquière A (1998) Genetic basis and mapping of the resistance to rice yellow mottle virus. I. QTLs identification and relationship between resistance and plant morphology. Theor. Appl. Genet 97, 1145–1154. [Google Scholar]
- Ayliffe M, Singh R. and Lagudah E. (2008) Durable resistance to wheat stem rust needed. Curr. Opin. Plant Biol 11, 187–192. [DOI] [PubMed] [Google Scholar]
- Baker EAG, Wegrzyn JL, Sezen UU et al. (2018) Comparative transcriptomics among four white pine species. G3: Genes - Genomes Genetics, 8, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Subramaniam R. and Dangl JL (2004) Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol 7, 391–399. [DOI] [PubMed] [Google Scholar]
- Bian Y, Yang Q, Balint-Kurti PJ, Wisser RJ and Holland JB (2014) Limits on the reproducibility of marker associations with southern leaf blight resistance in the maize nested association mapping population. BMC Genom, 15, 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello P, Gordon TR, Herms DA, Wood DL and Erbilgin N. (2006) Nature and ecological implications of pathogen-induced systemic resistance in conifers: a novel hypothesis. Physiol. Mol. Plant Pathol 68, 95–104. [Google Scholar]
- Boris KV, Ryzhova NN and Kochieva EZ (2012) NBS-ARC domain sequence variations in RX1 homologues of cultivated and wild potato species. Mol. Biol 46, 107–109. [PubMed] [Google Scholar]
- Bradley DJ, Gilbert GS and Parker IM (2003) Susceptibility of clover species to fungal infection: the interaction of leaf surface traits and environment. Am. J. Bot 90, 857–864. [DOI] [PubMed] [Google Scholar]
- Brodny U. (1986) The residual and interactive expressions of “defeated” wheat stem rust resistance genes. Phytopathology, 76, 546. [Google Scholar]
- Broman KW, Wu H, Sen S. and Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics, 19, 889–890. [DOI] [PubMed] [Google Scholar]
- Bruns E, Hood ME and Antonovics J. (2015) Rate of resistance evolution and polymorphism in long- and short-lived hosts. Evolution, 69, 551–560. [DOI] [PubMed] [Google Scholar]
- Cheng X, Peng J, Ma J, Tang Y, Chen R, Mysore KS and Wen J. (2012) NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol. 195, 71–84. [DOI] [PubMed] [Google Scholar]
- Collins A, Milbourne D, Ramsay L, Meyer R, Chatot-Balandras C, Oberhagemann P, De Jong W, Gebhardt C, Bonnel E. and Waugh R. (1999) QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol. Breed 5, 387–398. [Google Scholar]
- Corwin JA and Kliebenstein DJ (2017) Quantitative resistance: more than just perception of a pathogen. Plant Cell, 29, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepeau MW, Langley CH and Stevens KA (2017) From pine cones to read clouds: rescaffolding the megagenome of sugar pine (Pinus lambertiana). G3: Genes - Genomes - Genetics, 7, 1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Piot A, Liu B, Wilhite B, Weiss M. and Porth I. (2020) Functional and morphological evolution in gymnosperms: a portrait of implicated gene families. Evol. Appl 13, 210–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Puiu D, Crepeau MW, Stevens K, Salzberg SL, Langley CH and Neale DB (2019) Genomic architecture of complex traits in loblolly pine. New Phytol. 221, 1789–1801. [DOI] [PubMed] [Google Scholar]
- Debener T, Lehnackers H, Arnold M. and Dangl JL (1991) Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1, 289–302. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ and Innes RW (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol 7, 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowkiw A. and Bastien C. (2007) Presence of defeated qualitative resistance genes frequently has major impact on quantitative resistance to Melampsora larici-populina leaf rust in P. × interamericana hybrid poplars. Tree Genet. Genomes, 3, 261–274. [Google Scholar]
- Druka A, Potokina E, Luo Z. et al. (2008) Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia graminis f. sp. tritici in barley. Theor. Appl. Genet 117, 261–272. [DOI] [PubMed] [Google Scholar]
- Elfstrand M, Baison J, Lunden K. et al. (2020) Association genetics identifies a specifically regulated Norway spruce laccase gene, PaLAC5, linked to Heterobasidion parviporum resistance. Plant Cell Environ. 43(7), 1779–1791. [DOI] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJD and Coaker G. (2011) Plant NB-LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol 14, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman JB and Plomion C. (2014) Genome analysis LPmerge: an R package for merging genetic maps by linear programming. Bioinformatics, 30, 1623–1624. [DOI] [PubMed] [Google Scholar]
- Falk T, Herndon N, Grau E. et al. (2018) Growing and cultivating the forest genomics database, TreeGenes. Database, 2018, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH (1971) Current status of the gene-for-gene concept. Annu. Rev. Phytopathol 9, 275–296. [Google Scholar]
- Gebhardt C. and Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol 39, 79–102. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ibeas D, Martinez-Garcia PJ, Famula RA, Delfino-Mix A, Stevens KA, Loopstra CA, Langley CH, Neale DB and Wegrzyn JL (2016) Assessing the gene content of the megagenome: sugar pine (Pinus lambertiana). G3: Genes - Genomes - Genetics, 6(12), 3787–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Ohnishi T, Hamberger B, Seguin A. and Bohlmann J´ (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 157(4), 1677–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Webb CA and Hulbert SH (1997) Adult plant phenotype of the Rp1-DJ compound rust resistance gene in maize. Phytopathology, 87, 236–241. [DOI] [PubMed] [Google Scholar]
- Jakobsson M. and Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801–1806. [DOI] [PubMed] [Google Scholar]
- Jermstad KD, Eckert AJ, Wegrzyn JL, Delfino-Mix A, Davis DA, Burton DC and Neale DB (2011) Comparative mapping in Pinus: sugar pine (Pinus lambertiana Dougl.) and loblolly pine (Pinus taeda L.). Tree Genet. Genomes, 7, 457–468. [Google Scholar]
- Jombart T. (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405. [DOI] [PubMed] [Google Scholar]
- Jones JDG and Dangl JL (2006) The plant immune system. Nature, 444 (7117), 323–329. [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H. and Terauchi R. (2003) Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol 4, 383–391. [DOI] [PubMed] [Google Scholar]
- King JN, David A, Noshad D. and Smith J. (2010) A review of genetic approaches to the management of blister rust in white pines. Forest Pathol. 40, 292–313. [Google Scholar]
- Kinloch BB, Parks GK and Fowler CW (1970) White pine blister rust: simply inherited resistance in sugar pine. Science, 167, 193–195. [DOI] [PubMed] [Google Scholar]
- Kinloch BB (2003) White pine blister rust in North America: past and prognosis. Phytopathology, 93, 1044–1047. [DOI] [PubMed] [Google Scholar]
- Kinloch BB and Dupper GE (2002) Genetic specificity in the white pineblister rust pathosystem. Phytopathology, 93, 278–280. [DOI] [PubMed] [Google Scholar]
- Kinloch BB, Sniezko RA, Barnes GD and Greathouse TE (1999) A major gene for resistance to white pine blister rust in western white pine from the western cascade range. Phytopathology, 89, 861–867. [DOI] [PubMed] [Google Scholar]
- Kinloch BB, Sniezko RA and Dupper GE (2004) Virulence gene distribution and dynamics of the white pine blister rust pathogen in western North America. Phytopathology, 94, 751–758. [DOI] [PubMed] [Google Scholar]
- Kinloch BB Jr and Littlefield JL (1977) White pine blister rust: hypersensitive resistance in sugar pine. Can. J. Bot 55, 1148–1155. [Google Scholar]
- Kolpak SE, Sniezko RA and Kegley AJ (2013) Rust infection and survival of 49 Pinus monticola families at a field site six years after planting. Ann. Forest Res 51, 67–80. [Google Scholar]
- Kovalchuk A, Kerio S, Oghenekaro AO, Jaber E, Raffaello T. and¨ Asiegbu FO (2013) Antimicrobial defenses and resistance in forest trees: challenges and perspectives in a genomic era. Annu. Rev. Phytopathol 51, 221–244. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ and Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yun HS and Kwon C. (2012) Molecular communications between plant heat shock responses and disease resistance. Mol. Cells, 34, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Barrett J. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G. and Durbin R. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Luo LJ, Mei HW et al. (1999) A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol. Gen. Genet 261, 58–63. [DOI] [PubMed] [Google Scholar]
- Liu J-J and Ekramoddoullah AKM (2007) The CC-NBS-LRR subfamily in Pinus monticola: targeted identification, gene expression, and genetic linkage with resistance to Cronartium ribicola. Phytopathology, 97, 728–736. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Sniezko RA and Ekramoddoullah AKM (2011) Association of a Novel Pinus monticola Chitinase Gene (PmCh4B) with quantitative resistance to Cronartium ribicola. Phytopathology, 101, 904–911. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Sniezko RA, Zamany A, Williams H, Wang N, Kegley A, Savin DP, Chen H. and Sturrock RN (2017) Saturated genic SNP mapping identified functional candidates and selection tools for the Pinus monticola Cr2 locus controlling resistance to white pine blister rust. Plant Biotechnol. J 15, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Hammett C. and Sniezko RA (2013) Pinus monticola pathogenesis-related gene PmPR10–2 alleles as defense candidates for stem quantitative disease resistance against white pine blister rust (Cronartium ribicola). Tree Genet. Genomes, 9, 397–408. [Google Scholar]
- Lu H, Klessig DF, Kogel K-H, Koch A, Kang H-G, Steinbrenner J. and Dempsey MA (2017) MORC proteins: novel players in plant and animal health. Front. Plant Sci 8, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K. and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics, 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED and Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science, 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- McDonald BA and Linde C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol 40, 349–379. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M. and Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Lauter N, Steffenson B. and Wise RP (2011) Quantitative and qualitative stem rust resistance factors in barley are associated with transcriptional suppression of defense regulons. PLoS Genet. 7, e1002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumaran J, Manivel P, Kannan M, Jeyakanthan J. and Krishna R. (2011) A framework for classification of antifreeze proteins in over wintering plants based on their sequence and structural features. J. Bioinform. Seq. Anal 3, 70–88. [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M. and Narusaka Y. (2009) RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Nass HA, Pedersen WL, Mackenzie DR and Nelson RR (1981) The residual effects of some defeated powdery mildew Erysiphe-Graminis-F-Sp-Tritici resistance genes in isolines of winter wheat. Phytopathology, 71, 1315–1318. [Google Scholar]
- Neale DB and Kremer A. (2011) Forest tree genomics: growing resources and applications. Nat. Rev. Genet 12, 111–122. [DOI] [PubMed] [Google Scholar]
- Neale DB and Wheeler NC (2019) The Conifers. The conifers: genomes, variation and evolution. Switzerland: Springer. 10.1007/978-3-319-46807-5 [DOI] [Google Scholar]
- Nelson R, Wiesner-Hanks T, Wisser R. and Balint-Kurti P. (2018) Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet 19, 21–33. [DOI] [PubMed] [Google Scholar]
- Nesmith JCB, Wright M, Jules ES, McKinney ST, Nesmith JCB, Wright M, Jules ES and McKinney ST (2019) Whitebark and foxtail pine in Yosemite, Sequoia, and Kings Canyon National Parks: initial assessment of stand structure and condition. Forests, 10, 35. [Google Scholar]
- Nystedt B, Street NR and Wetterbom A. et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature, 497, 579–584. [DOI] [PubMed] [Google Scholar]
- Piot A, Prunier J, Isabel N, Klápště J, El-Kassaby YA, Villarreal Aguilar JC and Porth I. (2020) Genomic diversity evaluation of populus trichocarpa germplasm for rare variant genetic association studies. Front. Genet 10, 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC and Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. [DOI] [PubMed] [Google Scholar]
- Porth I, Hamberger B, White R. and Ritland K. (2011) Defense mechanisms against herbivory in Picea: sequence evolution and expression regulation of gene family members in the phenylpropanoid pathway. BMC Genom, 12, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada T, Gopal V, Cumbie WP, Eckert AJ, Wegrzyn JL, Neale DB, Goldfarb B, Huber DA, Casella G. and Davis JM (2010) Association mapping of quantitative disease resistance in a natural population of loblolly pine (Pinus taeda L.). Genetics, 186, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M. and Pritchard JK (2014) fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics, 197, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoettle AW, Sniezko RA, Kegley A. and Burns KS (2014) White pine blister rust resistance in limber pine: evidence for a major gene. Phytopathology, 104, 163–173. [DOI] [PubMed] [Google Scholar]
- Sniezko R, Smith J, Liu J-J and Hamelin R. (2014) Genetic resistance to fusiform rust in southern pines and white pine blister rust in white pines—a contrasting tale of two rust pathosystems—current status and future prospects. Forests, 5, 2050–2083. [Google Scholar]
- Sniezko RA, Kegley AJ and Danchok R. (2008) White pine blister rust resistance in North American, Asian and european species - results from artificial inoculartion trials in Oregon. Ann. Forest Res 51, 53–66. [Google Scholar]
- Sniezko RA, Johnson JS and Savin DP (2020) Assessing the durability, stability, and usability of genetic resistance to a non-native fungal pathogen in two pine species. PLANTS, PEOPLE, PLANET, 2, 57–68. [Google Scholar]
- St.Clair, D.A. (2010) Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol 48, 247–268. [DOI] [PubMed] [Google Scholar]
- Stevens KA, Wegrzyn JL, Zimin A. et al. (2016) Sequence of the sugar pine megagenome. Genetics, 204, 1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. and Butler D. (2017) R package ASMap: efficient genetic linkage map construction and diagnosis. J. Stat. Softw 79, 1–29.30220889 [Google Scholar]
- Tenhaken R, Levine A, Brisson LF, Dixon RA and Lamb C. (1995) Function of the oxidative burst in hypersensitive disease resistance. Proc. Natl Acad. Sci. USA, 92, 4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DL and Bergquist RR (1984) Inheritance of mature plant resistance to Helminthosporium maydis Race 0 in maize 1. Crop Sci. 24, 807–811. [Google Scholar]
- Tweneboah S. and Oh SK (2017) Biological roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in solanaceous crops. J. Plant Biotechnol 44, 1–11. [Google Scholar]
- Van Ooijen JW (2018) JoinMap 5, Software for the calculation of genetic linkage maps in experimental populations of diploid species. Wageningen, Netherlands: JoinMap, Kyazma B.V. [Google Scholar]
- Vázquez-Lobo A, De La Torre AR, Martínez-García PJ et al. (2017) Finding loci associated to partial resistance to white pine blister rust in sugar pine (Pinus lambertiana Dougl.). Tree Genet. Genomes, 13, 108. [Google Scholar]
- Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC and Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics, 136, 1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Hamann A, Spittlehouse D. and Carroll C. (2016) Locally downscaled and spatially customizable climate data for historical and future periods for North America. PLoS One, 11(6), e0156720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL, Keeling CI, Yuen MMS et al. (2015) Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J. 83, 189–212. [DOI] [PubMed] [Google Scholar]
- Wegrzyn JL, Staton MA, Street NR et al. (2019) Cyberinfrastructure to improve forest health and productivity: the role of tree databases in connecting genomes, phenomes, and the environment. Front. Plant Sci 10, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Zhao J, Fan S, Li L, Dai J. and Xu M. (2007) Mapping of genome-wide resistance gene analogs (RGAs) in maize (Zea mays L.). Theor. Appl. Genet 115, 501–508. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu F, Zhu S. and Li X. (2018) The maize NBS-LRR gene ZmNBS25 enhances disease resistance in rice and arabidopsis. Front. Plant Sci 9, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CW, Gonzalez-Lamothe R, Ewan RA, Rowland O, Yoshioka H,´ Shenton M, Ye H, O’Donnell E, Jones JDG and Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell, 18, 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND (1996) QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol 34, 479–501. [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Baek HN, Leung H. and Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell, 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Population structure based on PCA results. (a) PCA of SNP data with PC1 and PC2 represented by the X and Y axes. Clusters are differentiated by color. (b) Geographic location of genetic clusters across the species’ distribution range. (c) The number of clusters was selected using the silhouette algorithm in the R package factoextra testing 1–10 clusters with 100 bootstraps. The silhouette algorithm suggested three distinct clusters for our PCA. (d) The first five principal components explain most of the variation in the dataset.
Figure S2. Minor allele frequency versus effect size for all SNPs in this study.
Figure S3. Heatmap of correlations between phenotypes and environment. Color legend specifies whether correlation was positive or positive. Rows marked with “X” denote non-significant (P < 0.05) correlations.
Figure S4. Correlation between WPBR quantitative resistance and genetic clusters identified in Figure 1. Ancestry correlates with latitude and elevation; therefore no significant correlation with quantitative resistance was found for any of these variables (F(1, 273) = 1.5, P = 0.7).
Figure S5. Minor allele frequency distribution for all SNPs in this study.
Table S1. Results from the GWAS for WPBR quantitative disease resistance showing significant SNPs (FDR-corrected P < 0.05). Columns indicate the SNP marker, the scaffold position for each SNP, linkage group positions, and the linkage group ID for each SNP. Also included are additive and dominance effects (add_effect, dom_effect) for each SNP, F values from the F-test for additive and dominance effects (add_F, dom_F) and P-values for the F-test for additive and dominance effects (add_P, dom_P), minor allele frequencies (MAFs), structural and functional annotations based on version 1.5 of the sugar pine reference genome, and gene ontologies.
Table S2. Results from the GWAS for WPBR qualitative disease resistance (parental MGR) showing significant SNPs (FDR-corrected P < 0.05). Columns indicate the SNP marker, scaffold position for each SNP, and linkage group positions for each SNP. Also included are additive and dominance effects (add_effect, dom_effect) for each SNP, F values from the F-test for additive and dominance effects (add_F, dom_F) and P-values for the F-test for additive and dominance effects (add_P, dom_P), minor allele frequencies (MAFs), structural and functional annotations based on version 1.5 of the sugar pine reference genome, and gene ontologies.
Table S3. Consensus linkage map for 8703 SNPs distributed in 12 linkage groups in sugar pine. Variables include SNP marker, position (pos, in cM), linkage group (LG), and scaffold position in the sugar pine reference genome.
Table S4. Results of the QTL mapping for WPBR quantitative disease resistance showing SNPs from regions determined to be associated with QTLs. Variables include consensus linkage group (LG), position (pos, in cM), logarithm of the odds (LOD), SNP marker, scaffold ID, structural and functional annotations from version 1.5 of the sugar pine reference genome, and gene ontologies.
Table S5. Scaffolds associated with the major resistance gene Cr1 in sugar pine that contained SNPs mapped in our linkage map. Variables include scaffold ID, SNP ID, location of the SNP in the linkage group (cM), and linkage group number (LG).
Data Availability Statement
Sequencing raw reads are deposited in the NCBI SRA (https://www.ncbi.nlm.nih.gov/sra) under bioproject PRJNA174450.