Abstract
SecA is a dynamic protein that undergoes ATP-dependent membrane cycling to drive protein translocation across the Escherichia coli inner membrane. To understand more about this process, azide-resistant (azi) and signal sequence suppressor (prlD) alleles of secA were studied. We found that azide resistance is cold sensitive because of a direct effect on protein export, suggesting that SecA-membrane interaction is regulated by an endothermic step that is azide inhibitable. secG function is required for expression of azide-resistant and signal sequence suppressor activities of azi and prlD alleles, and in turn, these alleles suppress cold-sensitive and export-defective phenotypes of a secG null mutant. These remarkable genetic observations support biochemical data indicating that SecG promotes SecA membrane cycling and that this process is dependent on an endothermic change in SecA conformation.
Export of preproteins across the inner membrane of Escherichia coli has been studied extensively during the past decade. Both biochemical and genetic approaches have resulted in the identification of many if not all of the proteinaceous components of the translocation machinery (30, 36). Precursor proteins, synthesized with an amino-terminal signal peptide, associate with chaperones, such as SecB protein, which maintains them in an export-competent conformation (29). They are then targeted to the plasma membrane, where they associate with the translocase complex, which is composed of the membrane-dissociable SecA protein and the integral membrane protein, SecYEG, which are thought to form a translocation channel (5, 9, 14, 19). Central to preprotein assembly at the translocase complex is SecA protein, a 204-kDa homodimeric protein, which has been shown to bind the signal peptide and mature region of the preprotein, SecB protein, anionic phospholipids, and the amino-terminal portion of SecY protein (1, 4, 8, 15, 17, 20, 32). SecA regulates these diverse interactions and drives protein translocation by its ATPase activity (23, 34). ATP binding to SecA catalyzes the initial insertion of preprotein into the membrane, while hydrolysis promotes translocation across the membrane (31). Protein translocation appears to depend on the ability of SecA to undergo multiple cycles of membrane insertion and retraction, and such SecA-membrane cycling has been shown to depend on the function of the high-affinity ATP-binding domain of SecA (11, 28). This biochemical behavior of SecA has led to a model in which SecA has been hypothesized to act like a molecular ratchet, utilizing its membrane cycling activity to translocate proteins (12). Recently, however, it has been argued that protein translocation can occur under conditions in which SecA is permanently imbedded in the plasma membrane (6). In addition to SecA-membrane cycling, it has been found that SecG protein undergoes a topology inversion during protein translocation, and this event appears to be coupled to the insertion and retraction cycle of SecA protein, suggestive of a mechanistic linkage of these two processes (25).
Sodium azide is a known inhibitor of many ATPases, and azide-resistant mutants of Escherichia coli, denoted azi, have been found to be alleles of secA (13, 16, 26). Previous studies indicate that azide inhibits the translocation ATPase activity of SecA (26). Azide has been shown to trap SecA in the membrane-inserted state, as judged by the formation of a protease-resistant and membrane-protected 30-kDa fragment of SecA (35). In addition, previous genetic studies of signal sequence suppressor alleles of secA, denoted prlD, found that most such strains are altered in their azide sensitivity or resistance, thus indicating a strong interconnection between these two properties of SecA protein (16). Most prlD alleles are located in or adjacent to the ATP-binding domains of SecA, which govern SecA membrane cycling (16, 17, 23). In order to understand more about SecA function and its regulation, we have performed further genetic characterization of azi and prlD mutants.
The strains used in this study are described in Table 1, and where necessary they were constructed by P1 transduction (22). The growth medium employed in this study has been described previously (22). The concentrations of ampicillin, kanamycin, and tetracycline used were 100, 15, and 10 μg ml−1, respectively. Sodium azide was purchased from Mallinckrodt.
TABLE 1.
Characteristics of the bacterial strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| CK1801 | MC4100 Δ(uncB-uncC) | Carol Kumamoto |
| DG100 | KN370 leu::Tn10 | This study |
| DG100.2 | KN370 secG+ argG::Tn10 | This study |
| DG101 | KN370 prlD22 leu::Tn10 | This study |
| DG101.2 | KN370 prlD22 secG+ argG::Tn10 | This study |
| DG104 | KN370 prlD20 leu::Tn10 | This study |
| DG309 | KN370 azi-4 leu::Tn10 | This study |
| DG309.2 | KN370 azi-4 secG+ argG::Tn10 | This study |
| DG311 | KN370 prlD5 leu::Tn10 | This study |
| DG313 | KN370 prlD2 leu::Tn10 | This study |
| DG313.2 | KN370 prlD2 secG+ argG::Tn10 | This study |
| DG1801 | CK1801 ΔsecG::Kan | This study |
| DO168 | MC4100 leu::Tn10 | Laboratory stock |
| DO309 | MC4100 azi-4 | 26 |
| DO312 | MC4100 azi-7 | 26 |
| DO315 | MC4100 azi-6 | 26 |
| DO318 | MC4100 azi-9 | 26 |
| JH101 | STA14D prlD22 leu::Tn10 | 16 |
| JH104 | STA14D prlD20 leu::Tn10 | 16 |
| KB311 | STA14D prlD5 leu::Tn10 | 16 |
| KB313 | STA14D prlD2 leu::Tn10 | 16 |
| KB315 | STA14D prlD4 leu::Tn10 | 16 |
| KN370 | C600 recD1009 ΔsecG::Kan | 24 |
| LG800 | STA14D prlD43 | 16 |
| MC4100 | F−araD139 relA1 thi rpsL150 flB5301 Δ(argF-lac)U169 deoC7 ptsF25 rbsR | 26 |
| MM2 | MC4100 malE14-1 | 2 |
| MM100 | MM2 leu::Tn10 | This study |
| MM101 | MM2 prlD22 leu::Tn10 | This study |
| MM104 | MM2 prlD20 leu::Tn10 | This study |
| MM309 | MM2 azi-4 leu::Tn10 | This study |
| MM311 | MM2 prlD5 leu::Tn10 | This study |
| MM313 | MM2 prlD2 leu::Tn10 | This study |
| RV100 | MM100 ΔsecG::Kan | This study |
| RV101 | MM101 ΔsecG::Kan | This study |
| RV104 | MM104 ΔsegG::Kan | This study |
| RV309 | MM309 ΔsecG::Kan | This study |
| RV311 | MM311 ΔsecG::Kan | This study |
| RV313 | MM313 ΔsecG::Kan | This study |
| RV400 | STA14D leu::Tn10 ΔsecG::Kan | This study |
| RV401 | STA14D leu::Tn10 azi-4 ΔsecG::Kan | This study |
| RV402 | STA14D leu::Tn10 prlD5 ΔsecG::Kan | This study |
| RV403 | STA14D leu::Tn10 prlD20 ΔsecG::Kan | This study |
| RV404 | STA14D leu::Tn10 prlD22 ΔsecG::Kan | This study |
| RV405 | STA14D leu::Tn10 prlD43 ΔsecG::Kan | This study |
| STA14D | MC4100 lamB14D | 16 |
Azide resistance is cold sensitive.
Since previous studies showed that the insertion of SecA into the membrane involves a temperature-dependent unfolding of the protein (33) and azide prevents retraction of SecA from the membrane (35), we were interested in determining the effect of temperature on azide resistance. azi and prlD mutants that are normally azide resistant at 37°C were tested for their ability to form colonies on Luria-Bertani (LB) plates containing azide at various temperatures. Remarkably, none of the azi or prlD mutants that were azide resistant at 37 or 42°C were able to grow on LB plates containing 1 mM azide at 20°C, although growth was normal on LB plates (Table 2). The azi and prlD mutants showed reduced azide resistance at 30°C, where single-colony formation was inhibited at 1 mM azide and growth was blocked completely at 2 mM azide. These findings indicate that azide resistance is cold sensitive.
TABLE 2.
Azide resistance is cold sensitivea
| Allele | Growth at tempb:
|
|||
|---|---|---|---|---|
| 20°C | 30°C | 37°C | 42°C | |
| secA+ | − | − | − | − |
| prlD2 | − | +/− | + | + |
| prlD4 | − | +/− | + | + |
| prlD5 | − | +/− | + | + |
| prlD22 | − | +/− | + | + |
| azi-4 | − | +/− | + | + |
| azi-6 | − | +/− | + | + |
| azi-7 | − | +/− | + | + |
| azi-9 | − | +/− | + | + |
Strains were streaked on LB plates containing 1 mM sodium azide and incubated overnight at the indicated temperature. The following strains were used: DO168 (secA+), KB313 (prlD2), KB315 (prlD4), KB311 (prlD5), JH101 (prlD22), DO309 (azi-4), DO315 (azi-6), DO312 (azi-7), and DO318 (azi-9).
−, no growth; +, growth with single colonies; +/−, growth without single colonies. Parallel controls showed that all strains grew well at these temperatures on LB plates lacking sodium azide.
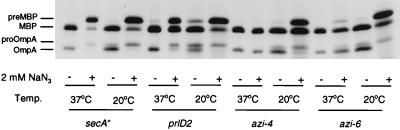
To determine whether the cold sensitivity of azide resistance correlates with reduction in the rate of protein secretion under these conditions, we determined the effect of temperature and azide on the rate of processing of maltose-binding protein (MBP) and OmpA in azi and prlD mutants. The rate of protein processing is a valid measure of the rate of protein secretion, given the topology of signal peptidase I (7). Even in the absence of azide, lowering the growth temperature of the prlD2 mutant to 20°C reduced the rate of protein secretion, particularly for OmpA (Fig. 1), indicating that protein export was somewhat cold sensitive in this case. Furthermore, while protein secretion in the prlD and azi mutants was substantially resistant to the effects of azide at 37°C, it was not resistant at 20°C. These data demonstrate that protein export in azi and prlD strains is phenotypically azide sensitive at low growth temperatures.
FIG. 1.
Analysis of protein secretion of azi and prlD mutants at low temperature in the presence or absence of azide. Strains (left to right: DO168, KB313, DO309, and DO315) were grown in M63 minimal medium (22) containing 0.4% glycerol, 0.4% maltose, and 20 μg (each) of 18 amino acids (lacking cysteine and methionine) ml−1 at 37°C until the mid-logarithmic phase, when portions of each culture were shifted to the indicated temperature. After 20 min, sodium azide was added to a final concentration of 2 mM to the indicated cultures. Five minutes later, a 0.5-ml aliquot of each culture was pulse-labeled with 10 μCi of Tran 35S-label (>1,000 Ci mmol−1; ICN) for 1 min, followed by the addition of an equal volume of ice-cold 10% trichloroacetic acid. MBP and OmpA were immunoprecipitated and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography as described previously (26). Three times more sample was loaded on the gel for the samples labeled at 20°C. The positions of the precursor and mature forms of MBP (preMBP and MBP, respectively) and OmpA (proOmpA and OmpA, respectively) are given.
Azide resistance is SecG dependent.
Since both SecG and azide have been suggested to affect SecA membrane cycling (25, 35), we were interested in studying the effect of secG function on azide resistance. Using P1 transduction, azi and prlD alleles were introduced into KN370 containing a secG deletion. Remarkably, all azi or prlD ΔsecG double mutants were unable to form colonies on LB plates containing 1 mM azide at 37°C (Table 3). To show that this result was directly due to the lack of secG function and was not an effect of the general strain background, these strains were transformed with plasmids containing secG, secE and secY, or secE, secY, and secG expressed from the trc promoter (10). An azide-resistant phenotype was recovered only in strains containing plasmids with secG, demonstrating the importance of this gene in azide resistance. Since overproduction of SecYE protein alone was insufficient to promote an azide-resistant phenotype, it seems unlikely that the involvement of secG in this case was indirect, for example, by promoting a higher level of activity of SecYE protein.
TABLE 3.
secG is required for azide resistance, while azi and prlD alleles suppress the cold sensitivity of ΔsecG strainsa
| Strain genotype | Growth atb:
|
||||
|---|---|---|---|---|---|
| 37°C with 1 mM azide
|
20°C with no plasmid | ||||
| No plasmid | secG | secEY | secEYG | ||
| ΔsecG secA+ | − | − | − | − | − |
| ΔsecG prlD2 | − | + | − | + | + |
| ΔsecG prlD5 | ND | ND | ND | ND | + |
| ΔsecG prlD20 | ND | ND | ND | ND | − |
| ΔsecG prlD22 | − | + | − | + | + |
| ΔsecG azi-4 | − | + | − | + | + |
To test for azide resistance, strains were streaked onto LB plates without (control) or with 1 mM sodium azide and incubated overnight at 37°C. To test for cold sensitivity, logarithmic-phase cultures were plated at a 106 dilution onto duplicate LB plates and incubated either overnight at 37°C (control) or for 3 days at 20°C. Colony formation was then scored. The following strains were used: DG100 (ΔsecG), DG313 (ΔsecG prlD2), DG311 (ΔsecG prlD5), DG104 (ΔsecG prlD20), DG101 (ΔsecG prlD22), and DG309 (ΔsecG azi-4). The plasmids used were pTrcG (secG+), pTrcHA-EY (secE+ secY+), and pTrcHA-EYG (secE+ secY+ secG+).
+, growth; −, no growth; ND, not done.
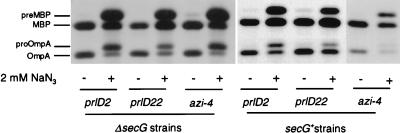
To determine whether the observed azide-sensitive phenotype of azi or prlD ΔsecG double mutants correlates with a reduced rate of protein export under these conditions, the rate of MBP and OmpA secretion was investigated. While azide addition caused a significant inhibition of MBP and OmpA secretion in the azi and prlD single mutants, more severe inhibition of protein export was noted for the azi or prlD ΔsecG double mutants (Fig. 2). These effects were not due to a decrease in SecA protein levels in any of these strains as monitored by pulse-labeling and immunoprecipitation or by Western blotting (up to 2 h in the presence of 2 mM azide [data not shown]). These data are consistent with the observed azide sensitivity of growth of the azi or prlD ΔsecG strains.
FIG. 2.
Analysis of the azide sensitivity of protein secretion of azi or prlD ΔsecG double mutants. Strains (from left to right: DG313, DG101, DG309, DG313.2, DG101.2, and DG309.2) were grown at 37°C, treated with sodium azide (where indicated), and radiolabeled, and MBP and OmpA were analyzed as described in the legend to Fig. 1.
Deletion of secG results in a cold-sensitive phenotype in certain strain backgrounds, such as C600 and W3110 (24). In some cases, the cold-sensitive phenotype is manifested only when the unc genes encoding F1F0-ATPase are deleted also (10). MC4100 ΔsecG mutants were not cold sensitive, whether they contained unc+ or ΔuncB-C alleles (results not shown), indicating that the status of the unc locus need not determine the cold-sensitive phenotype of secG mutants. The reason for such variation among different strain backgrounds is unclear. Because MC4100 ΔsecG derivatives are not cold sensitive, we tested azi and prlD derivatives of this strain for the dependence of azide resistance on secG function. Even in this strain background, we found that secG function affected the level of azide resistance, although not as severely as KN370 derivatives. azi and prlD MC4100 derivatives were able to form colonies on LB plates containing 3.5 mM azide, while their isogenic ΔsecG counterparts were only able to form colonies on LB plates containing 1 mM azide, with the exception of the prlD22 ΔsecG mutant, which formed colonies on LB plates containing up to 2 mM azide (results not shown).
Cold sensitivity caused by secG deletion can be suppressed by azi and prlD alleles.
Loss of secG function leads to cold sensitivity of growth and an accumulation of preproteins at low temperature (24). Overexpression of acidic phospholipids has been shown to suppress this phenotype (18), suggesting that the loss of secG function may relate to a defect in SecA-membrane binding or insertion (which requires anionic phospholipids [3, 33]). Since purified SecA proteins containing the azi and prlD mutations displayed increased membrane ATPase activity, even at 28°C (28a), indicating enhanced SecA-membrane interaction, we speculated that azi and prlD alleles may be able to suppress the cold sensitivity of secG mutants. To this end, the growth property of the azi or prlD ΔsecG derivatives was investigated. Nearly all of these double mutants were able to form colonies on LB plates at 20°C (Table 3), indicating that azi and prlD alleles suppressed the cold sensitivity caused by the secG mutation. One exception to this pattern of suppression was the prlD20 ΔsecG mutant, which was unable to form colonies at 20°C. The lack of suppression in this case seems to relate to the cold sensitivity of prlD20 strains (data not shown). Our findings are consistent with those of an earlier study showing that secA36, which leads to azide resistance, can suppress a secG defect in protein export at 20°C (21).
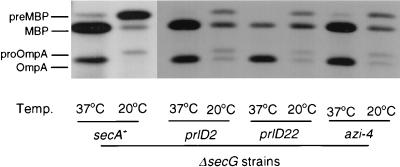
In order to determine whether the growth observed for azi or prlD ΔsecG double mutants at low temperatures correlates with an increase in the rate of protein export, secretion of MBP and OmpA was investigated. The azi or prlD ΔsecG mutants displayed an increased rate of protein secretion at 20°C compared to the isogenic ΔsecG parent (Fig. 3). These results suggest that a direct mechanism of suppression of the ΔsecG growth defect by these secA alleles is most probable.
FIG. 3.
Analysis of protein secretion of azi or prlD ΔsecG double mutants at low temperature. Strains (from left to right: DG100, DG313, DG101, and DG309) were grown at 37°C, shifted to the temperature indicated for 20 min, and subjected to radiolabeling, and MBP and OmpA were analyzed as described in the legend to Fig. 1, except that an equal amount of each sample was loaded on the gel.
Signal sequence suppressor activity of prlD alleles is secG dependent.
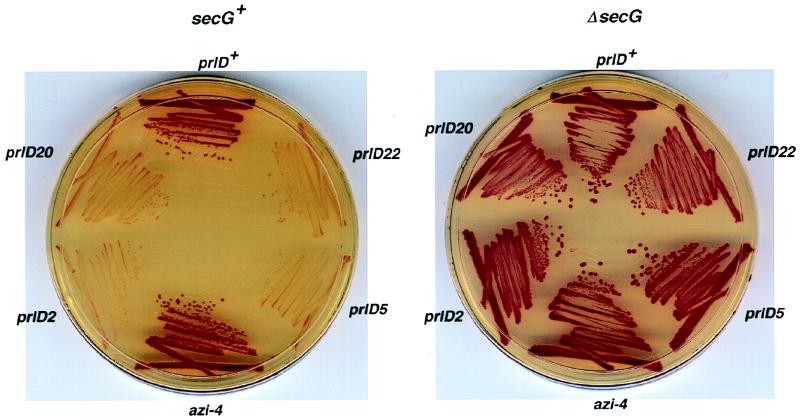
Since secG function is needed for azide resistance, we were interested whether it was also required for the signal sequence-suppressor activity of prlD alleles, since both of these properties would result in an increased demand on the protein translocation system. To this end, prlD or azi alleles were introduced into secG+ or ΔsecG derivatives of MM2 that contain the malE14-1 allele, which is a defect in the signal sequence of MBP (2). As expected, prlD secG+ strains were able to suppress the signal sequence defect, resulting in a Mal+ phenotype, while the azi-4 secG+ strain was Mal− (Fig. 4). Remarkably, all prlD ΔsecG double mutants were Mal−, indicating that secG function is required for prlD-mediated signal sequence suppression. In a second assay system that measures lamB14D signal sequence suppressor activity by the strain’s sensitivity to lambda phage adsorption and killing by cross-streaking, all prlD ΔsecG double mutants had a lambda-resistant phenotype, except for the prlD22 ΔsecG double mutant (prlD22 is the strongest signal sequence suppressor [16]), which was weakly lambda sensitive (data not shown). We conclude that secG function is needed for the expression of the signal sequence suppressor activity of prlD alleles.
FIG. 4.
secG function is required for signal sequence suppression by prlD alleles. prlD or azi MM2 derivatives with (secG+) or without (ΔsecG) secG function were tested for their ability to suppress the malE14-1 signal sequence mutation by overnight growth on maltose-tetrazolium plates at 37°C. Red colonies (Mal−) indicate little or no suppressor activity, while white colonies (Mal+) indicate good suppressor activity.
In this work, we have shown a remarkable genetic interaction between the SecA and SecG proteins. This conclusion is based on the fact that the azide-resistant and signal sequence-suppressor properties of azi and prlD mutants are SecG dependent, and yet azi and prlD alleles suppress the cold sensitivity of secG mutants. The requirement of SecG for expression of the azi and prlD phenotypes presumably relates to the ability of SecG to increase the pool of biochemically activated, SecYEG-bound SecA protein. Thus, in the azi or prlD secG double mutants, there is insufficient activated SecA protein to promote the azide-resistant and signal sequence suppression activities of SecA, which place an unusual demand on the translocation process. However, there is sufficient activated SecA protein under this circumstance to promote normal protein translocation at low temperature in the absence of secG function. The ability of these mutations to activate SecA protein in the absence of secG function as well as to promote suppression of signal sequence defects may relate to their predicted destabilization of the compact quaternary structure of SecA protein that is likely to require a conformational change to promote biochemical activation (based on the atomic structure of SecA) (16a).
A second important conclusion from our study is that azide resistance is cold sensitive. It is tempting to speculate that this cold sensitivity is another manifestation of the inherent cold sensitivity of the protein export process that has been observed previously (27) and that its biochemical basis rests on an endothermic transition of SecA conformation that has been noted previously (3, 33), and which is required to promote SecA membrane interaction and cycling. Presumably SecG, as well as perhaps other Sec components, normally helps to overcome this cold-sensitive step, which is azide inhibitable and which can be partially compensated for by azi and prlD mutations, thereby bypassing the strict requirement for SecG function. This step is likely to correspond to one in the membrane insertion-retraction cycle of SecA, consistent with the ability of azide to block SecA membrane retraction and SecG to promote SecA membrane cycling (25, 35). Further elucidation of this complex system will require biochemical studies that are currently under way.
Acknowledgments
We thank John Hunt for productive discussions on SecA structure and mechanism and Bill Wickner, Hajime Tokuda, and Tom Silhavy for provision of strains.
This work was supported by grant GM42033 from the National Institutes of Health to D.O.
REFERENCES
- 1.Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8164–8169. [PubMed] [Google Scholar]
- 2.Bedouelle H, Bassford P J, Fowler A V, Zabin I, Beckwith J, Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980;285:78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- 3.Breukink E, Demel R A, de Korte-Kool G, de Kruijff B. SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study. Biochemistry. 1992;31:1119–1124. doi: 10.1021/bi00119a021. [DOI] [PubMed] [Google Scholar]
- 4.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 5.Cabelli R J, Dolan K M, Qian L, Oliver D B. Characterization of membrane-associated and soluble states of SecA protein from wild-type and secA51(Ts) mutant strains of Escherichia coli. J Biol Chem. 1991;266:24420–24427. [PubMed] [Google Scholar]
- 6.Chen X, Xu H, Tai P. A significant fraction of functional SecA is permanently embedded in the membrane. J Biol Chem. 1996;271:29698–29706. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 7.Dalbey R, Lively M, Bron S, van Dijl J. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Blaauwen T, Terpetschnig E, Lakowicz J, Driessen A. Interaction of SecB with soluble SecA. FEBS Lett. 1997;416:35–38. doi: 10.1016/s0014-5793(97)01142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douville K, Price A, Eichler J, Economou A, Wickner W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 10.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG, and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Economou A, Pogliano J, Beckwith J, Oliver D, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 12.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 13.Fortin Y, Pheonix P, Drapeau G R. Mutations conferring resistance to azide in Escherichia coli occur primarily in the secA gene. J Bacteriol. 1990;172:6607–6610. doi: 10.1128/jb.172.11.6607-6610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanein D, Matlack K, Jungnickel B, Plath K, Kalies K, Miller K, Rapoport T, Akey C. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 15.Hartl F-U, Lecker S, Schiebel E, Hendrick J P, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 16.Huie J, Silhavy T J. Suppression of signal sequence defects and azide resistance in Escherichia coli commonly result from the same mutations in secA. J Bacteriol. 1995;177:3518–3526. doi: 10.1128/jb.177.12.3518-3526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Hunt, J., S. Weinkrauf, P. McNicholas, D. Oliver, and J. Deisenhofer. Unpublished data.
- 17.Kimura E, Akita M, Matsuyama S-I, Mizushima S. Determination of a region of SecA that interacts with presecretory proteins in Escherichia coli. J Biol Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- 18.Kontinen V P, Tokuda H. Overexpression of phosphotidylglycerophosphate synthase restores protein translocation in a secG deletion mutant of Escherichia coli at low temperature. FEBS Lett. 1995;364:157–160. doi: 10.1016/0014-5793(95)00378-m. [DOI] [PubMed] [Google Scholar]
- 19.Lill R, Cunningham K, Brundage L A, Ito K, Oliver D, Wickner W. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto G, Yoshihisa T, Ito K. SecY and SecA interact to allow SecA insertion and protein translocation across the Escherichia coli plasma membrane. EMBO J. 1997;16:6384–6393. doi: 10.1093/emboj/16.21.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 23.Mitchell C, Oliver D B. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama K-I, Hanada M, Tokuda H. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishiyama K-I, Suzuki T, Tokuda H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 26.Oliver D, Cabelli R, Dolan K, Jarosik G. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci USA. 1990;87:8227–8231. doi: 10.1073/pnas.87.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogliano K J, Beckwith J. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajapandi T, Oliver D. Integration of SecA protein into the Escherichia coli inner membrane is regulated by its amino-terminal ATP-binding domain. Mol Microbiol. 1996;20:43–51. doi: 10.1111/j.1365-2958.1996.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 28a.Ramamurthy, V., and D. Oliver. Unpublished observations.
- 29.Randall L, Toppings T, Hardy S, Pavlov M, Freistroffer D, Ehrenberg M. Binding of SecB to ribosome-bound polypeptides has the same characteristics as binding to full-length, denatured proteins. Proc Natl Acad Sci USA. 1997;94:802–807. doi: 10.1073/pnas.94.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schatz P, Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- 31.Schiebel E, Driessen A J M, Hartl F-U, Wickner W. DmH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 32.Snyders S, Ramamurthy V, Oliver D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J Biol Chem. 1997;272:11302–11306. doi: 10.1074/jbc.272.17.11302. [DOI] [PubMed] [Google Scholar]
- 33.Ulbrandt N, London E, Oliver D. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 34.van der Wolk J, Klose M, Breukink E, Demel R, de Kruijff R, Freudl R, Driessen A. Characterization of a Bacillus subtilis SecA mutant protein deficient in translocation ATPase and release from the membrane. Mol Microbiol. 1993;8:31–42. doi: 10.1111/j.1365-2958.1993.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 35.van der Wolk J P W, de Wit J G, Driessen A J M. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickner W, Leonard M. Escherichia coli preprotein translocase. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]