Abstract
The synthesis of the murein precursor lipid I is performed by MraY. We have shown that mraY is an essential gene for cell growth. Cells depleted of MraY first swell and then lyse. The expression of mraY DNA in vitro produces a 40-kDa polypeptide detectable by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
In Escherichia coli, the mra (murein region a [14]) cluster is located at 2 min on the genetic map. The region contains 16 open reading frames (ORFs), the functions of 14 of which have been identified (Fig. 1). The genes are involved in either cell division or murein synthesis or both (for reviews, see references 3, 10, and 19). The mraY and murG genes encode enzymes which together catalyze the formation of the lipid-linked disaccharide pentapeptide, lipid II [undecaprenyl-pyrophosphoryl-N-acetyl muramyl-(pentapeptide)-N-acetyl glucosamine], the precursor required for murein biosynthesis (8, 13, 15, 19). The first step of this two-stage process involves MraY, a membrane-bound translocase, which catalyzes the binding of UDP-MurNAc-(pentapeptide) to bactoprenol, a membrane-bound C55 isoprenoid lipid, to produce lipid I [undecaprenyl-pyrophosphoryl (PP)-MurNAc-pentapeptide] (8, 15, 20). The mraY mutant phenotype is not known. In this study, the phenotype of an mraY-null mutant is described. The proposed mraY ORF of 1,080 bp, identified by Ikeda et al., (7), is shown to complement an mraY-null allele. This ORF also produced a polypeptide of ∼40 kDa when it was expressed in an in vitro transcription and translation system.
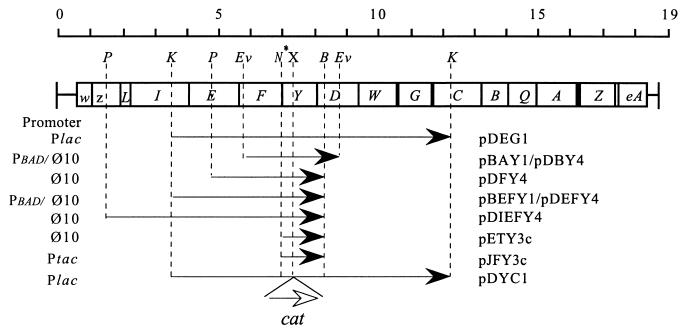
FIG. 1.
Organization of the mra region and the plasmids constructed for the purpose of this study. The upper part shows the ORFs in the mra region to scale in kilobase pairs. The boxed letters refer to the genes within this region: w, mraW; z, mraZ; L, ftsL; I, ftsI; E, murE; F, murF; Y, mraY; D, murD; W, ftsW; G, murG; C, murC; B, ddlB; Q, ftsQ; A, ftsA; Z, ftsZ; and eA, envA. Also shown are the cloned fragments with direction of expression as indicated by arrowheads. The types of promoter used to express these ORFs are listed at the left. P, PvuII; K, KpnI; Ev, EcoRV; N∗, NdeI; X, XmnI; and B, BglII. The NdeI site was introduced by PCR-directed mutagenesis.
Insertional inactivation of mraY.
An 8.4-kb KpnI fragment from Kohara phage λ6F3 (110) was cloned into pUC18 to create pDEG1 (Fig. 1) (1, 9). The mraY locus in pDEG1 was inactivated by the insertion of the chloramphenicol resistance gene (cat) into the XmnI site of mraY to make pDYC1 (Fig. 1). The disrupted mraY::cat construct was used to replace a wild-type mraY allele in the recD strain DSB1 [rodA(Am) recD::minitet Tetr Kanr Sup0], which has a duplication of the 2-min region (1). Chloramphenicol-resistant (Cmpr) colonies were isolated from cells transformed with a linear 9.3-kb KpnI restriction fragment from pDYC1. Southern blot analysis of the genomic DNA from one Cmpr clone showed that both the wild-type and the disrupted copy of mraY were present (Fig. 2). This strain was designated DBYC1. P1 lysates were grown on DBYC1 and used to transduce C600T (galK leu::Tn10 Tetr) to Cmpr. No Cmpr transductants were recovered. When C600T carrying pUC18 or pDEG1 was then transduced with the same P1 lysate, Cmpr transductants were recovered only from cells carrying pDEG1. The inability to transduce C600T (with or without pUC18) suggested that mraY may be an essential gene. The genetic linkage between leu::Tn10 and mraY::cat was determined by measuring the cotransduction frequency of the two markers. Of the Cmpr isolates screened, 78% were Tets Leu+, consistent with the proximity of leu and mraY.
FIG. 2.
Southern blot analysis of the genomic DNA from the partial diploid strains DSB1 (mraY wild type, lane 2) and DBYC1 (mraY::cat mutant, lane 3). Lanes 1 and 4 contain control DNAs mraY::cat (pDYC1) and mraY wild type (pDEG1), respectively. All of the DNAs were restricted with EcoRV, electrophoresed through agarose, and Southern blotted onto a nylon membrane. The blot was first probed with mraY (A) to show mraY::cat at 3.5 kb and mraY at 2.7 kb (lanes 1 and 4, respectively). Lane 2 shows strain DSB1 to have only wild-type mraY at 2.7 kb. Lane 3 shows both the wild-type and disrupted forms of mraY from DBYC1 DNA. Probing the same blot with cat (B) shows that only pDYC1 (lane 1) and DBYC1 (lane 3) contain the mraY::cat fragment at 3.5 kb.
Complementation analysis of mraY::cat.
The region required for the independent expression of mraY was identified by using several clones of mraY in the vector pT7-4 (17, 18), a multicopy plasmid lacking exogenous promoters. These plasmids were pDBY4, pDFY4, pDEFY4, and pDIEFY4 (Fig. 1). The expression of cloned DNA in these constructs is dependent on the presence of T7 RNA polymerase, usually supplied by an inducible (lacUV5) λ bacteriophage (λDE3, absent in the host strain, C600T). Using P1 transduction, we were able to introduce the mraY::cat allele into C600T carrying either pDEFY4 or pDIEFY4 but not when it was carrying the other pT7-4-based mraY clones, pDBY4 or pDFY4. Hara et al. (6) recently demonstrated that transcription of the first seven genes in the mra cluster is dependent on Pmra, a promoter located at the start of the mra region. Because complementation of mraY::cat was achieved only in multicopy number plasmids, it is possible that a weak promoter-like sequence which exists in the upstream DNA is sufficient to complement the null allele when present at a high copy number.
The mraY::cat phenotype.
To study the effects of MraY depletion in growing cells, a 2.7-kb EcoRV fragment containing mraY was cloned into the vector pBAD18 (5) to create pBAY1 (Fig. 1). Expression from PBAD can be induced or repressed with arabinose or glucose, respectively. A second clone, pBEFY1, was made by subcloning a 4.5-kb EcoRI/HindIII fragment from pDEFY4 into pBAD18 (Fig. 1). C600T containing either pBAD18 or the other plasmids was transduced with P1 lysates prepared from DBYC1. The only Cmpr transductants recovered contained either pBAY1 or pBEFY1. Cmpr isolates carrying pBAY1 were arabinose dependent for growth as expected, whereas Cmpr isolates carrying pBEFY1 were able to grow in the presence of either glucose or arabinose, as expected, since mraY can be expressed from PmraY independently of PBAD.
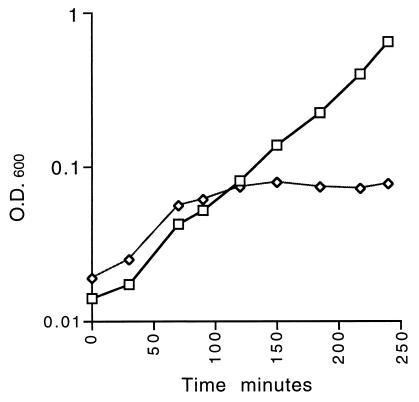
To achieve more-controlled regulation of expression of mraY from pBAY1 in strain DBYC2 (galK mraY::cat Cmpr), the plasmid copy number was reduced by introducing the pcnB::kan allele (11) by P1 transduction to give strain DBYC3 (as DBYC2 pcnB::Kan Kanr). DBYC3/pBAY1 was initially grown in Luria broth (LB) plus chloramphenicol with 0.2% arabinose at 37°C until the cells reached exponential phase. The method for culturing DBYC3/pBAY1 in either LB plus 0.2% arabinose (wt/vol) or LB plus 0.2% glucose (wt/vol) was as described earlier (1). After 80 min, the growth rate of the glucose-containing culture started to decrease, and growth stopped after 120 min (Fig. 3). The arabinose-containing culture continued to grow unaffected (Fig. 3). Cells from the arabinose-containing culture at 0 and 180 min were mainly short rods (Fig. 4). But, after 180 min in glucose, the cells were misshapen or had lysed (Fig. 4). Cells of C600T/pBAY1 cultured under the same conditions were indistinguishable from DBYC3/pBAY1 grown in 0.2% arabinose with respect to growth rate and cell morphology (data not shown).
FIG. 3.
Induction and repression of PBAD::mraY in the mraY-null mutant strain DBYC3/pBAY1 cultured in LB plus kanamycin supplemented with either 0.2% (vol/vol) arabinose (□) or 0.2% (vol/vol) glucose (◊) at 37°C. O.D., optical density.
FIG. 4.
Micrographs of DBYC3/pBAY1 grown in LB supplemented with either arabinose or glucose. (A and C) Cells from the arabinose-containing culture sampled at 0 and 180, respectively. (B and D) Cells from the glucose-containing culture sampled at 0 and 180 min, respectively. Bar, 5 μm.
The phenotype of the mraY::cat strain is very similar to that of mutants of peptidoglycan precursor genes (19). This suggests that synthesis of peptidoglycan is interrupted, presumably by a depletion in the pool of lipid I, causing cell death by autolysis. We conclude that mraY is an essential gene required for cell wall growth in E. coli.
Identification of MraY.
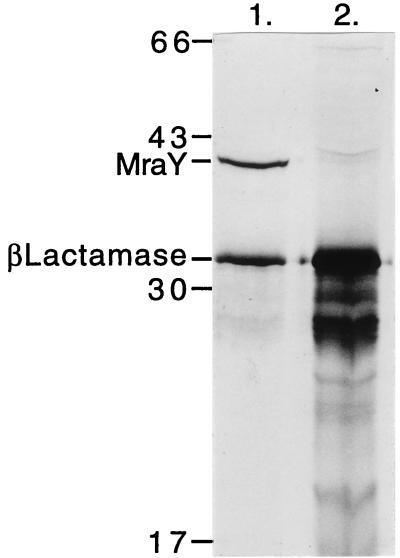
The MraY polypeptide has not been detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis even when overproduced in vivo (12), although increased levels of enzyme activity have been observed (2, 8). We failed to detect radiolabelled MraY from the T7 clones listed in Fig. 1 by SDS-PAGE. These included pETY3c, in which mraY was cloned in frame to the T7 gene 10 Shine-Dalgarno sequence by PCR-directed mutagenesis with primers DAV2 (5′ GGAGAATGGCATATGTTAGTTTGG 3′) and DAV3 (5′ CAATCAGATCTGCCGCCA 3′) (16). mraY was subcloned from pETY3c into pJF118EH (4) to create pJFY3c (Fig. 1). pJF118EH and pJFY3c DNA were used as the templates for in vitro translation. Analysis (SDS–10% PAGE) revealed that pJFY3c produced a peptide of ∼40 kDa (Fig. 5) which was absent in the pJF118EH control (Fig. 5). In agreement with this result, pJFY3c complemented the null allele (introduced by P1 transduction to C600T/pJFY3c), whereas pJF118EH did not.
FIG. 5.
SDS–10% PAGE analysis of the in vitro translation products (radiolabelled) produced by pJFY3c (lane 1) and pJF118EH (lane 2) after induction with 0.5 M isopropyl-β-d-thiogalactopyranoside. In lane 1 a unique polypeptide migrates as a 40-kDa peptide. This band is absent from the pJF118EH sample and is therefore presumed to be MraY.
Acknowledgments
This work was funded by a grant from the BBSRC.
REFERENCES
- 1.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 2.Brandish P E, Burnham M K, Lonsdale J T, Southgate R, Inukai M, Bugg T D H. Slow binding inhibition of phospho-N-acetylmuramyl-pentapeptide-translocase (Escherichia coli) by mureidomycin A. J Biol Chem. 1996;271:7609–7614. doi: 10.1074/jbc.271.13.7609. [DOI] [PubMed] [Google Scholar]
- 3.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 4.Furste J P, Pansegrau W, Frank R, Blocker H, Scholtz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 5.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara H, Yasuda S, Horiuchi K, Park J T. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda M, Wachi M, Ishino F, Matsuhashi M. Nucleotide sequence involving murD and an open reading frame orf-Y spacing murF and ftsW in Escherichia coli. Nucleic Acids Res. 1990;18:1058. doi: 10.1093/nar/18.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda M, Wachi M, Jung H K, Ishino F, Matsuhashi M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide:undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol. 1991;173:1021–1026. doi: 10.1128/jb.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 10.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1615–1626. [Google Scholar]
- 11.Masters M, Colloms M D, Oliver I R, He L, MacNaughton E J, Charters Y. The pcnB gene of Escherichia coli, which is required for ColE1 copy number maintenance, is dispensable. J Bacteriol. 1993;175:4405–4413. doi: 10.1128/jb.175.14.4405-4413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mengin-Lecreuix D M, Parquet C, Desviat L R, Plà J, Flouret B, Ayala J A, van Heijenoort J. Organization of the murE-murG region of Escherichia coli: identification of the murD gene encoding the d-glutamic-acid-adding enzyme. J Bacteriol. 1989;171:6126–6134. doi: 10.1128/jb.171.11.6126-6134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetyl glucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyakawa T, Matsuzawa H, Matsuhashi M, Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972;112:950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J T. The murein sacculus. In: Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 16.Rosenberg A H, Lade B N, Chui D-S, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 17.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 18.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1025–1034. [Google Scholar]
- 20.Wright A, Dankert M, Fennessey P, Robbins P W. Characterisation of a polyisoprenoid compound functional in O-antigen biosynthesis. Proc Natl Acad Sci USA. 1967;57:1789–1803. doi: 10.1073/pnas.57.6.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]