Abstract
The lemur family of protein kinases has gained much interest in recent years as they are involved in a variety of cellular processes including regulation of axonal transport and endosomal trafficking, modulation of synaptic functions, memory and learning, and they are centrally placed in several intracellular signalling pathways. Numerous studies have also implicated role of the lemur kinases in the development and progression of a wide range of cancers, cystic fibrosis, and neurodegenerative diseases. However, parallel discoveries and inaccurate prediction of their kinase activity have resulted in a confusing and misleading nomenclature of these proteins. Herein, a group of international scientists with expertise in lemur family of protein kinases set forth a novel nomenclature to rectify this problem and ultimately help the scientific community by providing consistent information about these molecules.
Subject terms: Cancer genetics, Tumour biomarkers, Alzheimer's disease, Protein transport, Phosphorylation
Here a group of scientists with expertise in lemur family of protein kinases propose a new nomenclature for lemur kinases.
Introduction
Lemur kinase protein family consists of four protein members, lemur tyrosine kinase (LMTK) 1A, LMTK1B, LMTK2 and LMTK3, encoded by three genes. Despite their misleading name, work to date (published and unpublished) has predominantly demonstrated that they are actually serine/threonine-protein kinases instead of tyrosine kinases.
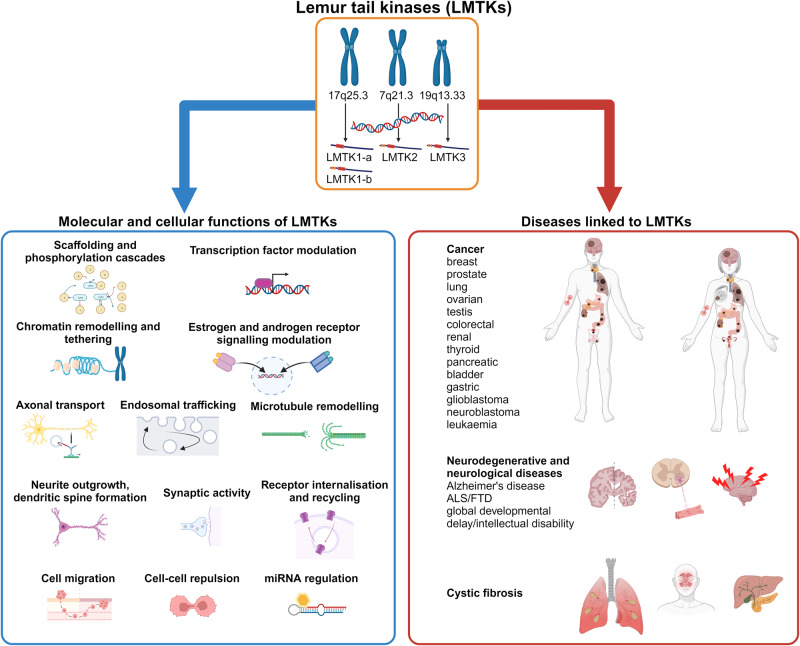
During the last decade, LMTKs have gained increasing attention due to their involvement in various diseases including cancer, cystic fibrosis, Alzheimer’s disease, amyotrophic lateral sclerosis/frontotemporal dementia and global developmental delay/intellectual disability1–6 (Fig. 1). Their association with cancer is the most studied to date as they have been implicated in a wide range of tumours including breast, prostate, lung, colorectal, renal, testis and ovarian, thyroid, pancreatic, bladder, gastric, glio- and neuroblastoma, and leukaemia7–9.
Fig. 1. Molecular and cellular functions, and diseases associated with lemur tail kinases (LMTKs).
LMTK1-a, LMTK1-b, LMTK2 and LMTK3 are involved in a wide range of cellular processes including gene expression regulation, protein phosphorylation cascades, intracellular transport, neuronal outgrowth, synaptic activity, and cell motility. LMTKs are also strongly implicated in the development and progression of a wide range of cancers, neurodegenerative and neurological diseases, and cystic fibrosis. ALS/FTD, amyotrophic lateral sclerosis/frontotemporal dementia; miRNA, microRNA.
Among them, LMTK2 has been identified as a susceptibility gene in prostate cancer and several other studies also highlighted that changes in LMTK2 and LMTK3 expressions can also be used as prognostic markers in colorectal, gastric, prostate and breast cancers4,5,10–12. These oncogenic changes in LMTK expression can also promote tumour growth by modulating drug resistance to avoid cytotoxic chemotherapy, promote tissue invasion and cell migration by modulating transcription factors, miRNA processing and chromatin remodelling13–19. In the case of prostate and breast cancer, LMTK2 and LMTK3 respectively can contribute to cancer development by interacting with the androgen receptor and oestrogen receptor α and modulating their functions4,20.
LMTKs are also extensively studied in the context of endosomal vesicle trafficking and axonal transport as they are present in endosomal membranes and bind to myosin VI and kinesin-1/kinesin light chain molecular motors, and are also involved in microtubule remodelling21–25. Specifically, LMTK1 is involved in axonal and dendritic outgrowth and maturation by regulating Rab11-positive endosome transport26–28. LMTK2 and LMTK3 also bind to and regulate the transport of several proteins which are all involved in synaptic functions, memory and learning such as α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-D-aspartic acid (NMDA) receptor subunits, potassium-chloride cotransporter 2, and the synaptic kinase cyclin-dependent kinase 5 (cdk5)/p3521,23,29,30. Apart from cdk5/p35, LMTKs also interact with protein phosphatase 1 to regulate glycogen synthase kinase-3β activity, which is also linked to synaptic functions15,31–35. Recently, both LMTK1 and LMTK2 have been linked to Alzheimer’s disease, amyotrophic lateral sclerosis/frontotemporal dementia and global developmental delay/intellectual disability, neurodegenerative and neurological diseases characterised by axonal transport defects and synaptic loss, suggesting that the role of LMTKs in intracellular transport is an important factor for the pathogenesis of these diseases1–3,21,36–38. LMTK2 also binds to and phosphorylates cystic fibrosis transmembrane conductance regulator (CFTR) to regulate its availability on the plasma membrane6. CFTR mutations cause cystic fibrosis and LMTK2 might be a potential drug target for the treatment of the disease. The presence and abundance of CFTR targeting LMTK2 in the apical membrane of airway epithelial cells is controlled by Rab11-positive endosomes in a transforming growth factor (TGF)-β1-dependent manner39. LMTK2 and TGF-β1 signalling is interwoven in complex signalling cascades as not only TGF-β1 affects LMTK2 signalling but TGF-β1 signalling is also influenced by LMTK2 regulatory network40 (Fig. 1).
All these findings direct the spotlight on LMTKs as potential disease drivers and therapeutic targets. However, their several alternative and misleading names cause scientific conundrum. Now that these kinases have gained growing attention, it is imperative to use a simple and unambiguous LMTK nomenclature to avoid further confusion. Following the ‘1st LMTKs international conference 2023’, the authors of this manuscript and members of this consortium set forth a novel lemur kinase nomenclature to rectify this problem and help the scientific community.
A brief history of lemur kinase nomenclature
The first lemur kinase family member was identified as a gene whose expression increases in mouse myeloid precursor cells during apoptosis. Sequence analysis suggested that it is a novel, putative tyrosine kinase since its kinase domain showed strong homology with other tyrosine kinases, and therefore, it was termed as apoptosis-associated tyrosine kinase (AATYK)41. Its human homologue was identified by the Kazusa cDNA sequencing project, an effort to identify novel human genes, and got the identification number KIAA064142. The human genes of the other two human family members were identified later under the identification numbers KIAA1079 and KIAA188343,44. Parallel with these discoveries, studies aiming to catalogue human protein kinases classified AATYK and the two newly identified genes as a new family of kinases. One of these studies named the family as AATYK and its members AATYK1 (the originally described AATYK), AATYK2 and AATYK345 while the other study named the family as Lmr consisting of Lmr1 (the originally described AATYK), Lmr2 and Lmr346. In this latter case, Manning and colleagues46 did not resolve what Lmr abbreviates for but others, in a later publication, referred to them as human lemur (Lmr) kinases47. The name lemur potentially originates from the visual similarity of the long carboxyl-terminal tail of the protein which makes it akin to the tail of the Madagascan lemurs. To increase the confusion, a human homologue of AATYK and a splice variant of KIAA0641 was identified in a yeast two-hybrid screen searching for p35 binding partners and was named as human AATYK short isoform-p35 binding polypeptide (hAATYKs-p35BP)48. This was the point when the earlier identified human KIAA1079 gene-encoded protein and its mouse homologue were identified by the Brautigan, Miller and Tadashi groups independently from each other34,49,50. Not surprisingly, these parallel discoveries resulted in three new names for the same protein, kinase/phosphatase/inhibitor-2 (KPI-2), Cdk5/p35-regulated kinase (Cprk) and brain-enriched kinase (BREK).
A few years later, Tomomura and co-workers identified two AATYK splice variants and renamed the original AATYK to AATYK1A, and the newly identified form AATYK1B and termed the other two lemur kinase family members as AATYK2, and AATYK351. Interestingly, the gene encoding AATYK had started to be referred to as AATK from 2008 without explaining the reason; see e.g., refs. 52,53. The year 2006 marks the first appearance of the now widely used LMTK acronym. It was mentioned in a paper describing an LMTK2 knockout mouse model, however, the authors did not explain the origin of this new name and after mentioning it as an alias they kept calling the gene and protein as Brek54. Since that year on, all the afore-mentioned names appeared in the literature but mainly usage of AATYK and LMTK dominated it, although some interesting hybrid names have also arisen such as AATKA and AATKB instead of AATYK1A and 1B55. To our surprise, after screening various gene and protein databases such as NCBI, Ensembl and UniProt we found several further LMTK aliases which have never appeared in the scientific literature according to our knowledge such as e.g., ‘CDK5-binding protein’, ‘protein phosphatase 1, regulatory subunit 77’ or TYKLM3. The origin of these aliases is unknown and they clearly do not make navigation easier in the already confusing sea of names and abbreviations of the lemur kinases (Table 1).
Table 1.
Standardised names for the lemur kinase family.
| Standardised gene and protein names | Other names |
|---|---|
|
Lemur tail kinase-1 LMTK1 |
apoptosis-associated tyrosine kinase (AATK, AATYK) apoptosis-associated tyrosine kinase-1 (AATYK1) lemur tyrosine kinase-1 (LMTK1) serine/threonine-protein kinase LMTK1 human AATYK short isoform-p35 binding polypeptide (hAATYKs-p35BP) KIAA0641 LMR1 CDK5-binding proteina brain apoptosis-associated tyrosine kinasea p35BPa protein phosphatase 1, regulatory subunit 77 (PPP1R77)a |
|
Lemur tail kinase-2 LMTK2 |
apoptosis-associated tyrosine kinase-2 (AATYK2) lemur tyrosine kinase-2 (LMTK2) brain-enriched kinase (BREK) Cdk5/p35-regulated kinase (Cprk) serine/threonine-protein kinase LMTK2 kinase/phosphatase/inhibitor-2 (KPI-2) serine/threonine-protein kinase KPI-2a KIAA1079 LMR2 protein phosphatase 1, regulatory subunit 100 (PPP1R100)a FLJ46659a KPI2a |
|
Lemur tail kinase-3 LMTK3 |
apoptosis-associated tyrosine kinase-3 (AATYK3) lemur tyrosine kinase-3 (LMTK3) serine/threonine-protein kinase LMTK3 KIAA1883 LMR3 TYKLM3a phosphatase 1, regulatory subunit 101 (PPP1R101)a |
aMarks those aliases which have never been referenced to in the scientific literature according to our knowledge.
Kinase specificity of LMTKs
As mentioned above, when LMTK1 was originally identified computational analysis of its kinase domain predicted that it is a tyrosine kinase41. However, experimental data conducted by several laboratories independently have proved that LMTK1, LMTK2 and LMTK3 solely phosphorylate serine and threonine residues and do not target tyrosine residue34,47,50,56. These findings strongly suggest that their original name is a misleading misnomer and that LMTKs are actually serine/threonine-specific kinases. Although we acknowledge the possibility that LMTKs might be dual-specific serine/threonine-tyrosine kinases there is no experimental data supporting this hypothesis to date. Despite of all the experimental data showing that LMTKs are serine/threonine kinases it might seem surprising that publications categorising kinases have still classified LMTKs as receptor tyrosine kinases until very recently with only one exception57. The reason behind it is that kinase family classifications into phylogenetic trees are based on sequence similarities of their catalytic domains and not on their function, however, we would like to emphasise again that LMTKs are serine/threonine-specific kinases.
The revised LMTK nomenclature
As a result of the ‘1st LMTKs international conference 2023’, all senior PIs agreed that a non-confusing single nomenclature must be adopted to facilitate research in the field. We constructed and are suggesting using the following rules and rationale.
The highly confusing reference to tyrosine kinase activity must be eliminated. At the same time, we suggest keeping the most widely used acronym LMTK. In this line, we embrace a recent suggestion to name these proteins ‘lemur tail kinases’ instead of ‘lemur tyrosine kinases’58. This naming approach has already been followed by some recent publications8,27.
Name lemur kinase genes and proteins identically in agreement with the HUGO Gene Nomenclature Guidelines and the International Protein Nomenclature Guidelines59 and https://www.ncbi.nlm.nih.gov/genome/doc/internatprot_nomenguide/. This avoids confusion that exists in the case of LMTK1 where the gene is named as AATK while the protein as LMTK1.
If new family members are identified, these must bear the name lemur tail kinase (LMTK) and the consecutive Arabic number. New subfamily members should get a Latin capital letter starting with ‘A’.
Functional protein-coding splice variants can get their own names but should be clearly distinguished from proteins encoded by different genes to avoid confusion with gene subfamily members using a hyphen and lowercase Latin letters starting with ‘a’. Therefore, we suggest using LMTK1-a and LMTK1-b instead of LMTK1A and LMTK1B for the LMTK1 splice variants.
We recommend that a statement needs to be included within the introduction section of publications referring to the former LMTK gene and protein names. For example, LMTK2 is a membrane-anchored serine/threonine kinase formerly known as kinase/phosphatase/inhibitor-2 (KPI-2), Cdk5/p35-regulated kinase (Cprk), brain-enriched kinase (BREK), apoptosis-associated tyrosine kinase-2 (AATYK2), KIAA1079 and LMR234,43,46,49–51.
Research groups supporting the use of this system for lemur kinase nomenclature include the following:
Neil A. Bradbury, Oana Caluseriu, Georgios Giamas, Shin-ichi Hisanaga, Heinz-Josef Lenz, Christopher C.J. Miller, Stephen J. Moss, Gábor M. Mórotz, Agnieszka Swiatecka-Urban.
Acknowledgements
This work was supported by the János Bolyai Research Fellowship of the Hungarian Academy of Sciences (BO/00277/23/5), the ÚNKP-23-5-SE-9 New National Excellence Programme of the Ministry for Culture and Innovation from the source of the National Research, Development and Innovation Fund, and the National Research, Development, and Innovation Office (NKFIH) of Hungary (OTKA-FK-146163) (to G.M.M), National Institutes of Health grants R01HL144539 and P50DK096373-11, and the Cystic Fibrosis Foundation SWIATE18G0 (to A.S-U.), MRC grant MR/R022666/1 and grants from the Alzheimer’s Society and ARUK (to C.C.J.M.). Figure 1 was created with BioRender.com.
Author contributions
All authors (G.M.M., N.A.B., O.C., S.H., C.C.J.M., A.S-U., H-J.L., S.J.M., G.G.) contributed to the design of this perspective. G.M.M. wrote and G.G. amended the first drafts of the manuscript. All authors edited the manuscript and approved the final version.
Peer review
Peer review information
Communications Biology thanks Krzysztof Pawłowski, Roland Dunbrack, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Isabelle Lucet and Christina Karlsson Rosenthal.
Competing interests
G.G. is the founder and chief scientific officer of Stingray Bio. G.G. is an Editorial Board Member for Communications Biology, but was not involved in the editorial review of, nor the decision to publish this article. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Neil A. Bradbury, Oana Caluseriu, Shin-ichi Hisanaga, Christopher C. J. Miller, Agnieszka Swiatecka-Urban, Heinz-Josef Lenz, Stephen J. Moss.
These authors jointly supervised this work: Gábor M. Mórotz, Georgios Giamas.
Contributor Information
Gábor M. Mórotz, Email: morotz.gabor@semmelweis.hu
Georgios Giamas, Email: g.giamas@sussex.ac.uk.
References
- 1.Vrabec K, et al. Differential expression of several miRNAs and the host genes AATK and DNM2 in leukocytes of sporadic ALS patients. Front. Mol. Neurosci. 2018;11:106. doi: 10.3389/fnmol.2018.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari R, et al. A genome-wide screening and SNPs-to-genes approach to identify novel genetic risk factors associated with frontotemporal dementia. Neurobiol. Aging. 2015;36:2904 e13–2904 e26. doi: 10.1016/j.neurobiolaging.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Kasbi G, et al. The diagnostic yield, candidate genes, and pitfalls for a genetic study of intellectual disability in 118 middle eastern families. Sci. Rep. 2022;12:18862. doi: 10.1038/s41598-022-22036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giamas G, et al. Kinome screening for regulators of the estrogen receptor identifies LMTK3 as a new therapeutic target in breast cancer. Nat. Med. 2011;17:715–719. doi: 10.1038/nm.2351. [DOI] [PubMed] [Google Scholar]
- 5.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 6.Luz S, et al. LMTK2 mediated phosphorylation regulates CFTR endocytosis in human airway epithelial cells. J. Biol. Chem. 2014;289:15080–15093. doi: 10.1074/jbc.M114.563742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditsiou A, et al. The multifaceted role of lemur tyrosine kinase 3 in health and disease. Open Biol. 2021;11:210218. doi: 10.1098/rsob.210218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendler F, Purice T-M, Simon T, Stebbing J, Giamas G. The LMTK-family of kinases: emerging important players in cell physiology and pathogenesis. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2021;1867:165372. doi: 10.1016/j.bbadis.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Cruz DF, Farinha CM, Swiatecka-Urban A. Unraveling the function of lemur tyrosine kinase 2 network. Front. Pharmacol. 2019;10:24. doi: 10.3389/fphar.2019.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, et al. Serum lemur tyrosine kinase 3 expression in colorectal cancer patients predicts cancer progression and prognosis. Méd. Oncol. 2013;30:754. doi: 10.1007/s12032-013-0754-x. [DOI] [PubMed] [Google Scholar]
- 11.Du M, et al. Integrated multi-omics approach to distinct molecular characterization and classification of early-onset colorectal cancer. Cell Rep. Med. 2023;4:100974. doi: 10.1016/j.xcrm.2023.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakatsuki T, et al. Prognostic role of lemur tyrosine kinase-3 germline polymorphisms in adjuvant gastric cancer in Japan and the United States. Mol. Cancer Ther. 2013;12:2261–2272. doi: 10.1158/1535-7163.MCT-12-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stebbing J, et al. LMTK3 is implicated in endocrine resistance via multiple signaling pathways. Oncogene. 2013;32:3371–3380. doi: 10.1038/onc.2012.343. [DOI] [PubMed] [Google Scholar]
- 14.Stebbing J, et al. LMTK3 confers chemo-resistance in breast cancer. Oncogene. 2018;37:3113–3130. doi: 10.1038/s41388-018-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, et al. LMTK3 represses tumor suppressor-like genes through chromatin remodeling in breast cancer. Cell Rep. 2015;12:837–849. doi: 10.1016/j.celrep.2015.06.073. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, et al. The kinase LMTK3 promotes invasion in breast cancer through GRB2-mediated induction of integrin beta(1) Sci. Signal. 2014;7:ra58. doi: 10.1126/scisignal.2005170. [DOI] [PubMed] [Google Scholar]
- 17.Conti A, et al. Lemur tyrosine kinase 2 (LMTK2) is a determinant of cell sensitivity to apoptosis by regulating the levels of the BCL2 family members. Cancer Lett. 2016;389:59–69. doi: 10.1016/j.canlet.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, L. et al. Lemur tyrosine kinase 2 has a tumor-inhibition function in human glioblastoma by regulating the RUNX3/Notch pathway. Biochim. Biophys. Acta BBA-Mol. Cell Res. 119509 10.1016/j.bbamcr.2023.119509 (2023). [DOI] [PubMed]
- 19.Jacob J, et al. LMTK3 escapes tumour suppressor miRNAs via sequestration of DDX5. Cancer Lett. 2016;372:137–146. doi: 10.1016/j.canlet.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 20.Shah K, Bradbury NA. Lemur tyrosine kinase 2, a novel target in prostate cancer therapy. Oncotarget. 2015;6:14233–14246. doi: 10.18632/oncotarget.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mórotz GM, et al. LMTK2 binds to kinesin light chains to mediate anterograde axonal transport of cdk5/p35 and LMTK2 levels are reduced in Alzheimer’s disease brains. Acta Neuropathol. Commun. 2019;7:73. doi: 10.1186/s40478-019-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chibalina MV, et al. and its interacting protein LMTK2 regulate tubule formation and transport to the endocytic recycling compartment. J. Cell Sci. 2007;120:4278–4288. doi: 10.1242/jcs.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, et al. BREK/LMTK2 is a myosin VI-binding protein involved in endosomal membrane trafficking. Genes Cells. 2008;13:483–495. doi: 10.1111/j.1365-2443.2008.01184.x. [DOI] [PubMed] [Google Scholar]
- 24.Sharma G, et al. Kinase activity of endosomal kinase LMTK1A regulates its cellular localization and interactions with cytoskeletons. Genes Cells. 2016;21:1080–1094. doi: 10.1111/gtc.12404. [DOI] [PubMed] [Google Scholar]
- 25.Cilibrasi C, et al. LMTK3 inhibition affects microtubule stability. Mol. Cancer. 2021;20:53. doi: 10.1186/s12943-021-01345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei R, et al. Isoform-dependent subcellular localization of LMTK1A and LMTK1B, and their roles in axon outgrowth and spine formation. J. Biochem. 2020;168:23–32. doi: 10.1093/jb/mvaa019. [DOI] [PubMed] [Google Scholar]
- 27.Hisanaga S-I, Wei R, Huo A, Tomomura M. LMTK1, a novel modulator of endosomal trafficking in neurons. Front. Mol. Neurosci. 2020;13:112. doi: 10.3389/fnmol.2020.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishino H, et al. The LMTK1-TBC1D9B-Rab11A cascade regulates dendritic spine formation via endosome trafficking. J. Neurosci. 2019;39:9491–9502. doi: 10.1523/JNEUROSCI.3209-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montrose K, Kobayashi S, Manabe T, Yamamoto T. Lmtk3-KO mice display a range of behavioral abnormalities and have an impairment in GluA1 trafficking. Neuroscience. 2019;414:154–167. doi: 10.1016/j.neuroscience.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 30.Smalley JL, et al. Isolation and characterization of multi-protein complexes enriched in the K-Cl co-transporter 2 from brain plasma membranes. Front. Mol. Neurosci. 2020;13:563091. doi: 10.3389/fnmol.2020.563091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gagnon KB, England R, Diehl L, Delpire E. Apoptosis-associated tyrosine kinase scaffolding of protein phosphatase 1 and SPAK reveals a novel pathway for Na-K-2C1 cotransporter regulation. Am. J. Physiol. Cell Physiol. 2007;292:C1809–C1815. doi: 10.1152/ajpcell.00580.2006. [DOI] [PubMed] [Google Scholar]
- 32.Manser C, Vagnoni A, Guillot F, Davies J, Miller CC. Cdk5/p35 phosphorylates lemur tyrosine kinase-2 to regulate protein phosphatase-1C phosphorylation and activity. J. Neurochem. 2012;121:343–348. doi: 10.1111/j.1471-4159.2012.07650.x. [DOI] [PubMed] [Google Scholar]
- 33.Manser C, et al. Lemur tyrosine kinase-2 signalling regulates kinesin-1 light chain-2 phosphorylation and binding of Smad2 cargo. Oncogene. 2012;31:2773–2782. doi: 10.1038/onc.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Brautigan DL. A novel transmembrane Ser/Thr kinase complexes with protein phosphatase-1 and inhibitor-2. J. Biol. Chem. 2002;277:49605–49612. doi: 10.1074/jbc.M209335200. [DOI] [PubMed] [Google Scholar]
- 35.Hendrickx A, et al. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Komaki K, et al. Lemur tail kinase 1 (LMTK1) regulates the endosomal localization of β-secretase BACE1. J. Biochem. 2021;170:729–738. doi: 10.1093/jb/mvab094. [DOI] [PubMed] [Google Scholar]
- 37.Bencze J, et al. Neuropathological characterization of Lemur tyrosine kinase 2 (LMTK2) in Alzheimer’s disease and neocortical Lewy body disease. Sci. Rep. 2019;9:17222. doi: 10.1038/s41598-019-53638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bencze J, et al. Lemur tyrosine kinase 2 (LMTK2) level inversely correlates with phospho-tau in neuropathological stages of Alzheimer’s disease. Brain Sci. 2020;10:68. doi: 10.3390/brainsci10020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruz DF, Mitash N, Farinha CM, Swiatecka-Urban A. TGF-beta1 augments the apical membrane abundance of lemur tyrosine kinase 2 to inhibit CFTR-mediated chloride transport in human bronchial epithelia. Front. Cell Dev. Biol. 2020;8:58. doi: 10.3389/fcell.2020.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz DF, Mitash N, Mu F, Farinha CM, Swiatecka-Urban A. Differential gene expression analysis reveals global LMTK2 regulatory network and its role in TGF-β1 signaling. Front. Oncol. 2021;11:596861. doi: 10.3389/fonc.2021.596861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaozza E, Baker SJ, Vora RK, Reddy EP. AATYK: a novel tyrosine kinase induced during growth arrest and apoptosis of myeloid cells. Oncogene. 1997;15:3127–3135. doi: 10.1038/sj.onc.1201575. [DOI] [PubMed] [Google Scholar]
- 42.Ishikawa K, et al. Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:169–176. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- 43.Kikuno R, et al. Prediction of the coding sequences of unidentified human genes. XIV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:197–205. doi: 10.1093/dnares/6.3.197. [DOI] [PubMed] [Google Scholar]
- 44.Nagase T, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes. XXI. The complete sequences of 60 new cDNA clones from brain which code for large proteins. DNA Res. 2001;8:179–187. doi: 10.1093/dnares/8.4.179. [DOI] [PubMed] [Google Scholar]
- 45.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 46.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Brautigan DL. Peptide microarray analysis of substrate specificity of the transmembrane Ser/Thr kinase KPI-2 reveals reactivity with cystic fibrosis transmembrane conductance regulator and phosphorylase. Mol. Cell. Proteom. 2006;5:2124–2130. doi: 10.1074/mcp.M600188-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Honma N, et al. Apoptosis-associated tyrosine kinase is a Cdk5 activator p35 binding protein. Biochem. Biophys. Res. Commun. 2003;310:398–404. doi: 10.1016/j.bbrc.2003.08.143. [DOI] [PubMed] [Google Scholar]
- 49.Kesavapany S, et al. Identification of a novel, membrane-associated neuronal kinase, cyclin-dependent kinase 5/p35-regulated kinase. J. Neurosci. 2003;23:4975–4983. doi: 10.1523/JNEUROSCI.23-12-04975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawa S, Fujimoto J, Tezuka T, Nakazawa T, Yamamoto T. Involvement of BREK, a serine/threonine kinase enriched in brain, in NGF signalling. Genes Cells. 2004;9:219–232. doi: 10.1111/j.1356-9597.2004.00714.x. [DOI] [PubMed] [Google Scholar]
- 51.Tomomura M, et al. Structural and functional analysis of the apoptosis-associated tyrosine kinase (AATYK) family. Neuroscience. 2007;148:510–521. doi: 10.1016/j.neuroscience.2007.05.048. [DOI] [PubMed] [Google Scholar]
- 52.Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Almgren M, Nyengaard JR, Persson B, Lavebratt C. Carbamazepine protects against neuronal hyperplasia and abnormal gene expression in the megencephaly mouse. Neurobiol. Dis. 2008;32:364–376. doi: 10.1016/j.nbd.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 54.Kawa S, et al. Azoospermia in mice with targeted disruption of the Brek/Lmtk2 (brain-enriched kinase/lemur tyrosine kinase 2) gene. Proc. Natl Acad. Sci. USA. 2006;103:19344–19349. doi: 10.1073/pnas.0603603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma S, Rubin BP. Apoptosis-associated tyrosine kinase 1 inhibits growth and migration and promotes apoptosis in melanoma. Lab. Investig. 2014;94:430–438. doi: 10.1038/labinvest.2014.13. [DOI] [PubMed] [Google Scholar]
- 56.Ditsiou A, et al. The structure-function relationship of oncogenic LMTK3. Sci. Adv. 2020;6:eabc3099. doi: 10.1126/sciadv.abc3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trenker R, Jura N. Receptor tyrosine kinase activation: from the ligand perspective. Curr. Opin. Cell Biol. 2020;63:174–185. doi: 10.1016/j.ceb.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bencze J, et al. Biological function of lemur tyrosine kinase 2 (LMTK2): implications in neurodegeneration. Mol. Brain. 2018;11:20. doi: 10.1186/s13041-018-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruford EA, et al. Guidelines for human gene nomenclature. Nat. Genet. 2020;52:754–758. doi: 10.1038/s41588-020-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]