Abstract
In this issue of Neuron, Boivin et al. (2021) show that a polyglycine-expanded protein, uN2CpolyG, is translated from an expansion of GGC repeats in the 5′ UTR of the NOTCH2NLC (Notch homolog 2 N-terminal-like C) gene, defining a new pathological mechanism for neuronal intranuclear inclusion diseases (NIID).
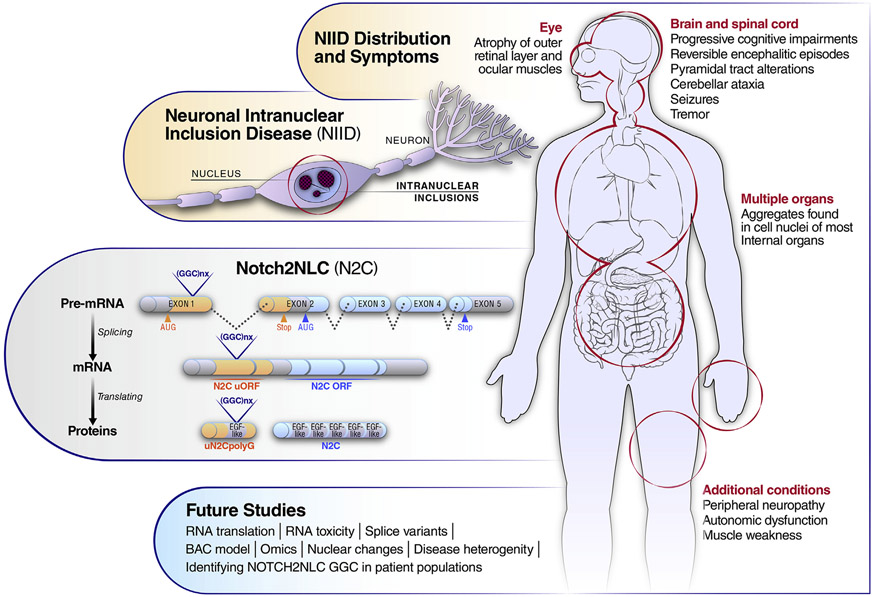
Neuronal intranuclear inclusion disease (NIID) is a rare sporadic or autosomal dominant multisystem neurodegenerative disease characterized by ubiquitin-immunopositive nuclear inclusions. Muscle and skin biopsies revealed eosinophilic intranuclear inclusions from NIID patients, and these were initially used to diagnose the disease. NIID presentation is highly variable but may include progressive cognitive impairments, tremor, reversible encephalitic episodes, alterations in the pyramidal tracts that innervate motor neurons of the spinal cord and brain stem, cerebellar ataxia, peripheral neuropathy, autonomic dysfunction, seizures, and muscle weakness, including the ocular muscles (Figure 1). There are also ocular changes: the outer retinal layer atrophies, but the inner retinal layer is preserved (Nakamura et al., 2020). The muscles of the eye are atrophied with bilateral miosis. NIID can affect the entire body, resulting in other symptoms, and intranuclear inclusions exist in all organs (Figure 1). Interestingly, repeat expansions in NIID are found in patients from Japan, China, and Malaysia but, to date, not in NIID patients of European descent, implying that there are two distinct forms and/or etiologies of NIID.
Figure 1.
Schematic of the features of NIID, production of uN2CpolyQ protein, and future studies proposed for NIID.
A key question in deciphering NIID is to determine the mechanism of formation of the multi-organ, filamentous, ubiquitin-immunopositive nuclear inclusions. In addition to ubiquitin, NIID nuclear inclusions contain p62/SQSTM1 (p62/sequestrome-1), the RNA-binding protein Sam68 (src-associated I mitosis, 68 kDa), MBNL (muscleblind-like 1), SUMO-1, OPTN (optineurin), dynamin-1, and FUS immunoreactivity. The inclusions do not contain TDP-43. Because both NIID and CAG/polyglutamine expansion disorders have intranuclear inclusions and some overlapping symptoms, patient samples were tested for CAG expansion but were negative. In an exciting discovery a few years ago, NIID was found to be caused by a non-coding, GGC repeat expansion in the 5′ UTR of the Notch homolog 2 N-terminal-like C (NOTCH2NLC) gene. This provided the first clue to the pathophysiologic mechanisms of NIID (Ishiura et al., 2019; Tian et al., 2019). The NOTCH2NLC mRNA levels were not altered in NIID patients compared to controls (Tian et al., 2019). Therefore, since the GGC expansion occurred in the 5’ UTR, the prevailing view was that the disease was caused by RNA-mediated toxicity or RAN translation unrelated to Notch2NLC protein.
In this issue of Neuron, Boivin et al. demonstrate that the GGC repeats are located in a short upstream open reading frame (uORF) distinct from the NOTCH2NLC ORF and result in production of a novel uN2CpolyG protein with an expanded polyglycine (Boivin et al., 2021). To characterize the uN2CpolyG protein, they designed constructs to express the uN2CpolyG protein in HEK293 cells and confirmed the identity of the protein by mass spectrometry. Impressively, Boivin et al. (2021) generated two antibodies that specifically detect the polyglycine-expanded uN2CpolyG protein in the neuronal intranuclear inclusions of NIID postmortem tissue, in primary mouse cortical neurons and in a mouse model intravenously transduced with an AAV vector directing expression of the protein. Both the uN2CpolyG (4D12) and the uN2CpolyG (4C4) antibodies bind p62 and ubiquitin-positive aggregates in NIID postmortem frontal cortex and cerebellum. Using an AAV serotype (PHP.eB) that crosses the blood-brain barrier, GFP-tagged uN2C with either a normal or an expanded repeat length was expressed in mice at 2 months of age. In this NIID model of the uN2C polyglycine expansion, the mice had shortened life-spans and a progressive motor disorder characterized by increased hindlimb clasping, decreased motor performance on rotarod and other behavior tests, and the appearance of neuronal inclusions immunopositive for p62 and with the uN2CpolyG 4D12 and 4C4 antibodies.
The relationship between the NOTCH2NLC transcripts and the uN2C protein remains to be investigated. Currently, two transcripts are reported to arise from NOTCH2NLC. The location of the GGC repeat expansion in the coding region of NOTCH2NLC transcript isoform 2 (NM_001364013.1) could produce a NOTCH2NLC protein with a polyglycine repeat of 236 amino acids. Boivin et al. (2021) suggest a 56-aa uN2C protein and NOTCH2NLC with 236 amino acids are produced (Figure 1). Several different NOTCH2NLC isoforms resulting from alternative slicing are likely to be discovered. Isoform-specific antibodies will be required to differentiate the transcripts and various protein products. Further studies are needed to determine whether non-ATG (RAN) translation and RNA gain of function with sense and antisense occur and contribute to NIID and related diseases (GGC repeat expansion in NOTCH2NLC).
An intriguing finding in NIID postmortem tissue is that, relative to controls, there is a consistent increase in nuclear size of pontine neurons harboring nuclear inclusions and a corresponding decrease in nuclear size of adjacent neurons (Uchihara et al., 2003). An increase in overall nuclear shape and size is not found in C9orf72 ALS/FTD, but alterations in nuclear pore complex and nucleocytoplasmic transport are important mechanisms underlying this disease and other DNA expansion disorders (Coyne and Rothstein, 2021). In a related disease, fragile-X-associated tremor/ataxia syndrome (FXTAS), the FMR polyG protein interacts with the nuclear lamina protein LAP2b, leading to disorganization of the nuclear lamina architecture in neurons derived from FXTAS patient induced pluripotent stem cells (iPSCs) (Sellier et al., 2017). Some NIID cases present with dystonia, and DYT1/TOR1A dystonia has nuclear pore abnormalities (Pappas et al., 2018). It will be interesting to determine whether the recent mechanism of nuclear pore deficits or changes in lamina architecture are also part of the pathogenesis of NIID.
One of the clues leading to the discovery of the GGC expansion in the 5′ UTR of the NOTCH2NLC gene was the striking overlap of symptoms and brain MRI abnormalities in patients with NIID to those with FXTAS, oculopharyngodistal myopathy (OPDM), and oculopharyngeal myopathy leukoencephalopathy (Ishiura et al., 2019), all of which are caused by DNA expansions. The overlap suggested that these diseases may share pathogenetic mechanisms. A plethora of reports in the last year identified an expanded GGC repeat in the 5′ UTR of the NOTCH2NLC gene in patients with essential tremor, Alzheimer’s disease (Jiao et al., 2020), frontotemporal dementia (FTD) (Jiao et al., 2020), Parkinson’s disease, multiple system atrophy, adult leukoencephalopathy, amyotrophic lateral sclerosis (ALS), and OPDM.
In NIID, the GGC repeat ranges from 66 to over 500; control patients have fewer than 43 repeats. To date, there is no association between the GGC repeat size and the severity or the age of onset of the disease (Tian et al., 2019). Also, evidence of anticipation for NIID is lacking, and whether somatic expansions occur in NIID is not known. The 5′ intermediate lengths of 42–58 GGC repeats in the 5′ UTR of the NOTCH2NLC gene may be associated with symptoms. OPDM3 has 128–198 GGC repeats, but the non-GAG/GGA/AGC/ACG repeat interruptions in OPDM3 are higher than those in NIID patients. Transcriptional silencing, GGC repeat length, repeat interruptions, and genetic heterogeneity are mechanisms that might impact the symptoms. This finding is not surprising as there is growing evidence that triplet repeat expansions in a single gene can cause multiple, apparently disparate diseases. The GGGGCC hexanucleotide repeat expansions (GGGGCC) in the non-coding region of C9ORF72 can lead to ALS and FTD, and these patients exhibit significant clinical heterogeneity. We now know that C9ORF72 has three transcripts and the start site for transcript 2 is upstream of the pathogenic GGGGCC repeat mutation sequence. When the C9ORF72 gene is mutated, the normal transcript 1 is not produced, and transcripts 1 and 3 can generate pathogenic dipeptide repeats. This leads to multiple, simultaneously occurring pathophysiologic mechanisms, including RNA sense and antisense gain of function, loss of function of the C9or72 protein due to haploinsufficiency, and RAN translation.
Boivin et al. (2021) demonstrate that expression of uN2CpolyG with a repeat expansion in adult mice is sufficient to produce neurological deficits and Purkinje cell loss, which, importantly, are progressive in nature. As our understanding of the disease moves forward, a NIID BAC transgenic mouse with the repeat expansion and all the upstream and downstream elements of the NOTCH2NLC gene will help elucidate the relative contributions of the possible pathophysiologic mechanisms. GGA somatic expansion, RAN translation, and RNA toxicity should be evaluated in NIID postmortem tissue, BAC transgenic model, and patient iPSCs models. Interestingly, the normal function of the uN2C protein is to interact with the Ku70 and Ku80 proteins and activate DNA repair. This suggests that this newly discovered protein uN2C may be a novel regulator of DNA damage response. The polyglycine-expanded form of uN2C protein had reduced binding to Ku70 and Ku80 proteins and a blockage of non-homologous end joining (NHEJ) repair. In Huntington’s disease, genetic modifiers that alter somatic CAG expansion or DNA damage repair have been found (Wheeler and Dion, 2021). DNA damage is common in many repeat expansion diseases. Boivin et al. (2021) examined DNA damage in their uN2CpolyG mouse model and NIID postmortem tissue and surprisingly found no evidence of gH2AX-positive DNA damage in NIID. Further investigation will be needed to determine whether DNA repair alterations occur in individuals with NIID. In addition, understanding how the uN2C protein is involved in NHEJ repair mechanisms will elucidate the mechanism of action of this newly discovered protein. Polyglycine-containing proteins have many interesting functions—initiation of infection, regulation of proteolysis, cancer susceptibility, and targeting of the protein to the outer envelope. Further analysis of patient samples, the current NIID mouse model, and the proposed BAC model, including other organs using unbiased omics, may identify pathways critical to understand the pathogenic mechanisms in NIID. Future studies are summarized in Figure 1.
Progress in long-read and wholegenome sequencing is accelerating the identification of a growing number of DNA expansion diseases. Currently, more than 50 inherited human diseases are known to be caused by DNA tandem repeats with various lengths and sequences. Identifying the genetic cause of these devastating diseases is a step forward. Hopefully, that knowledge will lead to the development of disease-modifying therapies for patients with these diseases. The common mechanisms across these diseases will provide new therapeutic strategies and drug interventions to treat this class of disease. These may target pathways that prevent repeat expansions, knock down specific alleles, target upstream pathways that trigger the disease onset, and identify combination drugs that target multiple pathways mediating disease progression.
REFERENCES
- Boivin M, Deng J, Pfister V, Grandgirard E, Oulad-Abdelghani M, Morlet B, Ruffenach F, Negroni L, Koebel P, Jacob H, et al. (2021). Translation of GGC repeat expansions into a toxic polyglycine protein in NIID defines a novel class of human genetic disorders: the polyG diseases. Neuron 109, this issue, 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, and Rothstein JD (2021). Nuclear lamina invaginations are not a pathological feature of C9orf72 ALS/FTD. Acta Neuropathol. Commun 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura H, Shibata S, Yoshimura J, Suzuki Y, Qu W, Doi K, Almansour MA, Kikuchi JK, Taira M, Mitsui J, et al. (2019). Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet 51, 1222–1232. [DOI] [PubMed] [Google Scholar]
- Jiao B, Zhou L, Zhou Y, Weng L, Liao X, Tian Y, Guo L, Liu X, Yuan Z, Xiao X, et al. (2020). Identification of expanded repeats in NOTCH2NLC in neurodegenerative dementias. Neurobiol. Aging 89, 142.e1–142.e7. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Tsunoda K, Mitsutake A, Shibata S, Mano T, Nagashima Y, Ishiura H, Iwata A, Toda T, Tsuji S, and Sawamura H (2020). Clinical Characteristics of Neuronal Intranuclear Inclusion Disease-Related Retinopathy With CGG Repeat Expansions in the NOTCH2NLC Gene. Invest. Ophthalmol. Vis. Sci 61, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas SS, Liang CC, Kim S, Rivera CO, and Dauer WT (2018). TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum. Mol. Genet 27, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. (2017). Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron 93, 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Wang JL, Huang W, Zeng S, Jiao B, Liu Z, Chen Z, Li Y, Wang Y, Min HX, et al. (2019). Expansion of Human-Specific GGC Repeat in Neuronal Intranuclear Inclusion Disease-Related Disorders. Am. J. Hum. Genet 105, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihara T, Tanaka J, Funata N, Arai K, and Hattori T (2003). Influences of intranuclear inclusion on nuclear size - morphometric study on pontine neurons of neuronal intranuclear inclusion disease cases. Acta Neuropathol. 105, 103–108. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, and Dion V (2021). Modifiers of CAG/CTG Repeat Instability: Insights from Mammalian Models. J. Huntington’s Dis 10, 123–148. [DOI] [PMC free article] [PubMed] [Google Scholar]