Abstract
Root architectural phenotypes are promising targets for crop breeding, but root architectural effects on microbial associations in agricultural fields are not well understood. Architecture determines the location of microbial associations within root systems, which, when integrated with soil vertical gradients, determines the functions and the metabolic capability of rhizosphere microbial communities. We argue that variation in root architecture in crops has important implications for root exudation, microbial recruitment and function, and the decomposition and fate of root tissues and exudates. Recent research has shown that the root microbiome changes along root axes and among root classes, that root tips have a unique microbiome, and that root exudates change within the root system depending on soil physicochemical conditions. Although fresh exudates are produced in larger amounts in root tips, the rhizosphere of mature root segments also plays a role in influencing soil vertical gradients. We argue that more research is needed to understand specific root phenotypes that structure microbial associations and discuss candidate root phenotypes that may determine the location of microbial hotspots within root systems with relevance to agricultural systems.
Keywords: Carbon rhizodeposition, lateral roots, number of axial roots, rhizosphere microbiome, root growth angle, rooting depth, root system architecture, soil redox potential, soil vertical gradients
Root architecture and soil vertical gradients create a diverse set of physicochemical conditions in which rhizosphere microbes live; however, these scaffolds of soil microbial ecology are rarely studied.
Introduction
Suboptimal water and nutrient availability are primary, pervasive constraints to plant production in terrestrial ecosystems (Lynch, 2022a). Water, nitrogen, and phosphorus availability are commonly limiting in natural ecosystems and in low-input agroecosystems. In high-input agroecosystems, intensive fertilization and irrigation cause environmental degradation, and, in non-irrigated systems, drought stress is common and increasingly problematic as a result of global climate change. Root system architecture (RSA), the physical arrangement of root organs in space and time, determines the efficiency of soil resource capture, and therefore represents a potential solution to these current agricultural challenges.
RSA phenotypes such as root growth angle, rooting depth, lateral root branching density and length, and axial root number are possible crop breeding targets because they have adaptive value for soil resource capture (Lynch et al., 2021), they are relatively easy to measure at high throughput (Trachsel et al., 2011; Colombi et al., 2015; Liu et al., 2021; Schneider et al., 2021; Lynch, 2022b), and, for some, genetic control has been described (Ruangsiri et al., 2021; Schneider et al., 2021; Waite and Dardick, 2021). Other RSA-related phenotypes with adaptive value are dimorphism, the capability of roots to produce contrasting levels of a given trait in the same individual, and plasticity, the capacity to alter the phenotype in response to environmental stimuli (e.g. water deficit or suboptimal nutrient availability) (Lynch, 2019). These phenotypes are more complex to measure, and their genetics less well understood (Schneider and Lynch, 2020), but they integrate temporal and spatial dimensions into RSA.
While microbial effects on RSA have been widely documented, especially under controlled conditions (Eichmann et al., 2021; Box 1), natural variation in root architecture of agricultural plants is rarely considered as an important factor determining root microbial associations. The influence of root phenotypes on microbial associations, and the synergies and trade-offs between plants and microbes when studying specific root architectural phenotypes with adaptive value for the capture of soil resources, have rarely been studied, especially under field conditions. Furthermore, the relative contribution of microbial feedbacks on the architectural phenotype of a plant under field conditions is unclear. Although plants under abiotic stress growing in artificial growth media recruit specific microbiomes that may help in acquiring more of the limiting resource (Castrillo et al., 2017), aspects such as the localization and rate of microbial consumption of root exudates, and the metabolic cost of maintaining beneficial rhizosphere associations are not well understood either in controlled conditions or in the field.
Box 1. Microbially driven changes in root architecture and relationship to nitrogen acquisition.
Several plant-growth promoting microorganisms (PGPM) cause changes in root architecture. Specifically, the production of lateral roots or the modification of the elongation rate of the primary root are usually reported in Arabidopsis (Arabidopsis thaliana) (Verbon and Liberman, 2016). The main mechanisms of these interactions are through the production of phytohormones by the microbial partner, or through the microbial modification in phytohormone perception by the plant (Frankenberger and Arshad, 2020; Verbon and Liberman, 2016). For example, several species of the soil fungus Trichoderma (Contreras-Cornejo et al., 2009; Garnica-Vergara et al., 2016) produce metabolites that interfere with auxin transporters in Arabidopsis and tobacco (Nicotiana tabacum). Also, Pseudomonas putida (Ortiz-Castro et al., 2020), Azospirillum brasilense (Bashan and de-Bashan, 2010), Bradyrhizobium japonicum (Torres et al., 2018), and Burkholderia phytofirmans (Zúñiga et al. 2013) produce indole-3-acetic acid that is, or becomes, active to induce lateral roots in their respective plant host. Furthermore, bacteria-grazing amoebas have been associated with increased levels of cytokinins under high concentrations of nitrate, and the cytokinins have been hypothesized to interact with increased free auxin product of the grazing of the amoeba in the rhizosphere of Lepidium sativum to increase lateral root branching density (Krome et al., 2010). Nitric oxide (NO) has been discussed as a possible regulator of lateral root branching density (Singh et al., 2019), which could then link NO to bacterial metabolism in the rhizosphere of crops. These are just a few examples of multiple PGPM reported in the literature to produce plant growth regulators that may participate in changes in root architecture and, therefore, indirectly in nitrogen uptake.
Although there is a notable amount of research on the potentialities and basic mechanisms of phytohormone-mediated plant–microbe interactions, the development of technologies based on such interactions in agroecosystems is still scarce. Usually, results observed in model plants such as Arabidopsis or on crops grown under controlled conditions are not reported equally under field conditions. Only a few reports demonstrate the utility of inoculation with PGPM to reduce nitrogen fertilization by causing changes in root architecture (see, for example, the example of Azospirillum brasilense and maize in Brazil by Hungria et al., 2022). This reveals a poor understanding of such interactions, and the several questions that remain open (Galindo-Castañeda et al., 2022; Verbon and Liberman, 2016). How and when do plant genetic determinants and plasticity in the production of new lateral roots interact with phytohormones produced (or modified) by microbes to control lateral root branching? If plasticity exists in lateral root branching patterns in response to nutrient patches in soil, is this only mediated by the plant directly, or is this a result of a feedback of root–microbe interactions in the rhizosphere? Possible cues to start changing root architectural patterns might originate from newly mineralized nitrogen, or just metabolized nitrogen-containing compounds by microbes. This is a fascinating, yet unexplored topic that merits further exploration at the field scale, using crops.
Here, we discuss RSA phenotypes that have adaptive value in crops (Lynch, 2022b) and how they might associate with microbes to favor specific microbial processes in the rhizosphere of crops interacting with vertical soil physicochemical gradients. We first present the context in which crop roots develop, the soil, and its vertical gradients. Next, we present facts and hypotheses regarding the interactions of RSA with carbon rhizodeposition, and then present primarily hypotheses on the rhizosphere microbial processes as determined by RSA (Fig. 1). We also present our perspective on pathogenic interactions with RSA. In this essay, we highlight recent advances in the understanding of microbial and root feedbacks as they relate to RSA (Box 2) and identify gaps resulting from a lack of integration of soil gradients and RSA in root–microbiome research. Further recommendations on how to move forward in this field are summarized in Box 3. Root growth rate and plasticity are important components of RSA that have been recently reviewed (Schneider and Lynch, 2020; Bonkowski et al., 2021). However, they cannot be considered simple architectural traits, but rather complex processes that go beyond the scope of the present essay.
Fig. 1.
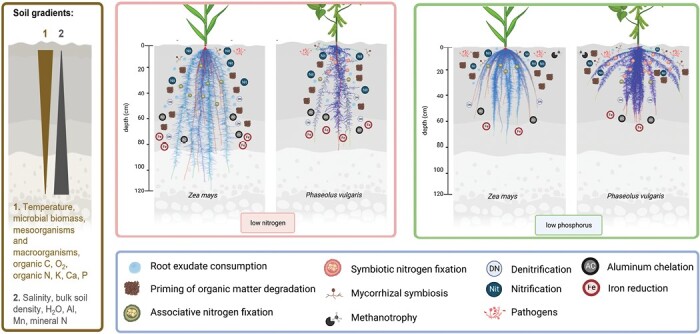
Hypothesized interactions of RSA with rhizosphere microbial processes (represented as icons) under limitation in a mobile (nitrate, Low N) and immobile (phosphorus, Low P) soil resource. Root models of the optimal root phenotypes to overcome each limitation for a cereal and a legume are depicted. The models are based on empirical observations (Lynch, 2019). Axial roots are in red and lateral roots in blue. The number and position of icons represent our hypothesized effects of the two RSA on the predominance and location of these rhizosphere processes along the soil profile and within the root system. Soil gradients are depicted to explain the hypothesized interactions. For example, shallow roots may favor nitrification, organic matter degradation, biodegradation of root exudates, and proliferation of fungal associations. In shallow roots, we hypothesize that nitrogen fixation would be performed mostly by bacteria with structures to protect nitrogenase from oxygen, and associative nitrogen fixation by a more diverse set of bacteria might be found in intermediate root systems. Deep roots would be associated with reductive microbial processes such as denitrification and ammonification, and manganese and iron reduction. Microbial aluminum chelation would be another process that might benefit the growing root tips in deep soil domains, although aluminum toxicity is also a problem in surface soil layers in tropical soils. More examples of similar analyses with RSA traits such as number of axial roots, dimorphism, and lateral root branching density can be found in ‘Interactions of root system architecture phenotypes with soil microbial processes’.
Box 2. Key developments in locating rhizosphere microbial interactions considering RSA.
In the following studies, RSA is integrated with rhizosphere processes relevant to microbial processes. In (A), RSA had a significant effect on rhizosphere microbial communities. Microbial inoculation was significantly associated with changes in RSA through an auxin-dependent mechanism in maize (B) and in an auxin-independent mechanism in A. thaliana (C). Studies (D–F) show that RSA is linked to changes in exudates in ways that are relevant to microbial processes.
A. Kawasaki et al. (2021): the magnitude of differences in microbial diversity at root tips and root bases of wheat or rice are comparable with the variation in root microbial diversity between different plant species growing in the same soil.
B. Hungria et al. (2022): inoculation of maize with Azospirillum brasilense strains Ab-V5 and Ab-V6 allowed the reduction of 25% of fertilization overall in maize, in a set of 30 field trials spanning 10 years and contrasting levels of productivity, soil quality, soil organic carbon content, and tropical and subtropical conditions. The results are attributed to increases in root branching density, root hair length, root length, and specific root length, and without an increase in root biomass by means of the production of indole acetic acid (IAA). However, the evaluations of RSA were made in pots of <1 liter, where RSA is restricted.
C. Gonin et al. (2023): propose a new auxin-independent, ethylene-mediated mechanism by which microbes change lateral root branching density of Arabidopsis and Selaginella moellendorffii, which also seems to work under high salinity and iron deficiency.
D. McKay Fletcher et al. (2020): show that RSA is a better predictor than citrate exudation in phosphorus uptake in wheat.
E. Bilyera et al. (2021): provide an example of a specific interaction of a secondary metabolite (benzoxazinoid) reducing β-glucosidase activity in the rhizosphere, especially in mature root systems, where there is a higher activity of this enzyme.
F. Hosseini et al. (2022): exemplify the differences in rhizosphere activity of lateral versus axial roots under drought, finding that drought negatively affects the rhizosphere volumes of axial roots but not of lateral roots of wheat, while also favoring the activity of β-glucosidase but not of acid phosphatase, and leucine aminopeptidase.
Box 3. Required tools, research questions, and prospective applications of integrating RSA with rhizosphere microbial processes.
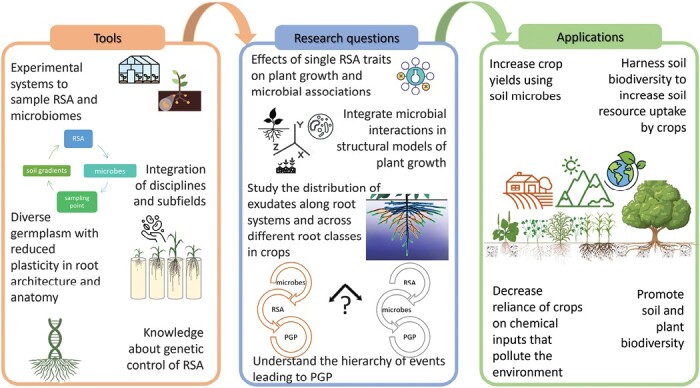
Research gaps in the study of relationships between RSA and rhizosphere microbes are presented in three categories: tools, research questions, and applications. We still lack tools to answer fundamental research questions to fulfill promising applications. PGP, plant growth promotion. Adapted from Galindo-Castañeda et al. (2022).
Vertical gradients in soil
Soil is a heterogenous and vertically stratified matrix in which changing physical and chemical properties create distinct niches that influence root development and microbial activity. The arrangement of soil particles and aggregates into a porous structure that determines the availability of oxygen, water, and nutrients changes with depth (Hartmann and Six, 2023). Accumulation and decomposition of organic matter in the topsoil lead to the biogenic formation of stable micro- and macroaggregates that form a highly interconnected soil pore network, facilitating air permeability and water infiltration. As soil depth increases, biogenic processes of aggregate formation are replaced by physicogenic processes in which soil particles are packed into prismatic aggregates that form smaller and less interconnected pores, restricting the flow of oxygen, water, and nutrients (Koestel et al., 2020; Pereira et al., 2021).
Vertical physicochemical gradients influence microbial properties (He et al., 2023). Topsoil domains characterized by greater diurnal and seasonal temperature variation, more oxygen, and less moisture favor aerobic microbes such as nitrifiers and methanotrophs as well as taxa able to tolerate fluctuating temperatures, desiccation, and solar radiation. Topsoil is also subjected to changes in pH due to fertilizer usage (Li et al., 2020); therefore, acid-tolerant microbes would be better adapted in soils used for intensive agriculture. In contrast, deeper soil domains show increasing anaerobicity and soil moisture, favoring anaerobic microbial processes such as denitrification or iron reduction. The pH of deep soil domains varies from alkaline in limestone-derived soils, to acidic in highly weathered soils (Hartemink et al., 2020). Plant nutrient availability also changes within the soil profile, with topsoils containing greater amounts of organic carbon and nitrogen as well as phosphorus, potassium, and calcium, while subsoils accumulate mineral nitrogen and have high bioavailability of manganese and aluminum (Lynch and Wojciechowski, 2015).
Agricultural practices such as tillage, fertilization, and irrigation often increase stratification. Although tillage homogenizes the topsoil, it also creates a plough pan that acts as a physical barrier for oxygen, water, nutrients, and possibly root penetration to deeper strata. Fertilizers, pesticides, and other amendments create local gradients that depend on solute mobility, for example with highly mobile mineral nitrogen accumulating in deeper strata and less mobile phosphorus residing in the topsoil (Lynch et al., 2021). These vertical physicochemical gradients determine the presence and activity of microbial communities at a given location and drive their associations with roots. Moreover, vertical gradients also create differences in natural horizontal gradients that exist in soils and that directly create the rhizosphere environment, generating another layer to the interaction of root architecture with rhizosphere microbial activities. For example, the vertical gradient in oxygen content might create differences between the horizontal oxygen gradient of the rhizosphere of a root tip compared with the rhizosphere at the base of the same root, which might be in a shallower position across the soil profile. The former horizontal gradient is expected to be less pronounced than the latter.
Carbon rhizodeposition and root system architecture
Carbon rhizodeposition includes root exudates, mucilage, shed root tissue, and debris from root loss (Dijkstra et al., 2021). The location of carbon rhizodeposition within the root system under actual field conditions as well as the duration and functionality of the exuded or deposited compounds in field soils is poorly understood (Schnepf et al., 2022; Zhang et al., 2023). Specifically for root exudates and mucilage, the metabolic cost of their production is still a matter of research (Wen et al., 2022). Understanding the spatiotemporal dynamics, rates of microbial consumption, and metabolic cost of carbon rhizodeposition is crucial to estimate the benefits of establishing plant–microbial associations to promote soil resource capture by plants (Galindo-Castañeda et al., 2022).
Root exudates are produced in greater concentrations in root tips and the elongation zone, but differences in spatial patterns have been found in the production of enzymes and other compounds within root systems depending on mutations (Bilyera et al., 2021), plant species (Razavi et al., 2016), and stress conditions (Liu et al., 2022; Smercina et al., 2021). Besides root exudates, other sources of carbon such as enzymes (Hosseini et al., 2022) and shedding of root tissue, especially from secondary root growth (Strock and Lynch, 2020), are found in the rhizosphere of mature roots. Traits such as root growth angle, rooting depth, and lateral root branching density determine the location and number of root tips, and therefore influence the abundance and location of root exudates and mucilage in reference to soil physicochemical gradients. Traits such as root diameter and the number of axial roots will modify rhizodeposition patterns around mature roots, as hypothesized in ‘Interactions of root system architecture phenotypes with soil microbial processes’. Anatomical phenotypes are also expected to interact with carbon rhizodeposition in mature root systems and with horizontal soil gradients (Galindo-Castañeda et al., 2022). For example, root diameter, an anatomical trait affected by thickness and biochemical composition of the outermost layer of each root, is linked to root exudation and the composition of the rhizosphere microbial communities (Zai et al., 2021). Determining the associations between root architecture and carbon rhizodeposition would be especially important to predict resource utilization and the metabolic cost of soil exploration (Schäfer et al., 2022).
The location of carbon rhizodeposition within root systems has important implications for resource capture and microbial activity in the rhizosphere in agricultural soils. The case of citrate exudate and phosphorus uptake by roots exemplifies this complexity. When the location and volume of rhizosphere hotspots and citrate-mediated phosphorus uptake are predicted using a model, contrasting results are found depending on whether such exudation occurs in the root tips only (in Vicia faba) or within the whole root system (in Brassica napus) (Schnepf et al., 2022). These results link root architecture with root exudation in silico in a homogeneous soil matrix. If such models were integrated with vertical soil gradients of phosphorus availability, plants allocating exudates in the topsoil would probably have greater acquisition of phosphorus solubilized by citrate. Not surprisingly, RSA is highly correlated with citrate solubilization and is a better predictor of phosphorus uptake than citrate exudation itself (McKay Fletcher et al., 2020).
If citrate or other exuded molecules are considered to estimate the extent of rhizosphere volumes, the extent and permanence of root exudates would vary depending on the root architecture and the rates at which microbes would consume the deposited carbon. This microbial degradation depends in large part on the depth at which these exudates are produced because the sorptive qualities of soils vary with depth, and because microenvironments change with depth (see ‘Vertical gradients in soil’). Therefore, root phenotypes such as lateral root branching density, root hairs, and root tips should be considered in models and estimations of rhizosphere volume and microbial activity.
The composition and permanence of root secreted compounds have effects on nutrient cycling and ecological interactions with microbes. On the one hand, plants encountering suboptimal availability of nitrogen and phosphorus change exudate composition (Smercina et al., 2021; Wen et al., 2022), and trigger changes in the nitrogen cycle in bulk soils (Liu et al., 2022). How these processes change in the rhizosphere, where there is a microenvironment different from the bulk soil (Schnepf et al., 2022), is unknown. On the other hand, the permanence of stress-responsive exudates in soils and the location in which they are produced are uncertain. A recent example of permanence of an exuded secondary metabolite is the production of benzoxazinoids, which mediate maize (Zea mays) defense against herbivores and seem to have a legacy and chemical fingerprint in field soils for a subsequent growth cycle (Gfeller et al., 2023). No studies are available about the location of exudation of benzoxazinoids or many other compounds important for ecological interactions within the root system.
Interactions of root system architecture phenotypes with soil microbial processes
Root architecture structures soil exploration in time and space and therefore regulates soil resource capture (Lynch, 2022b). Exploration of deep soil domains improves the capture of mobile resources such as water and nitrate in most environments, whereas topsoil foraging improves the capture of immobile resources such as phosphorus, potassium, and most other nutrients. Rooting depth is also related to biosequestration of atmospheric carbon, since carbon deposited in deep soil domains is degraded more slowly than that deposited in shallow soil (Kell, 2012). The extent and depth of root foraging for soil resources are also regulated by root anatomy, which determines the capacity of a root segment to acquire soil resources and transport them to the shoot (Lynch et al., 2021). Root anatomy also regulates the metabolic cost of soil exploration, which is an important constraint to soil foraging (Lynch and Wojciechowski, 2015). In the following paragraphs we provide hypotheses on how RSA modifies microbial processes in the rhizosphere, which are also summarized in Fig. 1 and Table 1. Recent research supporting the development of this integrative field is presented in Box 2.
Table 1.
Contrasting states of root architectural traits and the hypothesized effect on microbial habitat conditions
| Root trait | Trait states | Hypothesized effect on root exudation, physicochemical conditions, and habitat space for microbes | Hypothesized effect on microbial processes in the rhizosphere | Supporting evidence |
|---|---|---|---|---|
| Rooting angle and depth | Steep—deep roots versus shallow—shallow roots |
|
|
Not reported in the literature—nitrogen cycle in paddy rice as it relates to soil depth: Ishii et al. (2011). Soil compaction effects on rhizosphere microbiota in crops: Longepierre et al. (2021, 2022) |
| Number of axial roots | Many versus few |
|
|
Not reported in the literature—review on plant–microbe competence for nitrogen: Moreau et al. (2015) |
| Lateral root density | High versus low |
|
|
Schmidt et al. (2018); review on plant–microbe competence for nitrogen: Moreau et al. (2015) Studies on lateral root versus axial root: Saleem et al. (2016; Zai et al. (2021) |
| Lateral root length | Long versus short |
|
|
Not reported in the literature. Studies on lateral root versus axial root: Saleem et al. (2016); Zai et al. (2021); Kawasaki et al. (2021) |
Modified from Galindo-Castañeda et al. (2022).
Root growth angle and rooting depth
The growth angle of axial roots is perhaps one of the most important phenotypes determining the rhizosphere microbiome because axial roots are the scaffolds from which lateral roots emerge, therefore determining the depth at which most root biomass and exuding root tips are located. Such root allocation integrated with soil vertical gradients create diverse microbial niches (Fig. 1). Whether shallow roots or deep roots are associated with specific microbial processes in the rhizosphere remains largely unexplored. Mycorrhizal associations are more abundant in the basal segments of axial roots in common bean (Phaseolus vulgaris) under limiting phosphorus, but the symbiosis disappears with time due to secondary growth (Strock et al., 2018). We expect that shallow roots favor nitrification, organic matter degradation, biodegradation of root exudates, and proliferation of fungal associations; and nitrogen fixation would be carried out mostly by bacteria with structures to protect nitrogenase from oxygen. Perhaps associative nitrogen fixation by a more diverse set of bacteria might be found in intermediate root systems, where oxygen is reduced (Fig. 1). Deep roots would be associated with reductive microbial processes such as denitrification and ammonification, and manganese and iron reduction. Microbial aluminum chelation would be another process that might benefit growing root tips in deep soil domains, although aluminum toxicity is also a problem in surface soil layers in tropical soils.
Dimorphic root systems
An individual plant can have both shallow and deep roots, or have intermediate states between these two extremes. This can be advantageous when both immobile and mobile soil resources are limiting and the construction and maintenance cost must be minimized (Lynch, 2022b). In this case, diverging microbial communities might associate in the rhizosphere depending on the depth and root architecture. Carbon rhizodeposition produced in shallow soil domains would prime organic matter degradation and ammonia production, phosphorus solubilization, mycorrhization, and in some cases pathogenesis (see ‘Pathogenic associations and root system architecture’). Deep roots would meanwhile be surrounded by a less active microbial community participating in reductive reactions of metal and organic matter. Intermediate root systems with fanned root architectures might favor associative nitrogen fixation in the rhizosphere by protecting it from high oxygen concentrations that are present close to the surface.
Number of axial roots
For microbial associations, roots with more axial roots represent increased colonization area during root elongation, and perhaps increased carbon deposition through exudates and root debris. More axial roots also means more lateral roots (Strock et al., 2021), which are readily colonized by microbes. In crops such as maize and bean, expressing fewer axial roots is an adaptive phenotype under low nitrogen availability and water deficit, while being deleterious under low phosphorus (Saengwilai et al., 2014; Gao and Lynch, 2016; Rangarajan et al., 2018, 2022; Sun et al., 2018; Schäfer et al., 2022). Under soil impedance, an increased number of axial roots that can penetrate the hardpan in agricultural soils is beneficial, and overall enhances rooting depth and access to mobile resources (Strock et al., 2021). The location of axial roots deeper in compacted soils might favor microbial species involved in the reduction of nitrate, sulfate, or iron to interact in the rhizosphere. For example, soil compaction caused an increase in denitrifiers in the rhizosphere of peas (Pisum sativum) and wheat (Triticum aestivum) (Longepierre et al., 2022), and was associated with increased abundance of anaerobic prokaryotes and saprotrophic fungi, concomitant with decreases in aerobic prokaryotes and plant-associated fungi (Longepierre et al., 2021).
There are other indirect effects of increased axial root number in crops. First, with more axial roots, intra-plant competition for resources might create steeper horizontal gradients of resources in the rhizosphere given the more numerous active root segments at a given depth, which may create a more selective niche for the microbiome, with microbial communities showing perhaps a more diverse spectrum in resource usage. Under such conditions, competition for resources can also occur between plants and microbes (Hill and Jones, 2019). Secondly, plants with more axial roots must use carbon more efficiently, given the high metabolic cost of axial roots (Rangarajan et al., 2022). Therefore, carbon exudation in plants with many axial roots might be compromised, with direct consequences for the microbial commensals relying on such exudates. These processes have received little attention in the field of root microbiology.
Lateral root branching density
Plants have the capability to branch profusely in response to nutrient patches in soils (Schneider and Lynch, 2020), and this response may be enhanced by microbes given their ability to trigger lateral root branching (Chiu et al., 2022; Gonin et al., 2023; Verbon and Liberman, 2016). This seems beneficial to the microbes because it creates more opportunities to associate with roots and it may increase the production of fresh exudates to feed these microbes. However, the utility of a microbially mediated increase in lateral root branching for the plant is debatable, because under some conditions the metabolic cost of lateral roots exceeds the benefits they afford for resource capture. For example, the best root phenotypes of maize and bean to acquire mobile resources that are more abundant in deep soil domains such as nitrate and water have reduced lateral root branching density (LRBD) (Fig. 1). The reason is that the construction and maintenance costs of more lateral roots exceed the increase in nitrogen or water uptake due to intra-root competition for these limiting resources, and that reduced production of lateral roots permits more internal resources to be available for axial root elongation into deeper soil domains (Lynch, 2022b). Therefore, we call for critical assessments of the advantages of forcing plants to produce more lateral roots through microbial inoculation, especially under stress conditions where mobile resources are highly limiting. On the contrary, plants growing under phosphorus limitations may benefit from associations with microbes, causing increased LRBD (Chiu et al., 2022), because this phenotype increases phosphorus uptake through topsoil foraging (Jia et al., 2018; Rangarajan et al., 2022). Similar effects would be expected with other mobile and non-mobile resources within the soil profile. We call for more studies where inoculation experiments are accompanied by RSA assessments.
Seedling roots
The seed microbiome influences the seedling microbiome (Matsumoto et al., 2021; Walsh et al., 2021), which probably interacts with root phenotypes in seedling establishment. Young root systems have different architectures compared with their mature counterparts (Perkins and Lynch, 2021), which corresponds to different availabilities of soil resources in agricultural systems at each phenological stage (e.g. nitrogen fertilizer, water, and tillage). Not surprisingly, the seedling microbiome is different from those of mature root systems (Tannenbaum et al., 2020; Quiza et al., 2023), with soil microbes having stronger effects than the resident seed microbiome in the assembly of the seedling root microbiome (Rochefort et al., 2021). Conversely, most root microbiome research is frequently performed at the seedling stage and in small pots. Under these conditions, the seed microbiome might have a stronger legacy effect on the whole root microbiome as a result of the physical proximity of old and young roots. Therefore, the effects of microbial inoculation observed in small pots become irreproducible when scaling to field conditions. We call for studies with larger pots (mesocosms) with volumes comparable with those available to field-grown plants when field evaluation of seedlings is not possible.
Pathogenic associations and root system architecture
Abiotic stress is a nearly universal constraint in terrestrial ecosystems, and plants generally experience multiple abiotic stress conditions concurrently (Lynch, 2022a). These stresses reduce plant health, growth, and fitness, and hence change their sensitivity to biotic stress. The range of potential interactions of abiotic and biotic stresses is complex. In some cases, abiotic stress decreases sensitivity to biotic stress, as is the case, for example, for nitrogen deficiency reducing the severity of obligate fungal pathogens and some foliar-feeding insects by reducing the nutritional quality of the host plant (Marschner, 1995). In other cases, resistance to biotic and abiotic stress shares a mechanistic basis, as is the case for silicon nutrition regulating plant response to pathogens, insects, and heavy metal toxicity (Etesami and Jeong, 2018). However, the general case is that abiotic stress weakens defense mechanisms and increases susceptibility to biotic stress (Suzuki et al., 2014). Root architectural phenotypes capable of improving the capture of soil resources, thereby alleviating water and nutrient deficit stress, which are globally the most important abiotic stress factors (Lynch, 2022a), should therefore have an indirect benefit for resistance to biotic stress. A recent in silico study showed that root architecture directly affects plant growth in response to root loss (as occurs in response to many soil pests and pathogens) under nutrient stress, with some architectural phenotypes being resilient to root loss under nitrogen deficiency, or even increasing plant growth in response to root loss (Schäfer et al., 2022).
Identifying RSA phenotypes that reduce the deleterious effects of root disease should be a target of plant breeding. Pathogen colonization can occur in both axial and lateral roots, causing root loss or diminishment of root functions (Galindo-Castañeda et al., 2019). Crops are less sensitive to lateral root loss than to axial root loss (Schäfer et al., 2022), and lateral roots are more sensitive to pathogen colonization. Therefore, plants with an RSA with a greater length ratio of axial to lateral roots might be more resilient to pathogen attack than plants with lower ratios.
Root growth angles that locate larger proportions of the root biomass in intermediate and deep soil domains rather than shallow domains may be beneficial to avoid pathogen colonization of fungi and pathogens that remain from previous seasons (Fig. 1). Deep or intermediate roots would be less exposed to the initial inoculant and, in general, hypoxic conditions reduce a wide range of soil-borne pathogens (Lopes et al., 2022). There are very few field studies focusing on RSA and root pathogens, but phenotypes with deeper roots had increased disease resistance in alfalfa (Mattupalli et al., 2019) and in durum wheat (Alahmad et al., 2020; Saad et al., 2023) for example.
To reduce deleterious effects of pathogen colonization, timing is important. Plants need time to develop resistance to certain pathogens. Therefore, adaptations to evade pathogen colonization at the beginning of the growth cycle might be advantageous. Seminal roots with steep root growth angles to reduce encounters with pathogens, rapid proliferation of lateral roots that might act as traps for pathogens to trigger resistance responses in the plant and facilitate the expression of immunity, and plasticity to rapidly replace lost roots with new lateral or axial roots could be examined as possible architectural phenotypes to reduce soil-borne disease in roots.
Conclusion
RSA drives the location and environmental conditions in which root microbial associations occur in agricultural soils. However, RSA and soil vertical gradients are rarely included in root microbiome studies, creating a significant gap in our understanding of the mechanisms of root–microbe associations in real soils. Moreover, RSA has adaptive value for soil resource uptake, and plants naturally balance their carbon economy to allocate roots in soil domains where resources are more abundant. This allocation affects microbial processes in the rhizosphere in ways that are poorly understood. Therefore, efforts to select microbiomes that could benefit crops should include better ways to assess the mechanisms underlying root–microbiome associations using experimental systems that allow measurement of RSA and that account for soil vertical gradients. In this way, plants could express their real adaptations to acquire soil resources while also associating with soil microbes.
Research on the location of root carbon rhizodeposition within root systems is needed to understand microbial processes in the rhizosphere under realistic heterogenous soil conditions. We have provided several hypotheses regarding the effects of the interaction of RSA and soil vertical gradients on rhizosphere microbial associations and processes that merit experimental investigation (Table 1). Notably, research on the interactions of rhizosphere microbial processes with RSA is yet to be developed. We have further proposed a research agenda that will progressively lead to a more sustainable use of plant germplasm, soil microbes, and soil resources (Box 3). Methodological tools such as accessible experimental systems, sampling methods to capture the biodiversity of the rhizosphere in root systems, germplasm with contrasting but stable root phenotypes, and understanding of the genetic control of RSA, including plasticity, are urgent matters that will help achieve the goals of modern agriculture (Box 3).
Contributor Information
Tania Galindo-Castañeda, Department of Environmental Systems Service, ETH Zürich, 8092 Zurich, Switzerland.
Martin Hartmann, Department of Environmental Systems Service, ETH Zürich, 8092 Zurich, Switzerland.
Jonathan P Lynch, Department of Plant Science, The Pennsylvania State University, University Park, PA 16802, USA.
Miriam Gifford, University of Warwick, UK.
Author contributions
All authors contributed equally to this paper.
Conflict of interest
The authors declare no conflicts of interest.
Funding
Support for TGC and MH comes from the Swiss National Science Foundation (project 310030_207952), Horizon 2020 MSCA (project 839235), and Horizon H2020 (project 101000371).
References
- Alahmad S, Kang Y, Dinglasan E, Mazzucotelli E, Voss-Fels KP, Able JA, Christopher J, Bassi FM, Hickey LT.. 2020. Adaptive traits to improve durum wheat yield in drought and crown rot environments. International Journal of Molecular Sciences 21, 5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan Y, de-Bashan LE.. 2010. How the plant growth-promoting bacterium Azospirillum promotes plant growth—a critical assessment. Advances in Agronomy 108, 77–136. [Google Scholar]
- Bilyera N, Zhang X, Duddek P, et al. 2021. Maize genotype-specific exudation strategies: an adaptive mechanism to increase microbial activity in the rhizosphere. Soil Biology and Biochemistry 162, 108426. [Google Scholar]
- Bonkowski M, Tarkka M, Razavi BS, Schmidt H, Blagodatskaya E, Koller R, Yu P, Knief C, Hochholdinger F, Vetterlein D.. 2021. Spatiotemporal dynamics of maize (Zea mays L) root growth and its potential consequences for the assembly of the rhizosphere microbiota. Frontiers in Microbiology 12, 619499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SH, et al. 2017. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Roszak P, Orvošová M, Paszkowski U.. 2022. Arbuscular mycorrhizal fungi induce lateral root development in angiosperms via a conserved set of MAMP receptors. Current Biology 32, 4428–4437. [DOI] [PubMed] [Google Scholar]
- Colombi T, Kirchgessner N, Le Marié CA, York LM, Lynch JP, Hund A.. 2015. Next generation shovelomics: set up a tent and REST. Plant and Soil 388, 1–20. [Google Scholar]
- Contreras-Cornejo HA, Macías-Rodríguez L, Cortés-Penagos C, López-Bucio J.. 2009. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiology 149, 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra FA, Zhu B, Cheng W.. 2021. Root effects on soil organic carbon: a double-edged sword. New Phytologist 230, 60–65. [DOI] [PubMed] [Google Scholar]
- Eichmann R, Richards L, Schäfer P.. 2021. Hormones as go-betweens in plant microbiome assembly. The Plant Journal 105, 518–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etesami H, Jeong BR.. 2018. Silicon (Si): review and future prospects on the action mechanisms in alleviating biotic and abiotic stresses in plants. Ecotoxicology and Environmental Safety 147, 881–896. [DOI] [PubMed] [Google Scholar]
- Frankenberger WT, Arshad M.. 2020. Phytohormones in soils: microbial production and function. Boca Raton, FL: CRC Press. [Google Scholar]
- Galindo-Castañeda T, Brown KM, Kuldau GA, Roth GW, Wenner NG, Ray S, Schneider H, Lynch JP.. 2019. Root cortical anatomy is associated with differential pathogenic and symbiotic fungal colonization in maize. Plant, Cell & Environment 42, 2999–3014. [DOI] [PubMed] [Google Scholar]
- Galindo-Castañeda T, Lynch JP, Six J, Hartmann M.. 2022. Improving soil resource uptake by plants through capitalizing on synergies between root architecture and anatomy and root-associated microorganisms. Frontiers in Plant Science 13, 827369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Lynch JP.. 2016. Reduced crown root number improves water acquisition under water deficit stress in maize (Zea mays L). Journal of Experimental Botany 67, 4545–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnica-Vergara A, Barrera-Ortiz S, Muñoz-Parra E, Raya-González J, Méndez-Bravo A, Macías-Rodríguez L, Ruiz-Herrera LF, López-Bucio J.. 2016. The volatile 6-pentyl-2H-pyran-2-one from Trichoderma atroviride regulates Arabidopsis thaliana root morphogenesis via auxin signaling and ETHYLENE INSENSITIVE 2 functioning. New Phytologist 209, 1496–1512. [DOI] [PubMed] [Google Scholar]
- Gfeller V, Waelchli J, Pfister S, et al. 2023. Plant secondary metabolite-dependent plant–soil feedbacks can improve crop yield in the field. eLife 12, e84988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonin M, Salas-González I, Gopaulchan D, Frene JP, Roden S, Van de Poel B, Salt DE, Castrillo G.. 2023. Plant microbiota controls an alternative root branching regulatory mechanism in plants. Proceedings of the National Academy of Sciences, USA 120, e2301054120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartemink AE, Zhang Y, Bockheim JG, Curi N, Silva SHG, Grauer-Gray J, Lowe DJ, Krasilnikov P.. 2020. Soil horizon variation: a review. Advances in Agronomy 160, 125–185. [Google Scholar]
- Hartmann M, Six J.. 2023. Soil structure and microbiome functions in agroecosystems. Nature Reviews. Earth & Environment 4, 4–18. [Google Scholar]
- He L, Sun X, Li S, Zhou W, Chen Z, Bai X.. 2023. The vertical distribution and control factor of microbial biomass and bacterial community at macroecological scales. The Science of the Total Environment 869, 161754. [DOI] [PubMed] [Google Scholar]
- Hill PW, Jones DL.. 2019. Plant–microbe competition: does injection of isotopes of C and N into the rhizosphere effectively characterise plant use of soil N? New Phytologist 221, 796–806. [DOI] [PubMed] [Google Scholar]
- Hosseini SS, Lakzian A, Razavi BS.. 2022. Reduction in root active zones under drought stress controls spatial distribution and catalytic efficiency of enzyme activities in rhizosphere of wheat. Rhizosphere 23, 100561. [Google Scholar]
- Hungria M, Barbosa JZ, Rondina ABL, Nogueira MA.. 2022. Improving maize sustainability with partial replacement of N fertilizers by inoculation with Azospirillum brasilense. Agronomy Journal 114, 2969–2980. [Google Scholar]
- Ishii S, Ikeda S, Minamisawa K, Senoo K.. 2011. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes and Environments 26, 282–292. [DOI] [PubMed] [Google Scholar]
- Jia X, Liu P, Lynch JP.. 2018. Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. Journal of Experimental Botany 69, 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A, Dennis PG, Forstner C, Raghavendra AKH, Richardson AE, Watt M, Mathesius U, Gilliham M, Ryan PR.. 2021. The microbiomes on the roots of wheat (Triticum aestivum L.) and rice (Oryza sativa L) exhibit significant differences in structure between root types and along root axes. Functional Plant Biology 48, 871–888. [DOI] [PubMed] [Google Scholar]
- Kell DB. 2012. Large-scale sequestration of atmospheric carbon via plant roots in natural and agricultural ecosystems: why and how. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestel J, Larsbo M, Jarvis N.. 2020. Scale and REV analyses for porosity and pore connectivity measures in undisturbed soil. Geoderma 366, 114206. [Google Scholar]
- Krome K, Rosenberg K, Dickler C, Kreuzer K, Ludwig-Müller J, Ullrich-Eberius C, Scheu S, Bonkowski M.. 2010. Soil bacteria and protozoa affect root branching via effects on the auxin and cytokinin balance in plants. Plant and Soil 328, 191–201. [Google Scholar]
- Li Q, Li A, Yu X, et al. 2020. Soil acidification of the soil profile across Chengdu Plain of China from the 1980s to 2010s. The Science of the Total Environment 698, 134320. [DOI] [PubMed] [Google Scholar]
- Liu S, Barrow CS, Hanlon M, Lynch JP, Bucksch A.. 2021. DIRT/3D: 3D root phenotyping for field-grown maize (Zea mays). Plant Physiology 187, 739–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Evans SE, Friesen ML, Tiemann LK.. 2022. Root exudates shift how N mineralization and N fixation contribute to the plant-available N supply in low fertility soils. Soil Biology and Biochemistry 165, 108541. [Google Scholar]
- Longepierre M, Feola Conz R, Barthel M, Bru D, Philippot L, Six J, Hartmann M.. 2022. Mixed effects of soil compaction on the nitrogen cycle under pea and wheat. Frontiers in Microbiology 12, 822487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longepierre M, Widmer F, Keller T, Weisskopf P, Colombi T, Six J, Hartmann M.. 2021. Limited resilience of the soil microbiome to mechanical compaction within four growing seasons of agricultural management. ISME Communications 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes EA, Canedo EJ, Gomes VA, Vieira BS, Parreira DF, Neves WS.. 2022. Anaerobic soil disinfestation for the management of soilborne pathogens: a review. Applied Soil Ecology 174, 104408. [Google Scholar]
- Lynch JP. 2019. Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytologist 223, 548–564. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 2022a. Edaphic stress interactions: important yet poorly understood drivers of plant production in future climates. Field Crops Research 283, 108547. [Google Scholar]
- Lynch JP. 2022b. Harnessing root architecture to address global challenges. The Plant Journal 109, 415–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Strock CF, Schneider HM, Sidhu JS, Ajmera I, Galindo-Castañeda T, Klein SP, Hanlon MT.. 2021. Root anatomy and soil resource capture. Plant and Soil 466, 21–63. [Google Scholar]
- Lynch JP, Wojciechowski T.. 2015. Opportunities and challenges in the subsoil: pathways to deeper rooted crops. Journal of Experimental Botany 66, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. 1995. Mineral nutrition of higher plants. San Diego: Academic Press. [Google Scholar]
- Matsumoto H, Fan X, Wang Y, et al. 2021. Bacterial seed endophyte shapes disease resistance in rice. Nature Plants 7, 60–72. [DOI] [PubMed] [Google Scholar]
- Mattupalli C, Seethepalli A, York LM, Young CA.. 2019. Digital imaging to evaluate root system architectural changes associated with soil biotic factors. Phytobiomes Journal 3, 102–111. [Google Scholar]
- McKay Fletcher DM, Ruiz S, Dias T, Petroselli C, Roose T.. 2020. Linking root structure to functionality: the impact of root system architecture on citrate-enhanced phosphate uptake. New Phytologist 227, 376–391. [DOI] [PubMed] [Google Scholar]
- Moreau D, Pivato B, Bru D, Busset H, Deau F, Faivre C, Matejicek A, Strbik F, Philippot L, Mougel C.. 2015. Plant traits related to nitrogen uptake influence plant–microbe competition. Ecology 96, 2300–2310. [DOI] [PubMed] [Google Scholar]
- Ortiz-Castro R, Campos-García J, López-Bucio J.. 2020. Pseudomonas putida and Pseudomonas fluorescens influence Arabidopsis root system architecture through an auxin response mediated by bioactive cyclodipeptides. Journal of Plant Growth Regulation 39, 254–265. [Google Scholar]
- Pereira MG, Loss A, Batista I, Melo TR, Neto ECS, Pinto LASR.. 2021. Biogenic and physicogenic aggregates: formation pathways, assessment techniques, and influence on soil properties. Revista Brasileira de Ciencia do Solo 45, e0210108. [Google Scholar]
- Perkins AC, Lynch JP.. 2021. Increased seminal root number associated with domestication improves nitrogen and phosphorus acquisition in maize seedlings. Annals of Botany 128, 453–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiza L, Tremblay J, Pagé AP, Greer CW, Pozniak CJ, Li R, Haug B, Hemmingsen SM, St-Arnaud M, Yergeau E.. 2023. The effect of wheat genotype on the microbiome is more evident in roots and varies through time. ISME Communications 3, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan H, Hadka D, Reed P, Lynch JP.. 2022. Multi-objective optimization of root phenotypes for nutrient capture using evolutionary algorithms. The Plant Journal 111, 38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan H, Postma JA, Lynch JP.. 2018. Co-optimization of axial root phenotypes for nitrogen and phosphorus acquisition in common bean. Annals of Botany 122, 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi BS, Zarebanadkouki M, Blagodatskaya E, Kuzyakov Y.. 2016. Rhizosphere shape of lentil and maize: spatial distribution of enzyme activities. Soil Biology and Biochemistry 96, 229–237. [Google Scholar]
- Rochefort A, Simonin M, Marais C, Guillerm-Erckelboudt A-Y, Barret M, Sarniguet A.. 2021. Transmission of seed and soil microbiota to seedling. mSystems 6, e0044621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangsiri M, Vejchasarn P, Saengwilai P, et al. 2021. Genetic control of root architectural traits in KDML105 chromosome segment substitution lines under well-watered and drought stress conditions. Plant Production Science 24, 512–529. [Google Scholar]
- Saad A, Christopher J, Martin A, McDonald S, Percy C.. 2023. Fusarium pseudograminearum and F. culmorum affect the root system architecture of bread wheat. The Crop Journal 11, 316–321. [Google Scholar]
- Saengwilai P, Tian X, Lynch JP.. 2014. Low crown root number enhances nitrogen acquisition from low nitrogen soils in maize (Zea mays L). Plant Physiology 166, 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Law AD, Moe LA.. 2016. Nicotiana roots recruit rare rhizosphere taxa as major root-inhabiting microbes. Microbial Ecology 71, 469–472. [DOI] [PubMed] [Google Scholar]
- Schäfer ED, Owen MR, Postma JA, Kuppe C, Black CK, Lynch JP.. 2022. Simulating crop root systems using OpenSimRoot. Methods in Molecular Biology 2395, 293–323. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Nunan N, Höck A, Eickhorst T, Kaiser C, Woebken D, Raynaud X.. 2018. Recognizing patterns: spatial analysis of observed microbial colonization on root surfaces. Frontiers in Environmental Science 6, 61. [Google Scholar]
- Schneider HM, Lor VSN, Hanlon MT, et al. 2021. Root angle in maize influences nitrogen capture and is regulated by calcineurin B-like protein (CBL)-interacting serine/threonine-protein kinase 15 (ZmCIPK15). Plant, Cell & Environment 45, 837–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider HM, Lynch JP.. 2020. Should root plasticity be a crop breeding target? Frontiers in Plant Science 11, 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf A, Carminati A, Ahmed MA, et al. 2022. Linking rhizosphere processes across scales: opinion. Plant and Soil 478, 5–42. [Google Scholar]
- Singh BN, Dwivedi P, Sarma BK, Singh GS, Singh HB.. 2019. A novel function of N-signaling in plants with special reference to Trichoderma interaction influencing plant growth, nitrogen use efficiency, and cross talk with plant hormones. 3 Biotech 9, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smercina DN, Bowsher AW, Evans SE, Friesen ML, Eder EK, Hoyt DW, Tiemann LK.. 2021. Switchgrass rhizosphere metabolite chemistry driven by nitrogen availability. Phytobiomes Journal 5, 88–96. [Google Scholar]
- Strock CF, Lynch JP.. 2020. Root secondary growth: an unexplored component of soil resource acquisition. Annals of Botany 126, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strock CF, Morrow de la Riva L, Lynch JP.. 2018. Reduction in root secondary growth as a strategy for phosphorus acquisition. Plant Physiology 176, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strock CF, Rangarajan H, Black CK, Schäfer ED, Lynch JP.. 2021. Theoretical evidence that root penetration ability interacts with soil compaction regimes to affect nitrate capture. Annals of Botany 129, 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Gao Y, Lynch JP.. 2018. Large crown root number improves topsoil foraging and phosphorus acquisition. Plant Physiology 177, 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R.. 2014. Abiotic and biotic stress combinations. New Phytologist 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Tannenbaum I, Kaur J, Mann R, Sawbridge T, Rodoni B, Spangenberg G.. 2020. Profiling the Lolium perenne microbiome: from seed to seed. Phytobiomes Journal 4, 281–289. [Google Scholar]
- Torres D, Benavidez I, Donadio F, et al. 2018. New insights into auxin metabolism in Bradyrhizobium japonicum. Research in Microbiology 169, 313–323. [DOI] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP.. 2011. Shovelomics: high throughput phenotyping of maize (Zea mays L) root architecture in the field. Plant and Soil 341, 75–87. [Google Scholar]
- Verbon EH, Liberman LM.. 2016. Beneficial microbes affect endogenous mechanisms controlling root development. Trends in Plant Science 21, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite JM, Dardick C.. 2021. The roles of the IGT gene family in plant architecture: past, present, and future. Current Opinion in Plant Biology 59, 101983. [DOI] [PubMed] [Google Scholar]
- Walsh CM, Becker-Uncapher I, Carlson M, Fierer N.. 2021. Variable influences of soil and seed-associated bacterial communities on the assembly of seedling microbiomes. The ISME Journal 15, 2748–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, White PJ, Shen J, Lambers H.. 2022. Linking root exudation to belowground economic traits for resource acquisition. New Phytologist 233, 1620–1635. [DOI] [PubMed] [Google Scholar]
- Zai X, Luo W, Bai W, Li Y, Xiao X, Gao X, Wang E, Wei G, Chen W.. 2021. Effect of root diameter on the selection and network interactions of root-associated bacterial microbiomes in Robinia pseudoacacia L. Microbial Ecology 82, 391–402. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bilyera N, Fan L, Duddek P, Ahmed MA, Carminati A, Kaestner A, Dippold MA, Spielvogel S, Razavi BS.. 2023. The spatial distribution of rhizosphere microbial activities under drought: water availability is more important than root-hair-controlled exudation. New Phytologist 237, 780–792. [DOI] [PubMed] [Google Scholar]
- Zúñiga A, Poupin MJ, Donoso R, Ledger T, Guiliani N, Gutiérrez RA, González B.. 2013. Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Molecular Plant-Microbe Interactions 26, 546–553. [DOI] [PubMed] [Google Scholar]


