Abstract
The genomic sequences of Methanococcus jannaschii and Methanobacterium thermoautotrophicum contain a structurally uncommon seryl-tRNA synthetase (SerRS) sequence and lack an open reading frame (ORF) for the canonical cysteinyl-tRNA synthetase (CysRS). Therefore, it is not clear if Cys-tRNACys is formed by direct aminoacylation or by a transformation of serine misacylated to tRNACys. To address this question, we prepared SerRS from two methanogenic archaea and measured the enzymatic properties of these proteins. SerRS was purified from M. thermoautotrophicum; its N-terminal peptide sequence matched the sequence deduced from the relevant ORF in the genomic data of M. thermoautotrophicum and M. jannaschii. In addition, SerRS was expressed from a cloned Methanococcus maripaludis serS gene. The two enzymes charged serine to their homologous tRNAs and also accepted Escherichia coli tRNA as substrate for aminoacylation. Gel shift experiments showed that M. thermoautotrophicum SerRS did not mischarge tRNACys with serine. This indicates that Cys-tRNACys is formed by direct acylation in these organisms.
Aminoacyl-tRNA formation is a crucial process in the cell, ensuring translation of the genetic mRNA with the required exquisite accuracy. Aminoacyl-tRNA synthetases catalyze the esterification of an amino acid onto their cognate tRNA; this direct process is the major route of aminoacyl-tRNA formation. However, there is also an indirect route, relying on tRNA-dependent transformation of an amino acid incorrectly charged to tRNA (3, 7). Aminoacyl-tRNA synthetases are highly conserved in evolution; the amino acid sequences and structures of a synthetase specific for a certain amino acid normally show a high degree of conservation in different organisms (4). This aided the task of gene assignments in the analysis of genomic sequences. However, there are two exceptions. The known genomic sequences of two archaea, Methanococcus jannaschii (2) and Methanobacterium thermoautotrophicum (15), lack recognizable open reading frames (ORFs) for lysyl- and cysteinyl-tRNA synthetases. We now know that some archaea and some bacteria contain an unusual class I lysyl-tRNA synthetase structurally unrelated to the canonical class II lysyl-tRNA synthetase present in most bacteria and eukaryotes (8, 9). However, it is still unknown how Cys-tRNACys is formed. tRNA-dependent amino acid transformation (7) may be a route involving misacylation of tRNACys with serine by seryl-tRNA synthetase (SerRS) and subsequent thiolation of the tRNA-bound serine in a reaction similar to the formation of selenocysteinyl-tRNASec (1). Here we report experiments which show that M. thermoautotrophicum SerRS does not misacylate tRNACys. Thus, it is unlikely that amino acid transformation is the pathway for Cys-tRNACys formation in methanogenic archaea.
MATERIALS AND METHODS
General.
M. thermoautotrophicum Marburg (DSM 2133) was grown anaerobically at 65°C on 80% H2–20% CO2–0.1% H2S as previously described (14). The cells were harvested anaerobically and stored at −80°C until use. tRNA was purified from frozen M. thermoautotrophicum or Methanococcus maripaludis cells by standard procedures (19), except that DEAE-cellulose chromatography was included as a final step; 20 mg of tRNA was obtained from 50 g of cell mass. Escherichia coli tRNA was purchased from Sigma. Sodium dodecyl sulfate (SDS)-gel electrophoresis was performed as described previously (6), and gels were stained with Coomassie brilliant blue (12). Protein concentrations were determined with the assay kit from Bio-Rad. Proteins were prepared for N-terminal sequencing (performed by the Keck Biotechnology Resource Laboratory, Yale University) by blotting from an SDS-gel onto an Immobilon-P membrane (Millipore) (17). Standard molecular biology methods were as described previously (13).
SerRS assay.
Enzyme activity was measured as described previously for other SerRS enzymes (11). The reaction mixture contained 0.1 M HEPES (pH 8.0), 10 mM magnesium acetate, 10 mM KCl, 10 mM dithiothreitol, 1 mM ATP, 50 mM serine, 3.6 mM 3[H]serine (specific activity, 19.7 Ci/mmol), and tRNA (1 mg/ml). Incubations were performed at 60°C for the M. thermoautotrophicum enzyme and at 37°C for the M. maripaludis or E. coli SerRS. One unit of enzyme activity is defined as 1 pmol of serine charged per min per mg of protein.
Purification of SerRS from M. thermoautotrophicum cells.
All steps described were performed at 4°C. Frozen M. thermoautotrophicum cells (10 g) were resuspended in 30 ml of buffer A (50 mM Tris-HCl [pH 7.6], 10 mM magnesium chloride, and 2 mM dithiothreitol) plus 100 U of RNase-free DNase (Boehringer Mannheim), disrupted by passing the suspension twice through a French pressure cell at 1.5 MPa, and centrifuged at 100,000 × g for 60 min. The resulting S-100 extract was diluted with an equal volume of buffer A and applied to a Q-Sepharose FF HiLoad column. After the column was washed with 50 ml of buffer A the SerRS-containing protein fraction was eluted with 0.2 M NaCl in buffer A. The eluate was pooled, dialyzed against buffer A, and adjusted to 1.5 M potassium acetate before being separated by hydrophobic interaction chromatography on phenyl-Sepharose HP HiLoad 16/10, developed with a decreasing potassium acetate gradient (1.5 to 0 M), and washed with buffer A. SerRS eluted in the absence of potassium acetate and the protein fractions were pooled and applied to a MonoQ 5/5 column; SerRS eluted at 0.2 M NaCl. The proteins were eluted with a linear salt gradient from 0 to 0.4 M NaCl. Active fractions were pooled, concentrated, and further separated by gel filtration on Superose 12 in buffer A.
Cloning of the M. maripaludis serS gene.
An M. maripaludis genomic λ Zap Express library was screened with 32P-labeled oligonucleotides. The oligonucleotide sequences were from regions of the M. jannaschii and M. thermoautotrophicum serS gene with high conservation in known serS genes. A clone which contained the complete serS coding region was isolated and sequenced. The clone included 8 bp upstream of the serS ATG start codon at the 5′ end and the gene for a 50S ribosomal protein downstream. A forward primer with an NdeI site at the ATG start codon and a backward primer a few base pairs downstream of the stop codon containing a BlpI site were used in a PCR on DNA isolated from this clone. The PCR product was cut with restriction enzymes and directly cloned into expression vector pET11a (Invitrogen). This plasmid (pET11a-SerS) was sequenced and transformed into BL21(DE3) for expression of the protein.
Expression and isolation of M. maripaludis SerRS.
E. coli BL21(DE3) cells containing pET11a-SerS were grown at 30°C in Luria-Bertani medium with 100 μg of ampicillin per ml. Expression of the protein was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 5 h, the cells were collected by centrifugation and resuspended in 25 mM HEPES (pH 7.2)–10 mM KCl–5 mM dithiothreitol–4 mM β-mercaptoethanol–10% glycerol. The cells were disrupted by sonication, and a cell-free S-100 fraction was obtained by centrifugation for 1 h at 100,000 × g. To separate the expressed archaeal protein from the E. coli SerRS, the S-100 fraction was applied to a MonoQ HR 10/10 column previously equilibrated with 50 mM HEPES (pH 7.2)–10 mM magnesium chloride–2 mM β-mercaptoethanol. The proteins were eluted from the column with a linear gradient from 0 to 0.5 M NaCl. Fractions containing M. maripaludis SerRS, eluting around 0.2 M NaCl, were pooled, adjusted to 1.5 M ammonium sulfate, and loaded onto phenyl-Sepharose HiLoad. The column was developed with a linear gradient from 750 to 0 mM ammonium sulfate. The M. maripaludis SerS was eluted from the column at 0.66 M ammonium sulfate, while the E. coli SerRS was eluted at 0.20 M.
tRNA separation on acid-urea gels and Northern blot hybridization.
Aminoacyl-tRNA for separation on acidic polyacrylamide-urea gels was recovered from charging-reaction mixtures by extraction with phenol (equilibrated with 10 mM sodium acetate [pH 5.0]–50 mM NaCl) followed by ethanol precipitation. Aliquots of charged and uncharged tRNA were resuspended in sample buffer (0.1 sodium acetate [pH 5.0], 8 M urea, 0.05% bromophenol blue, 0.05% xylene cyanol) and fractionated on a 6.5% polyacrylamide gel as described previously (18). tRNA was transferred onto a Nytran membrane (Schleicher & Schuell) and fixed to the membrane by baking at 70°C for 2 to 3 h. Northern hybridization was carried out for 12 h at 42°C, as described previously (18), with 5′-32P-labeled oligonucleotides complementary to positions 25 to 49 of M. thermoautotrophicum tRNACys or positions 25 to 46 of tRNASer (mixed probe of three tRNASer sequences).
Phylogenetic tree analysis.
The phylogenetic tree was generated from complete SerRS sequences available in GenBank or Swissprot. The tree was based on maximum-likelihood analysis of quartets of aligned amino acid sequences by using the Puzzle program (16).
Nucleotide sequence accession number.
The nucleotide sequence of the M. maripaludis serS gene has been deposited in GenBank under accession no. AF009822.
RESULTS
Purification of M. thermoautotrophicum SerRS.
Examination of the serS ORF in the M. jannaschii and M. thermoautotrophicum genomic sequence revealed a protein which showed the poorest alignment with all known SerRS enzymes. Therefore, we decided to purify SerRS from M. thermoautotrophicum, to correlate it to the ORF by microsequencing its amino-terminal region, and to study its biochemical properties. SerRS was purified from frozen M. thermoautotrophicum cells by standard column chromatographic techniques, using seryl-tRNA formation of unfractionated M. thermoautotrophicum tRNA as the assay (see Materials and Methods) (Table 1). After purification on four columns, the enzyme was about 860-fold purified. The protein fraction was judged by SDS-gel electrophoresis to be over 90% pure; the SerRS protein had a mass of 62 kDa. The SerRS preparation acylated unfractionated M. thermoautotrophicum tRNA to a level of 140 pmol of serine per A260 unit of tRNA. This level of tRNASer is similar to what is found in E. coli. The amino-terminal sequence of the gel-purified SerRS was established to be MKFKLKGIIKLSK. This is identical to the first 13 amino acids of the deduced sequence of ORF Mt1076 (15). Thus, this ORF encodes SerRS.
TABLE 1.
Purification of M. thermoautotrophicum SerRS
| Fraction | Total amt of protein (mg) | Sp act (pmol min−1 mg−1) | Total activity (pmol) | Enrichment factor (fold) | Yield (%) |
|---|---|---|---|---|---|
| S-100 | 307.4 | 7.6 | 2,334 | ||
| Q-Sepharose | 44.1 | 43.3 | 1,905 | 5.7 | 81.6 |
| Phenyl-Sepharose | 4.62 | 109 | 504 | 14.3 | 21.6 |
| MonoQ 5/5 | 0.61 | 417 | 254 | 55 | 10.9 |
| Superose12 | 0.016 | 6,530 | 105 | 859 | 4.5 |
Cloning and expression of serS from M. maripaludis.
To provide another example of this uncommon SerRS we decided to clone serS, the gene encoding this enzyme, from the mesophilic archaeon M. maripaludis. A genomic clone was isolated by screening a genomic λ Zap Express library with degenerate oligonucleotides designed from the conserved regions of the serS gene from M. thermoautotrophicum and M. jannaschii. The nucleotide sequence was determined; the serS ORF also encoded a less common SerRS. The serS gene was subcloned into pET11a for expression of SerRS in E. coli. The expressed protein was mostly insoluble at 37°C, but soluble and active SerRS could be obtained by expression at a lower temperature (30°C). The expressed archaeal protein was separated from the E. coli SerRS by phenyl-Sepharose column chromatography. The partially purified SerRS charged M. maripaludis tRNA to a level of 120 pmol of serine/A260 unit of tRNA.
Serylation of various tRNAs by the archaeal SerRS enzymes.
The ability of SerRS from both methanogens and from E. coli to charge homologous and heterologous unfractionated tRNA samples was tested (Table 2). E. coli SerRS charged neither M. thermoautotrophicum nor M. maripaludis tRNA. While the M. jannaschii and M. thermoautotrophicum tRNASer species contain the identity elements required for recognition by E. coli SerRS, the presence of archaea-specific base modifications (see, e.g., reference 10) could be the reason for the lack of charging of M. thermoautotrophicum and M. maripaludis tRNA by E. coli SerRS. However, both archaeal SerRS enzymes charged E. coli tRNA in addition to archaeal tRNA. The M. thermoautotrophicum SerRS charged E. coli tRNA to a lesser extent (72 pmol per A260 unit) than did the E. coli enzyme (160 pmol per A260 unit), whereas M. maripaludis SerRS recognized E. coli tRNA well. All archaeal tRNASer isoacceptors are equal in length and contain 16 bases in their variable loop; two E. coli serine isoacceptors have longer variable loops (18 and 21 bases). Possibly, the longer tRNASer species are not substrates for the M. thermoautotrophicum enzyme.
TABLE 2.
Heterologous charging by SerRS
| SerRS source | Charging of:
|
||
|---|---|---|---|
| E. coli tRNA | M. maripaludis tRNA | M. thermoautotrophicum tRNA | |
| E. coli | ++a | − | − |
| M. maripaludis | ++ | ++ | + |
| M. thermoautotrophicum | + | ++ | ++ |
E. coli tRNA was charged to 160 pmol/A260 unit by E. coli SerRS.
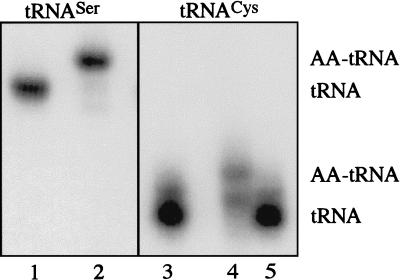
We then attempted to determine whether M. thermoautotrophicum SerRS is able to mischarge M. thermoautotrophicum tRNACys with serine. The M. thermoautotrophicum genomic sequence predicts the presence of three tRNASer and one tRNACys species. As in other organisms, the tRNASer species have a long variable loop whereas the tRNACys is 16 nucleotides shorter. Thus, these tRNAs should appear as separate bands on gel electrophoresis (18). We acylated unfractionated M. thermoautotrophicum tRNA with serine and separated charged and uncharged tRNA by acidic polyacrylamide-urea gel electrophoresis (Fig. 1). The positions of tRNACys and tRNASer in the charged and uncharged forms were identified by Northern hybridization with appropriate 32P-labeled oligonucleotides. Half the gel was hybridized with the tRNASer probe (Fig. 1), and the other was hybridized with the tRNACys oligonucleotide (Fig. 1). A major portion of the tRNASer was acylated, as shown by the lower gel mobility (upper band in Fig. 1, lane 2). However, M. thermoautotrophicum tRNACys was not misacylated (Fig. 1, compare lanes 3 and 4) by partially purified homologous SerRS (leading to some degradation of tRNA). However, the M. thermoautotrophicum tRNA could be charged with cysteine by purified E. coli CysRS (Fig. 1, lane 4). Hence, the M. thermoautotrophicum SerRS does not mischarge tRNACys with serine.
FIG. 1.
Northern blot analysis of unfractionated M. thermoautotrophicum tRNA charged with purified M. thermoautotrophicum SerRS. Charged and uncharged tRNA was separated (18), and the blots were probed with a tRNASer and tRNACys probe. Lanes: 1 and 3, uncharged tRNA; 2 and 5, tRNA charged with serine by M. thermoautotrophicum SerRS; 4, tRNA charged with cysteine by E. coli CysRS. AA, amino acid.
DISCUSSION
The serS genes in the three methanogenic archaea M. thermoautotrophicum, M. maripaludis, and M. jannaschii display only low similarity to serS genes present in bacteria, eucarya, and even some archaea (Archaeoglobus fulgidus, Pyrococcus furiosus, Pyrococcus horikoshii, Pyrobaculum aerophilum, and Haloarcula marismortui). The genomic annotated sequence alignment had raised some doubt about the correct identification of this gene in these organisms. As a class II aminoacyl-tRNA synthetase, SerRS is defined by the presence of three sequence motifs (4, 5). Figure 2 shows a sequence alignment of these motifs from a number of SerRS enzymes of eukaryotic, bacterial, and archaeal origins. As can be seen, the overall similarity between the SerRS from the methanogenic archaea and SerRS proteins from other organisms is lower. In addition, motif II has a gap which is absent in other SerRS proteins, including those from the other archaea Haloarcula marismortui, Pyrococcus horikoshii, and Archaeoglobus fulgidus. The compilation also reveals some sequence insertions elsewhere in the sequence of the methanogenic archaeal enzymes (data not shown).
FIG. 2.
Alignment of motifs 1, 2, and 3 (4) from a number of representative SerRSs. The sequences (accession numbers) are from Homo sapiens (X91257) (Hsa), Saccharomyces cerevisiae (X04884) (Sce), Arabidopsis thaliana (Z70313) (Ath), Drosophila melanogaster (Y14823) (Dme), Escherichia coli (X04017) (Eco), Bacillus subtilis (D26185) (Bsu), Thermus aquaticus (sp:P34945) (Taq), Haloarcula marismortui (X91007) (Hma), Archaeoglobus fulgidus (AE000962) (Afu), Pyrococcus horikoshii (AB009490) (Pho), Methanococcus jannaschii (U67550) (Mja), Methanobacterium thermoautotrophicum (AF009823) (Mth), and Methanococcus maripaludis (AF009822) (Mma). They were aligned with the Clustal program (16), and the motif regions are presented.
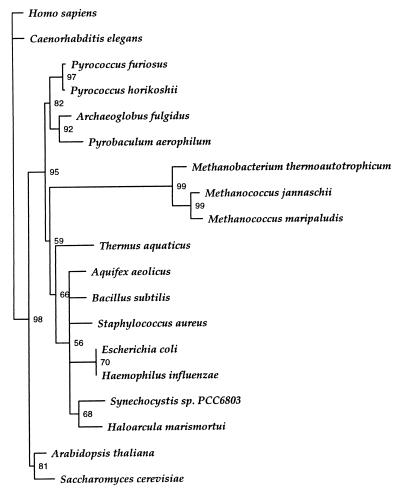
However, as demonstrated above, this enzyme is the active SerRS in these methanogenic archaea. An evolutionary tree (Fig. 3), based on maximum-likelihood methods, furthermore reveals that the SerRS of the methanogens cluster in a distinct clade separate from all other SerRSs including those of other archaea. It may also be pertinent that based on our current sequence knowledge, SerRS from the archaeon Haloarcula marismortui does not cluster with any of the archaea.
FIG. 3.
Phylogenetic analysis of SerRS alignments by maximum likelihood analysis, using the Puzzle program (16).
The absence of a recognizable cysteinyl-tRNA synthetase in the genomes of M. thermoautotrophicum and M. jannaschii, taken together with the pronounced difference of their SerRS sequence (see above), lent credence to the notion that in these organisms Cys-tRNACys might be produced by a tRNA-dependent thiolation of Ser-tRNACys resembling the synthesis of selenocysteinyl-tRNASec (1). Our in vitro experiments with purified SerRS do not support this idea. However, if serylation of tRNACys required proteins in addition to M. thermoautotrophicum SerRS, an unlikely scenario based on our current knowledge of aminoacyl-tRNA formation, we would not have detected mischarging.
ACKNOWLEDGMENTS
H.-S. Kim and Ute C. Vothknecht contributed equally to this work.
We are indebted to W. Gardner and W. Lin for providing a M. maripaludis library and cells; S. Cusack, K. W. Hong, M. Ibba, R. Leberman, and G. Olsen for discussions; R. Vaidyanathan and U. L. RajBhandary for help; and R. K. Thauer and W. Whitman for encouragement. We thank S. Fitz-Gibbon and J. Miller for sharing the unpublished Pyrobaculum aerophilum SerRS sequence.
This work was supported by grants from NIGMS (GM22864 and GM55674).
REFERENCES
- 1.Baron C, Böck A. The selenocysteine-inserting tRNA species: structure and function. In: Söll D, RajBhandary U, editors. tRNA: structure, biosynthesis and function. Washington, D.C: ASM Press; 1995. pp. 529–544. [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage Ar R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter F J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Curnow A W, Hong K W, Yuan R, Kim S I, Martins O, Winkler W, Henkin T M, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusack S. Eleven down and nine to go. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 5.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 6.Fling S P, Gregerson D S. Peptide and protein molecular weight determination by electrophoresis using a high-molarity Tris buffer system without urea. Anal Biochem. 1986;155:83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- 7.Ibba M, Curnow A W, Söll D. Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem Sci. 1997;22:39–42. doi: 10.1016/s0968-0004(96)20033-7. [DOI] [PubMed] [Google Scholar]
- 8.Ibba M, Bono J L, Rosa P A, Söll D. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi. Proc Natl Acad Sci USA. 1997;94:14383–14388. doi: 10.1073/pnas.94.26.14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibba M, Morgan S, Curnow A W, Pridmore D R, Vothknecht U C, Gardner W, Lin W, Woese C R, Söll D. A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- 10.Kowalak J A, Dalluge J J, McCloskey J A, Stetter K O. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 11.Lenhard B, Filipić S, Landeka I, Škrtić I, Söll D, Weygand-Durašević I. Defining the active site of yeast seryl-tRNA synthetase. J Biol Chem. 1997;272:1136–1141. doi: 10.1074/jbc.272.2.1136. [DOI] [PubMed] [Google Scholar]
- 12.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric-focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis R. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Schönheit P, Moll J, Thauer R K. Growth parameters (K2, mmax, Ys) of Methanobacterium thermoautotrophicum. Arch Microbiol. 1980;127:59–65. doi: 10.1007/BF00403508. [DOI] [PubMed] [Google Scholar]
- 15.Smith D R, Dourette-Stamm L A, Deboughery C, Lee H, Dubois J, Alderedge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qui D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani M, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7153. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobwin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets—procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varshney U, Lee, C. P. C P, Rajbhandary U L. Direct analysis of aminoacylation levels of transfer RNAs in vivo—application to studying recognition of Escherichia coli initiator transfer RNA mutants by glutaminyl-transfer RNA-synthetase. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 19.Zubay G. The isolation and fractionation of soluble ribonucleic acids. J Mol Biol. 1962;4:347–356. [Google Scholar]