Abstract
The virulence of Campylobacter fetus, a bacterial pathogen of ungulates and humans, is mediated in part by the presence of a paracrystalline surface layer (S-layer) that confers serum resistance. The subunits of the S-layer are S-layer proteins (SLPs) that are secreted in the absence of an N-terminal signal sequence and attach to either type A or B C. fetus lipopolysaccharide in a serospecific manner. Antigenic variation of multiple SLPs (encoded by sapA homologs) of type A strain 23D occurs by inversion of a promoter-containing DNA element flanked by two sapA homologs. Cloning and sequencing of the entire 6.2-kb invertible region from C. fetus 23D revealed a probable 5.6-kb operon of four overlapping genes (sapCDEF, with sizes of 1,035, 1,752, 1,284, and 1,302 bp, respectively) transcribed in the opposite direction from sapA. The four genes also were present in the invertible region of type B strain 84-107 and were virtually identical to their counterparts in the type A strain. Although SapC had no database homologies, SapD, SapE, and SapF had predicted amino acid homologies with type I protein secretion systems (typified by Escherichia coli HlyBD/TolC or Erwinia chrysanthemi PrtDEF) that utilize C-terminal secretion signals to mediate the secretion of hemolysins, leukotoxins, or proteases from other bacterial species. Analysis of the C termini of four C. fetus SLPs revealed conserved structures that are potential secretion signals. A C. fetus sapD mutant neither produced nor secreted SLPs. E. coli expressing C. fetus sapA and sapCDEF secreted SapA, indicating that the sapCDEF genes are sufficient for SLP secretion. C. fetus SLPs therefore are transported to the cell surface by a type I secretion system.
Campylobacter fetus is a gram-negative pathogen that causes infertility and infectious abortion in sheep and cattle and extraintestinal infections in immunocompromised humans (35, 55). Similar to many bacteria (54), wild-type C. fetus has a paracrystalline surface layer (S-layer) composed of S-layer proteins (SLPs) (23, 25). SLPs are the most abundant proteins in C. fetus, constituting as much as 10 to 15% of the total cell protein (19, 50). The C. fetus S-layer inhibits binding of complement factor C3b and therefore results in resistance to phagocytosis and to complement-mediated killing by normal or immune serum (13). Mutants lacking the S-layer are significantly less virulent in animal models than are those expressing the S-layer (11, 49).
Two types of SLPs exist (A and B), based on their specific binding to serotype A or B lipopolysaccharide. However, within each of the types are a number of SLP variants that range in size from 97 to 149 kDa. In C. fetus 23D, SLPs are encoded by a family of eight sapA homologs (26). A single C. fetus cell has the ability to change the type of SLP that it expresses by the recA-dependent inversion of a DNA segment containing a unique outward-facing sapA promoter (22). The minimum invertible DNA segment is 6.2 kb in size and is flanked by sapA homologs, although larger and more complex inversions allow expression of alternate sapA homologs (24, 31).
The majority of bacterial SLPs have N-terminal signal sequences and are secreted via the type II (sec-dependent) secretion pathway (15). The SLPs encoded by C. fetus (SapA homologs) and Caulobacter crescentus (RsaA) lack N-terminal signal sequences and therefore are probably secreted by a different mechanism (15). C terminally truncated versions of C. fetus and C. crescentus SLPs are not secreted, suggesting that the secretion signal lies in the C terminus of the protein (6, 8, 14). Furthermore, the C terminus of C. crescentus RsaA is sufficient to allow secretion of heterologous proteins from C. crescentus, and recent mutational studies on its C terminus have begun to delineate sequences important for RsaA secretion (7). SLPs lacking N-terminal signal sequences recently were identified in Serratia marcescens (38) and Campylobacter rectus (62).
The type I pathway uses C-terminal secretion signals on the targeted protein for secretion from gram-negative bacteria. Proteins secreted by this pathway include Escherichia coli α-hemolysin and other bacterial RTX toxins and proteases from Erwinia chrysanthemi, S. marcescens, and Pseudomonas aeruginosa (51, 61). The secretion apparatus is composed of three proteins homologous to HlyB, HlyD, and TolC of E. coli or PrtDEF of E. chrysanthemi. These three proteins form a transmembrane complex by which a C-terminal signal in the cognate protein is recognized, initiating its secretion from the cytoplasm directly into the extracellular medium. The HlyB homologs are cytoplasmic membrane proteins that are members of the ABC transporter family and are responsible for recognizing the C-terminal secretion signal on the protein to be transported (46, 61). HlyD-like proteins also localize to the cytoplasmic membrane but belong to the MFP (membrane fusion protein) family (18) and probably facilitate the direct interaction of the cytoplasmic and outer membranes during the secretion process. TolC homologs reside in the outer membrane and appear to form a pore as the outer component of the transport machinery (42). Recently, SLP-transporting type I systems have been characterized in C. crescentus and S. marcescens (2, 38). In C. crescentus, the ABC (RsaD) and MFP (RsaE) components are encoded by DNA immediately downstream of the SLP structural gene (rsaA), while the outer membrane component has not been identified (2). The S. marcescens SLP (SlaA) is secreted by the LipBCD type I transporter and thus shares this pathway with the S. marcescens extracellular lipase, LipA (38).
To investigate whether the invertible region contains genes involved in the expression, antigenic variation, or secretion of C. fetus SLPs, we cloned and sequenced the invertible regions from type A strain 23D and type B strain 84-107. Since each DNA sequence predicted four genes (sapCDEF), three of which were homologous to genes encoding type I secretion proteins, we created a mutation in sapD and showed that this mutant did not produce or secrete SLPs. Coexpression of the sapA and sapCDEF genes in E. coli showed that the sapCDEF genes are sufficient to allow secretion of SapA from the bacterial cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. C. fetus strains were grown at 37°C under microaerobic conditions in a GasPak jar using a CampyPak Plus gas generator (BBL Microbiology Systems, Cockeysville, Md.) on brucella agar (Difco Laboratories, Detroit, Mich.) containing antibiotics at the following concentrations: 7-U/ml polymyxin B, 10-μg/ml vancomycin, 10-μg/ml trimethoprim lactate, 15-μg/ml nalidixic acid (designated PVNT), and 40-μg/ml kanamycin (PVNTK) for kanamycin-resistant strains. Strains were also grown in brucella broth containing the above concentrations of PVNT under microaerobic conditions at 37°C. E. coli strains were grown on LB plates or broth (52) supplemented with trimethoprim lactate (10 μg/ml), kanamycin (40 μg/ml), tetracycline (15 μg/ml), or ampicillin (50 μg/ml) when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| C. fetus | ||
| 23D | sapA+sapCDEF+ | 48 |
| 23B | ΔsapAp | 48 |
| 97-205 | 23D sapD::aphA | This study |
| 84-107 | sapB+sapCDEF | 27 |
| E. coli | ||
| C600 | 3 | |
| DH5αMCR | 37 | |
| S17-1 | 53 | |
| Plasmids | ||
| pIR100 | sapCDEF in pAMP1 | This study |
| pBGYC1 | sapA in pACYC184 | This study |
| pIR13 | sapD in pBluescript | This study |
| pIR131 | sapD::aphA in pBluescript | This study |
| pILL570 | ReppBR322 mob Spcr | 43 |
| pILL131 | pIR131 insert in pILL570 | This study |
| pILL600 | ReppBR322 aphA | 44 |
DNA and protein techniques.
Restriction enzymes, the Klenow fragment of E. coli DNA polymerase I, and T4 DNA ligase were used as suggested by the manufacturer, either New England Biolabs (Beverly, Mass.), or Promega (Madison, Wis.). The sequences of the invertible regions from strains 23D and 84-107 were obtained by primer walking or direct sequencing of PCR products by using an ABI 377 (PE Applied Biosystems, Foster City, Calif.) automated sequencer by the Vanderbilt University Cancer Center Core Laboratory, and oligonucleotides were synthesized by the Vanderbilt University Molecular Biology Core Laboratory. DNA sequence analysis was done by using the GCG sequence analysis programs (17). Database similarity searches were performed by using the BLAST algorithms maintained by the National Center for Biotechnology Information (Bethesda, Md.). Searches of the PROSITE and MotifDic libraries for protein motifs were done by using the MotifFinder e-mail server (motiffinder@genome.ad.jp). Parsimony analysis of protein sequences was performed by using PAUP 3.1 (Smithsonian Institution, Washington, D.C.) with 1,000 bootstrap replicates.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Whole-cell lysates and water extracts of strains 23D, 23B, and 97-205 were prepared by previously described methods (50), and protein concentrations were assayed by using the Pierce BCA Protein Reagent Assay (Pierce, Rockford, Ill.). Protein samples were analyzed by sodium dodecyl sulfate–10%-polyacrylamide gel electrophoresis, and SLPs were detected by immunoblotting (1:10,000 dilution) by using polyclonal rabbit serum against C. fetus SLPs, generated by Cocalico Biologicals (Reamstown, Pa.). The secondary antibody (1:2,000 dilution) was goat anti-rabbit immunoglobulin G-2 alkaline phosphatase (Boehringer Mannheim, Indianapolis, Ind.).
Cloning of the invertible region.
Clones representing the invertible region from type A strain 23D were isolated as follows. First, the entire invertible region was amplified by PCR by using primer 2754 (59), which binds in the conserved sequences at the 5′ end of each flanking sapA homolog (see Fig. 1). This 6.2-kb product was subcloned into pAMP1 (Gibco-BRL, Gaithersburg, Md.) to yield pIR100, and the pIR100 insert was used as a probe to identify genomic clones for DNA sequence analysis. Clones were isolated from a genomic library (26) constructed in λZAPII (Stratagene, La Jolla, Calif.) from random fragments resulting from partial AluI digestion of chromosomal DNA from wild-type C. fetus 23D (48). Three λ clones (IR12, IR13, and IR15) hybridizing to the pIR100 insert were selected and characterized by restriction digestion, PCR, and DNA sequence analysis. A λ clone (IR20) covering a gap at the right end of the invertible region was identified by using as a probe a fragment released from pIR100 by NdeI digestion, corresponding to nucleotides 4715 to 5345 of the invertible region sequence. Plasmids (pIR12, pIR13, pIR15, and pIR20) derived from these purified λ clones were recovered by following the in vivo excision protocol recommended by the manufacturer (Stratagene). No clones were isolated for a region that represented a small gap between pIR13 and pIR12. We therefore amplified this region from C. fetus 23D chromosomal DNA by using primers A5598 (5′ TGTATCGTTTATGCTGCG; nucleotide positions 1677 to 1694) and C2968 (5′ CCGTCCGGAAGTCTTAGTATC; positions 2963 to 2983). The resulting 1,305-bp PCR product was cloned into pT7Blue (Novagen, Madison, Wis.), and the DNA sequences of three independent subclones were determined.
FIG. 1.
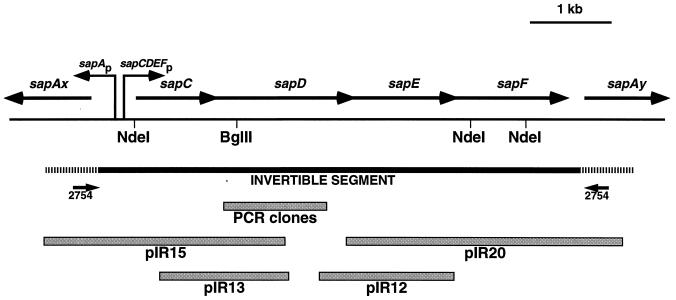
Schematic representation of the sapA invertible region showing the sapCDEF genes, the locations of the divergent sapA promoter and the putative sapCDEF promoter(s), and the clones (pIR15, pIR13, pIR12, and pIR20) from which the invertible region sequence was determined. The hatched areas represent the ca. 600-bp conserved regions at the 5′ ends of sapA homologs flanking the invertible region (here designated sapAx and sapAy), at which recombination occurs as the basis of sapA homolog rearrangements during SLP antigenic variation (22, 24).
The sequence of the invertible region from type B strain 84-107 was compiled from a series of PCR products amplified by using primers derived from the sequence of the 23D invertible region. Automated direct DNA sequencing of each PCR product was performed on both strands as described above.
Construction of a C. fetus sapD mutant.
Clone pIR13 was digested with BglII and ligated with the aphA (Kmr) cassette excised from pILL600 by BamHI digestion, yielding pIR131. To create a mobilizable plasmid for conjugation, the pIR131 insert was released by BamHI and AccI digestion and ligated with ClaI/BamHI-digested pILL570. The resulting plasmid (pILL131) was transformed into E. coli S17-1, which was mated with C. fetus 23D by previously described methods (14). Transconjugants were recovered after 3 days of growth on plates containing PVNTK.
Southern hybridization.
Chromosomal DNAs from C. fetus 23D and 97-205 were prepared by using the cetyltrimethylammonium bromide method (1). The DNA was digested with NdeI, and 1 μg was electrophoresed on a 0.7% agarose gel and then transferred to a nylon membrane (MSI, Westborough, Mass.). The membrane was hybridized with DNA probes labeled by using the Renaissance nonradioactive chemiluminescence kit supplied by NEN Research Products (Boston, Mass.). The membrane was probed with either BamHI-digested pIR13, BamHI-digested pILL570, or the aphA cassette from SmaI-digested pILL600 and subsequently exposed on Biomax MR scientific imaging film from Eastman Kodak (Rochester, N.Y.) for 1 h.
Primer extension analysis.
The levels of sapA mRNA in wild-type and sapD mutant C. fetus were determined by primer extension analysis as described previously (30), with the following modifications. Total cellular RNA was prepared from C. fetus 23D (wild type) or 97-205 (sapD mutant) grown for 48 h on brucella agar plates by the hot-phenol method (63). Reverse transcription was initiated by using primer 2754 (59), which binds 117 nucleotides downstream of the sapA transcriptional initiation site.
Serum susceptibility assay.
C. fetus 23D (SapA+), 23B (SapA−), and 97-205 (sapD::aphA) were harvested from 48-h plate cultures and assayed for killing by normal human serum (NHS) by previously described methods (12), with the following modifications. Dilutions of C. fetus cells (10−4 to 10−7) were incubated in a 37°C 5% CO2 environment for 60 min in the presence of 40% pooled NHS or heat-inactivated NHS (HINHS). Samples then were plated on tryptic soy agar with 5% sheep blood (BBL Microbiology Systems), and CFU were counted after 72 h. Net killing represented the difference between counts of cells incubated in HINHS and NHS as previously described (12).
E. coli SapA secretion assay.
To construct E. coli strains for assaying the secretion of SapA, we created an E. coli strain containing C. fetus sapA and sapCDEF on compatible plasmids. First, the sapA-containing EcoRI fragment of pBG1 (10) was subcloned into the EcoRI site of pACYC184 to create pBGYC1. Next, we transformed E. coli C600 with pBGYC1 and either pIR100 (sapCDEF) or pSPORT1 (vector control; Gibco-BRL). This pair of C600 strains (pSPORT1 plus pBGYC1 or pIR100 plus pBGYC1) was grown to mid-log phase, and the cells were harvested by centrifugation. The supernatants were passaged through 0.2-μm-pore-size filters (Micro Filtration Systems, Dublin, Calif.) to rid the supernatants of residual bacterial cells. Trichloroacetic acid (TCA) precipitation of proteins was performed as previously described (28), and the proteins were resolved by Western blot analysis with rabbit anti-SLP serum as described above.
Nucleotide sequence accession numbers.
The DNA sequence of the type A invertible region has been deposited in the GenBank database under accession no. AF027405. The DNA sequence of the 84-107 invertible region has been deposited in the GenBank database under accession no. AF071883.
RESULTS
Cloning of the type A invertible region.
Since bacterial genes involved in similar functions are often clustered, we sought to characterize the 6.2-kb invertible region between two sapA homologs. We first amplified the 6.2-kb fragment by PCR and subcloned the product into pAMP1 to yield pIR100. Next, we used this subcloned fragment as a probe to isolate a series of overlapping plasmid clones derived from a C. fetus genomic library constructed in λZAPII. Four of these, designated pIR15, pIR13, pIR12, and pIR20, represented the majority of the invertible region (Fig. 1) and were subjected to DNA sequence analysis. We were unable to isolate a clone that bridged a small gap between pIR13 and pIR12; this segment was amplified from C. fetus 23D genomic DNA by PCR using appropriate primers and subcloned into pT7Blue. To avoid the potential problem of PCR-induced errors, we sequenced three independent subclones, and in each case, the sequence was the same.
Analysis of type A invertible region features.
The DNA sequence of the 6,229-bp invertible region from strain 23D predicted four open reading frames (ORFs), which we designated sapCDEFA. The sapC gene began 596 bp from the initiation codon of the oppositely oriented upstream sapA homolog (Fig. 1). The sapC ORF was 1,035 bp in length and was immediately followed by the sapD, sapE, and sapF ORFs, which were 1,752, 1,284, and 1,302 bp long, respectively. Each gene in this cluster had a typical ribosome binding site, and they overlapped the preceding genes by 14 bp (sapC/D), 1 bp (sapD/E), and 11 bp (sapE/F). The sapF gene ended 287 bp upstream of the sapA homolog located downstream. The 74-bp sequences preceding the ATG codons initiating translation of the sapA homologs flanking sapCDEF were identical to each other (Fig. 1). These conserved segments have previously been noted upstream of the three characterized sapA homologs and may play a role in the inversion of this DNA segment (22, 24, 60). As a potential component of this mechanism, sequences resembling χ (RecBCD recognition) sites were present at positions 31 to 38 and 6192 to 6199. Several potential ς70-like promoters were noted 44 to 243 bp upstream of sapC. These putative sapCDEF promoters were oriented in the opposite direction from the sapA promoter, with the two −35 regions separated by 200 to 380 bp. Due to the overlapping nature of the sapCDEF genes and the lack of other putative promoters, it is likely that they are cotranscribed. No putative transcriptional terminators were evident within the invertible region.
Similarities of SapCDEF to other proteins.
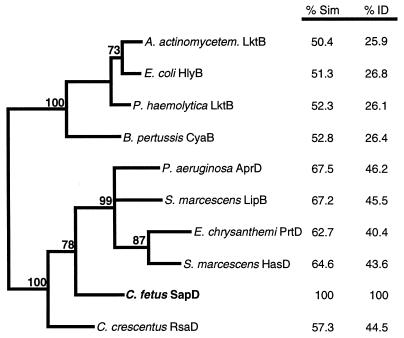
The sapC ORF predicted a protein product of 344 amino acids (aa) (39.7 kDa) that had no significant similarities in the nonredundant database maintained by the National Center for Biotechnology Information. A search by the MotifFinder server also did not reveal any recognizable protein motifs in SapC, and it did not have an N-terminal sequence suggestive of extracytoplasmic localization. In contrast, the products of the sapDEF genes had high similarities to proteins encoding type I secretion systems. SapD (584 aa, 64.0 kDa) was found to be related to the ABC family of transporters, especially those that are involved in translocation by the type I secretion systems of proteases, lipases, hemolysins, and leukotoxins across the envelopes of gram-negative bacterial pathogens. The degree of similarity found between SapD and these proteins was between 68% (with P. aeruginosa AprD) and 50% (Actinobacillus actinomycetemcomitans LktB; Fig. 2). In addition, SapD contained two motifs found in such proteins, an ATP/GTP binding site (GPSAAGKS; Walker box A) and a peptide (LSGGQRQRVALA) that is a signature sequence for ABC transporters. The SapE protein (428 aa, 47.9 kDa) was similar to the MFP proteins of type I transporters, typified by P. aeruginosa AprE (52% similar) and E. coli HlyD (49% similar). SapF (434 aa, 49.4 kDa) was related to the outer membrane component of type I systems such as P. aeruginosa AprF (45% similar), E. chrysanthemi PrtF (41% similar), and E. coli TolC (47% similar). The high similarity of each of the SapD, SapE, and SapF proteins to each of the proteins of type I secretion systems suggested that these proteins are involved in the secretion of C. fetus SLPs. Since the ABC component of type I transporters is responsible for substrate recognition (46), phylogenetic analysis of these proteins (SapD and homologs) is most likely to be informative with respect to the relationships between families of type I transporters. Parsimony and bootstrap analyses supported the classification of the C. fetus and C. crescentus SLP transporters on phylogenetic branches adjacent to the protease transporters but distinct from these and other type I secretion apparatuses (Fig. 2). Parsimony analysis of the MFPs (SapE and homologs) resulted in a similar tree, supporting this conclusion (data not shown). Phylogeny based on the type I outer membrane proteins showed few significant distinctions between subfamilies of transporters (data not shown).
FIG. 2.
Phylogram, generated by parsimony analysis, demonstrating the relatedness of the cytoplasmic membrane ABC proteins of type I secretion systems. Percentages of amino acid similarity (% Sim) and identity (% ID) with the C. fetus protein are shown at the right. The bold numbers adjacent to the phylogram branches indicate the percentages of 1,000 bootstrap replicates supporting the clustering of those branches. Branches without bootstrap values were clustered in less than 70% of the bootstrap replicates.
Invertible region features from type B strain 84-107.
To determine whether the features found in the invertible region of type A strain 23D also were conserved in a type B strain, we sequenced the invertible region from strain 84-107. The invertible region from this strain was 6,217 bp in length and was 99.1% identical to the invertible region from strain 23D. Similar to the conserved regions at each end of the type A invertible region, the first 67 nucleotides of the type B sequence were an exact inverse repeat of the last 67 nucleotides. However, these 67-bp segments at the ends of the type B invertible region were only 68.7% identical to the analogous sequences of the type A invertible region. Similar to the type A sequence, the type B invertible region sequence predicted a probable sapCDEF operon of 5.6 kb (designated sapCDEFB). The putative sapCDEFB promoter sequences and ribosome binding sites were identical to their counterparts in the 23D invertible region (data not shown). Most of the differences between the invertible regions of 23D and 84-107 were found in the noncoding regions upstream and downstream of sapCDEF, with the coding regions sharing 99.8% nucleotide identity. Due to the high sequence similarity between the two invertible regions, the SapC, SapD, SapE, and SapF proteins from the type A and B strains were nearly identical, sharing 99.7, 100, 99.5, and 100% amino acid identity, respectively.
Construction of a sapD mutant.
To test the hypothesis that genes in the C. fetus invertible region encode proteins responsible for SLP secretion, we constructed a derivative of type A strain 23D containing an insertional mutation in sapD. Since homologs of SapD (ABC transporters) are essential components of type I secretion systems (5, 41, 46), C. fetus mutants lacking SapD would be expected to be secretion deficient. To generate a sapD mutation, we chose a unique BglII site 224 bp from the upstream end of the gene (Fig. 1). The aphA cassette from pILL600 was inserted into this site, and transformants were selected on plates containing kanamycin. Plasmid DNA from kanamycin-resistant E. coli colonies was analyzed by EcoRI and HindIII digestion, and the aphA cassette was found to have been incorporated such that sapCDEF and the antibiotic resistance gene were transcribed in the same direction. This new clone was designated pIR131. The sapD insert containing the resistance fragment was subcloned into suicide vector pILL570 to yield pILL131, which was transformed into E. coli S17-1. Transformants were selected from trimethoprim-kanamycin plates, and verification of pILL131 was made by digestion with HindIII. E. coli S17-1 containing IncP DNA transfer functions and pILL131 was used as the conjugation donor, and wild-type C. fetus 23D was the recipient. Approximately 5,000 transconjugants were recovered (frequency of 4 × 10−6 transconjugants per recipient), 6 of which were picked for further screening. Chromosomal DNAs extracted from these six strains were digested with NdeI for Southern analysis by using hybridization probes for sapD, aphA, and pILL570. The hybridization results indicated that one of these strains (97-205) was derived from a double-crossover event in which only the mutagenized sapD allele was incorporated into the chromosome (data not shown). The remaining five strains produced results consistent with single crossovers in which the pILL570 vector also was introduced into the chromosome (data not shown). PCR experiments using sapD- and aphA-specific primers confirmed the Southern hybridization data (not shown). These results indicate that a strain containing an insertional mutation in sapD was successfully constructed.
Properties of a C. fetus sapD mutant.
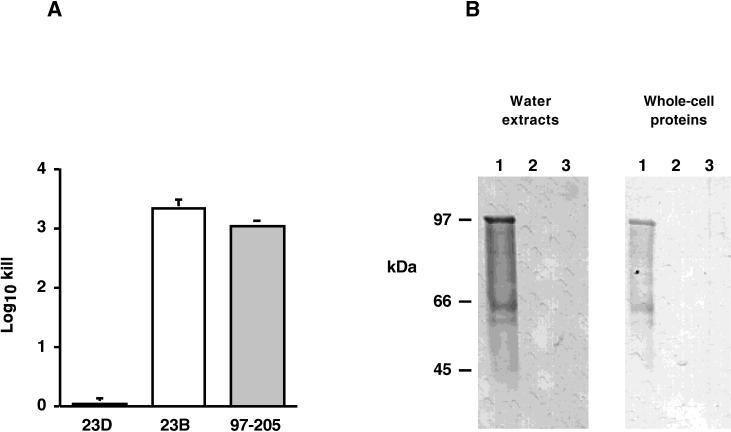
We next sought to determine whether the C. fetus sapD mutant 97-205 was able to export SLPs and to assemble a functional S-layer on its surface. First, we tested the abilities of the wild-type and sapD mutant strains to resist complement-mediated killing, a phenotype consistent with the presence of the S-layer. Suspensions of cells were exposed either to NHS or to HINHS, and the extent of complement-mediated killing was determined. As expected, S+ strain 23D was completely resistant to killing (Fig. 3A). In contrast, as expected, there was approximately 3 log10 killing of strain 23B, which is unable to express an S-layer (48, 59) (Fig. 3A). Results for strain 97-205, the sapD mutant, were nearly identical to those for 23B and are consistent with the absence of a functional S-layer on the surface of sapD mutant strain 97-205.
FIG. 3.
Phenotypes of C. fetus wild-type (23D), spontaneous S− mutant (23B), and sapD mutant (97-205) cells. (A) Serum susceptibilities of C. fetus strains. Serial dilutions of 103 to 105 CFU/ml were suspended in either 40% NHS or HINHS (56°C, 30 min) for 60 min. The amount of killing relative to that of the time zero inoculum was determined and is presented as mean log10 killing ± the standard error of the mean. (B) Western blot of C. fetus strains. Cell surface water extracts and whole-cell lysates were probed with polyclonal serum against C. fetus SLPs. Lanes: 1, strain 23D (S+); 2, strain 23B (S−); 3, strain 97-205 (sapD mutant). The positions of molecular mass markers are shown at the left.
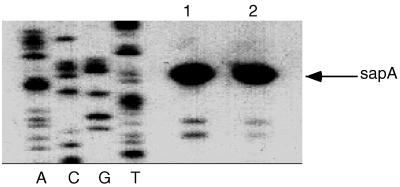
To understand the basis for the serum susceptibility of the sapD mutant, we performed immunoblot analysis to detect the presence of SLPs on the cell surface. SLPs can readily be removed from the surfaces of S+ cells by washes with distilled water (50). Therefore, water washes and whole-cell proteins of wild-type and sapD mutant cells were analyzed by immunoblotting with polyclonal antiserum against SLPs. SLPs were detected in both whole-cell samples and water washes of the wild-type strain, as expected for cells expressing a functional S-layer (Fig. 3B, lane 1), whereas no SLPs were detected in S− strain 23B (Fig. 3B, lane 2). However, SLPs were detected neither extracellularly nor in whole-cell samples of the sapD mutant, (Fig. 3B, lane 3). Thus, disruption of sapD by the insertion of an antibiotic resistance cassette had the effect of eliminating SLP expression altogether, as well as SLP secretion. It could not be ascertained from these experiments whether the effect of the sapD mutation on the inability to detect SLPs in the cytoplasm was due to a regulatory effect on SLP gene expression or whether full-length SLPs were simply degraded in the absence of a functional secretion apparatus. Primer extension experiments indicated that wild-type levels of sapA mRNA were present in strain 97-205 (Fig. 4), showing that potential regulatory effects of the sapD mutation on SapA expression did not occur through diminished sapA transcription.
FIG. 4.
Primer extension analysis of sapA-specific mRNA in wild-type and sapD mutant C. fetus strains. The levels of sapA transcription were determined by reverse transcription of standardized 40-μg RNA samples by using primer 2754 (59). The sequencing ladder was generated by using primer 2754 and plasmid pIR15, which contains the sapA promoter and 5′ region (Fig. 1). Lanes: 1, strain 23D (wild type); 2, strain 97-205 (sapD mutant). The +1 site for strain 23D is identical to that determined previously (59).
Secretion of SapA by E. coli expressing sapCDEF.
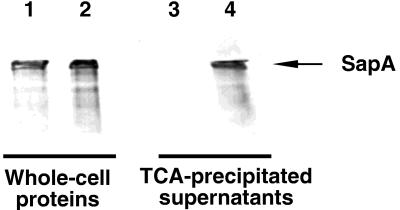
We had cloned the PCR-amplified sapCDEF-containing invertible region (pIR100) for use as a hybridization probe with which to identify genomic clones as described above. To determine whether sapCDEF permits the secretion of SapA, we tested the ability of E. coli strains carrying pIR100 to mediate the specific secretion of C. fetus SapA (Fig. 5). E. coli C600 strains were isolated that contained pBGYC1, a pACYC184-derived plasmid containing C. fetus sapA, and either pIR100 (sapCDEF) or pSPORT1 (vector control). SapA secretion was assayed by using immunoblots to indicate the appearance of SapA in filtered, TCA-precipitated culture supernatants prepared from these strains. As expected (10), both E. coli strains produced SapA (Fig. 5, whole-cell proteins). Secretion of SapA was detected only in supernatants from E. coli cells expressing sapCDEF (Fig. 5, lane 4). These results indicate that the C. fetus sapCDEF genes are sufficient to allow E. coli to secrete SapA.
FIG. 5.
Secretion of SapA from E. coli expressing C. fetus sapCDEFA, detected by immunoblotting with antiserum to C. fetus SLPs. One microgram of whole-cell proteins (lanes 1 and 2) or the amount of TCA-precipitated protein present in 250 μl of culture supernatant (lanes 3 and 4) was analyzed. Lanes: 1 and 3, C600(pSPORT1)(pBGYC1); 2 and 4, C600(pIR100)(pBGYC1).
Conserved features of SLP C termini.
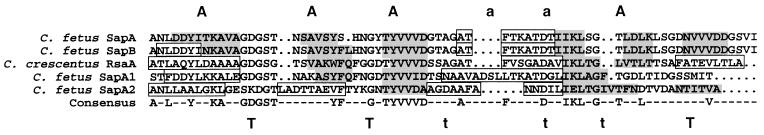
Proteins that are transported by type I secretion systems do not have the N-terminal signal peptides that are conserved in proteins exported by type II systems, instead relying on signals that are located at their extreme C termini (51, 61). However, these signals tend to have little primary sequence homology, and their exact structures have been difficult to elucidate. In an attempt to define candidate sequences or structures for C-terminal secretion signals in bacterial SLPs, we aligned the C-terminal 70 aa of four C. fetusSLPs for which the sequences are known (SapA, SapA1, SapA2, and SapB), as well as RsaA, the SLP from C. crescentus (9). Several conserved peptides were evident (Fig. 6). The sequences GDGS(T/G), GxTYVV(V/I)D, and DxxIKLxG were each present in at least four of the five SLPs (Fig. 6). The DVIV motifs implicated in protease secretion (33) were not present per se at the extreme C terminus of any of the SLPs, although a similar sequence (DGSVI) was found at the C termini of the SapA and SapB proteins (Fig. 6). Similar to type I secreted toxins and related exoproteins (56, 58), the SLPs had one to four hydroxylated residues (S or T) within the C-terminal 10 amino acids.
FIG. 6.
Conserved primary and secondary structures of the C termini of four C. fetus SLPs (SapA, SapA1, SapA2, and SapB) and RsaA, the SLP of C. crescentus, predicted by GCG PILEUP and by the algorithm of Garnier et al. (32). The consensus line indicates amino acid residues that were conserved in at least three of the five SLPs. Secondary-structure features shown are regions of predicted α-helix (boxed), β-sheet (light shading), random coil (no shading), and strong or weak amphipathicity (A or a, respectively) and strong or weak turn-forming residues (T or t, respectively).
To investigate whether the C-terminal domains of these SLPs also could form similar secondary structures, we analyzed each of these peptides by using the programs of Garnier et al. (32). The predicted C-terminal secondary structures (α-helix, β-sheet, amphipathic peptides, and turn-forming residues) were then superimposed on the alignment of the five C-terminal peptides. Four of these peptides (SapA, SapA1, SapB, and RsaA) consisted of segments predicted to form a sheet-sheet-helix-sheet structure, with turn-forming residues separating the domains (Fig. 6). The C-terminal domain of SapA2 was predicted to form a helix-sheet-helix-sheet structure. Furthermore, the sheet-forming regions of all five proteins, as well as the most N-terminal helix-forming region of SapA2, were predicted to be amphipathic. Taken together, these results suggest that the C termini of four C. fetus SLPs and one C. crescentus SLP contain conserved sequences and secondary structures that are candidates for secretion signals.
Since SapC did not have any database homologies, yet sapC was situated immediately upstream of sapDEF, we also compared the C terminus of SapC to the conserved SLP C termini. The C terminus of SapC did not at all resemble the C termini of the other proteins (data not shown); therefore, it is likely that SapC is not secreted by the C. fetus type I system.
DISCUSSION
Three genes in the sapA invertible region (sapDEFA) and in the homologous sapB invertible region (sapDEFB) encode proteins that are homologous to type I transporters, typified by the E. coli α-hemolysin and S. marcescens LipB lipase secretion systems. The sapDEF genes are adjacent to the sapA homologs that encode the proteins that are substrates for the C. fetus type I secretion system. Genes encoding type I secretion systems of other bacteria often show such a clustered arrangement (51, 61), although the gene encoding the outer membrane component is not necessarily adjacent (2, 4, 51). The first gene, sapC, predicts a protein that has no database homologies, no amino acid motifs, and no other recognizable features such as signal sequences or helix-turn-helix structures. Furthermore, since the C-terminal 100 aa bore little resemblance to those of the SLPs and proteases, SapC does not appear to be another substrate for the SapDEF secretion apparatus. Therefore, the function of SapC is unclear.
Some aspects of type I secretion systems have been well characterized, and studies by a number of researchers have yielded the following model of type I secretion. Three proteins form the secretion apparatus. ABC transporters homologous to SapD (e.g., HlyB and PrtD) localize to the cytoplasmic membrane, probably as homodimers (41, 61), and serve a number of functions in the initial stages of the transport process. ATP hydrolysis by the ABC protein plays a role in energizing the secretion process (40). Recognition of the C terminus of the secreted protein by the ABC protein sequesters, in turn, the MFP and the outer membrane proteins, completing the assembled apparatus (46). The MFP is located mostly within the cytoplasmic membrane but is thought also to associate with the outer membrane and mediate its juxtaposition with the cytoplasmic membrane (18). The outer membrane protein (TolC homolog) may serve as a pore through which the secreted protein is extruded (42). The secreted proteins are devoid of multiple cysteine residues and therefore lack disulfide bridges during their transit through both membranes. The high degree of conservation between each of the C. fetus transport proteins and each constituent of other type I transporters strongly suggests that the secretion mechanism of SLPs is similar to that shown for other proteins. However, the phylogram generated from comparisons of the ABC components of the secretion apparatus (Fig. 2) shows that the C. fetus and C. crescentus systems represent a third family of type I transporter, divergent from the toxin or protease transporters.
Analysis of the sequence of the C terminus of E. coli α-hemolysin revealed a potential signal consisting of a charged region, followed by an amphipathic α-helix, and a number of hydroxylated amino acid residues at the extreme C terminus of the protein (56). The importance of these structures, as well as of individual amino acid residues was supported by mutational analyses (16, 39, 56). Similar features are found in the C termini of other exoproteins that can be secreted by the HlyBD secretion apparatus (58). In contrast, these features are not universally found in other proteins secreted by type I systems. For example, in proteases, a C-terminal amino acid motif of DVIV is involved in secretion (33, 64). This exact motif is found in neither type I-secreted toxins nor SLPs. The remainder of the C termini among type I-secreted proteases, lipases, and the S. marcescens heme-binding protein HasA share few structural similarities, although amphipathic α-helices have been proposed as components of secretion signals of some of these (21, 56, 58, 64). Nevertheless, many of these proteins can be secreted by heterologous type I transporters (5, 21, 45, 47, 57), suggesting the conservation of secretion signals within divergent C termini.
It is likely that the secretion signals of C. fetus SLPs are located at the C termini of the proteins, for the following reasons. None of the four characterized SLPs has an N-terminal signal sequence (10, 26, 27, 60). A 50-kDa C terminally truncated form of SapA was not secreted by C. fetus, suggesting that the signal directing its transport was located at the C terminus of the protein (14). Furthermore, E. coli K-12 strains are unable to secrete SapA (10; this work), suggesting that proteins different than the E. coli type II (sec) apparatus are necessary for its transport. Now we have shown that type I secretion proteins are responsible for transporting C. fetus SLPs to the cell surface, and with the exception of colicin V (34), the signals recognized by type I transporters are found at the extreme C terminus of the secreted protein (61). The SLP of C. crescentus, which also is secreted by a type I system, contains a secretion signal within its C-terminal 242 aa (7). Alignment of the C-terminal 70 aa of four C. fetus and one C. crescentus SLPs revealed a surprisingly large number of conserved amino acid residues (Fig. 6). In fact, the C termini of C. fetus SapA and SapB appeared to be more closely related to that of C. crescentus RsaA than to the C termini of the C. fetus SapA1 and SapA2 proteins. These conserved residues, as well as predicted β-sheet and α-helical regions, provide attractive targets for mutational analyses in determining the important components of SLP C-terminal secretion signals. The availability of several related C. fetus SapA homologs with somewhat different C termini allows the testing of a variety of hypotheses regarding the secretion signals and offers advantages to comparing the structures of type I-secreted proteins from heterologous bacterial species.
The mutation of sapD had an inhibitory effect on SapA synthesis by C. fetus 97-205, although this is most likely related to the inability of the sapD mutant to export SLPs. Since SLPs are the most abundant protein produced by C. fetus, their accumulation in the cytoplasm would probably be deleterious to growth or viability. The detection of wild-type levels of sapA mRNA in the sapD mutant precludes the possibility that a mutation occurred (in either sapD or another locus) that ablated transcription from the sapA promoter. We therefore consider two main possibilities for the lack of detection of SLPs in the C. fetus sapD mutant. First, nonsecreted SLPs may simply be degraded. Smit and colleagues were unable to detect derivatives of C. crescentus RsaA resulting from disruption of the C-terminal secretion signal, presumably due to the degradation of the secretion-deficient protein (7). A similar phenomenon previously had been noted for nonsecreted E. coli α-hemolysin (16), P. aeruginosa AprA, or hybrids of AprA with Pseudomonas fluorescens lipase (20, 21, 36). Therefore, the disruption of the secretion process of these proteins may necessitate their degradation. Second, the accumulation of SLPs within the cytoplasm may have an inhibitory effect on their own synthesis at a posttranscriptional level. It seems desirable for the expression of such an abundant protein to be tightly regulated, and in fact, this occurs with the SLPs of Thermus thermophilus (29). Although the T. thermophilus SLPs are secreted by the general secretory pathway (type II secretion), recent studies have shown that the C terminus of the T. thermophilus SLP is able to bind to the 127-bp 5′ untranslated region (UTR) of its own message and inhibit SLP synthesis (29). The C. fetus sapA mRNA has a highly structured 114-bp 5′ UTR, and it is possible that a similar repression of SLP synthesis occurs in C. fetus. If C. fetus SLPs can inhibit their own synthesis in such a manner, then it is probably also dependent on the C-terminal half of the protein, since the expression of a 50-kDa N-terminal (secretion-deficient) fragment of SapA is tolerated (14). We cannot rule out either of these hypotheses, and further studies, such as pulse-chase labeling of SLPs to examine protein turnover and genetic studies with SLPs truncated at the C terminus, will address these possibilities.
That sapCDEF are sufficient for the secretion of C. fetus SLPs was demonstrated by the ability of these genes to direct the transport of SapA from E. coli. Therefore, it appears that, similar to other type I systems, three proteins (SapDEF) are probably necessary and sufficient for the secretion of C. fetus SLPs. In contrast to the inability to detect nonsecreted SLPs in C. fetus, large quantities of SapA are present in E. coli lacking sapCDEF. This difference may have been due to the 5′ UTR of sapA in the clone used in this secretion experiment. The sapA gene encoded by pBGYC1 is not expressed from its native promoter and has only 23 bp of the 114-bp 5′ UTR found in C. fetus sapA homologs (10). The ability to reconstruct the C. fetus SLP secretion system in E. coli now facilitates genetic studies intended to define the interactions between SapA and the SapDEF transport apparatus and to investigate the possible autoregulation of sapA.
We have characterized the SapDEF system that is responsible for the secretion of C. fetus SLPs and shown that it is a related but somewhat divergent member of the type I secretion family. Future studies intended to define the C-terminal secretion signals of C. fetus SLPs and their recognition by the SapDEF apparatus, as well as by heterologous type I transporters, may provide new insight into transporter-substrate interactions within these systems.
ACKNOWLEDGMENT
This work was supported in part by R01A124145 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 2.Awram P, Smit J. The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J Bacteriol. 1998;180:3062–3069. doi: 10.1128/jb.180.12.3062-3069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 4.Binet R, Wandersman C. Cloning of the Serratia marcescens hasF gene encoding the Has ABC exporter outer membrane component: a TolC analogue. Mol Microbiol. 1997;22:265–273. doi: 10.1046/j.1365-2958.1996.00103.x. [DOI] [PubMed] [Google Scholar]
- 5.Binet R, Wandersman C. Protein secretion by hybrid bacterial ABC-transporters: specific functions of the membrane ATPase and the membrane fusion protein. EMBO J. 1995;14:2298–2306. doi: 10.1002/j.1460-2075.1995.tb07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingle W H, Le K D, Smit J. The extreme N-terminus of the Caulobacter crescentus S-layer protein directs export of passenger proteins from the cytoplasm but is not required for secretion of the native protein. Can J Microbiol. 1996;42:672–684. doi: 10.1139/m96-092. [DOI] [PubMed] [Google Scholar]
- 7.Bingle W H, Nomellini J F, Smit J. Linker mutagenesis of the Caulobacter crescentus S-layer protein: toward a definition of an N-terminal anchoring region and a C-terminal secretion signal and the potential for heterologous protein secretion. J Bacteriol. 1997;179:601–611. doi: 10.1128/jb.179.3.601-611.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingle W H, Smit J. Alkaline phosphatase and a cellulase reporter are not exported from the cytoplasm when fused to large N-terminal portions of the Caulobacter crescentus S-layer protein. Can J Microbiol. 1994;40:777–782. doi: 10.1139/m94-122. [DOI] [PubMed] [Google Scholar]
- 9.Bingle W H, Walker S G, Smit J. Definition of form and function for the S-layer of Cavlobacter crescentus. In: Beveridge T L, Koval S F, editors. Advances in paracrystalline bacterial surface layers. New York, N.Y: Plenum Press; 1993. pp. 181–194. [Google Scholar]
- 10.Blaser M J, Gotschlich E C. Surface array protein of Campylobacter fetus: cloning and gene structure. J Biol Chem. 1990;265:14529–14535. [PubMed] [Google Scholar]
- 11.Blaser M J, Pei Z. Pathogenesis of Campylobacter fetus infections: critical role of high-molecular-weight S-layer proteins in virulence. J Infect Dis. 1993;167:372–377. doi: 10.1093/infdis/167.2.372. [DOI] [PubMed] [Google Scholar]
- 12.Blaser M J, Smith P F, Kohler P A. Susceptibility of Campylobacter isolates to the bactericidal activity in human serum. J Infect Dis. 1985;151:227–235. doi: 10.1093/infdis/151.2.227. [DOI] [PubMed] [Google Scholar]
- 13.Blaser M J, Smith P F, Repine J E, Joiner K A. Pathogenesis of Campylobacter fetus infections: failure of encapsulated Campylobacter fetus to bind C3b explains serum and phagocytosis resistance. J Clin Invest. 1988;81:1434–1444. doi: 10.1172/JCI113474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaser M J, Wang E, Tummuru M K R, Washburn R, Fujimoto S, Labigne A. High-frequency S-layer protein variation in Campylobacter fetus revealed by sapA mutagenesis. Mol Microbiol. 1994;14:453–462. doi: 10.1111/j.1365-2958.1994.tb02180.x. [DOI] [PubMed] [Google Scholar]
- 15.Boot H J, Pouwels P H. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol Microbiol. 1996;21:1117–1123. doi: 10.1046/j.1365-2958.1996.711442.x. [DOI] [PubMed] [Google Scholar]
- 16.Chervaux C, Holland I B. Random and directed mutagenesis to elucidate the functional importance of helix II and F-989 in the C-terminal secretion signal of Escherichia coli hemolysin. J Bacteriol. 1996;178:1232–1236. doi: 10.1128/jb.178.4.1232-1236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinh T, Paulsen I T, Saier M H., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubreuil J D, Kostrzynska M, Austin J W, Trust T J. Antigenic differences among Campylobacter fetus S-layer proteins. J Bacteriol. 1990;172:5035–5043. doi: 10.1128/jb.172.9.5035-5043.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duong F, Lazdunski A, Murgier M. Protein secretion by heterologous bacterial transporters: the C-terminus secretion signal of the secreted protein confers high recognition specificity. Mol Microbiol. 1996;21:459–470. doi: 10.1111/j.1365-2958.1996.tb02555.x. [DOI] [PubMed] [Google Scholar]
- 21.Duong F, Soscia C, Lazdunski A, Murgier M. The Pseudomonas fluorescens lipase has a C-terminal secretion signal and is secreted by a three-component bacterial ABC-exporter system. Mol Microbiol. 1994;11:1117–1126. doi: 10.1111/j.1365-2958.1994.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 22.Dworkin J, Blaser M J. Generation of Campylobacter fetus S-layer protein diversity utilizes a single promoter on an invertible DNA segment. Mol Microbiol. 1996;19:1241–1253. doi: 10.1111/j.1365-2958.1996.tb02469.x. [DOI] [PubMed] [Google Scholar]
- 23.Dworkin J, Blaser M J. Molecular mechanisms of Campylobacter fetus surface layer protein expression. Mol Microbiol. 1997;26:433–440. doi: 10.1046/j.1365-2958.1997.6151958.x. [DOI] [PubMed] [Google Scholar]
- 24.Dworkin J, Blaser M J. Nested DNA inversion as a paradigm of programmed gene rearrangement. Proc Natl Acad Sci USA. 1997;94:985–990. doi: 10.1073/pnas.94.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dworkin J, Shedd O L, Blaser M J. Nested DNA inversion of Campylobacter fetus S-layer genes is recA dependent. J Bacteriol. 1997;179:7523–7529. doi: 10.1128/jb.179.23.7523-7529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dworkin J, Tummuru M K R, Blaser M J. A lipopolysaccharide-binding domain of the Campylobacter fetus S-layer protein resides within the conserved N terminus of a family of silent and divergent homologs. J Bacteriol. 1995;177:1734–1741. doi: 10.1128/jb.177.7.1734-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dworkin J, Tummuru M K R, Blaser M J. Segmental conservation of sapA sequences in type B Campylobacter fetus cells. J Biol Chem. 1995;270:15093–15101. doi: 10.1074/jbc.270.25.15093. [DOI] [PubMed] [Google Scholar]
- 28.Felmlee T, Pellett S, Lee E-Y, Welch R A. Escherichia coli hemolysin is released extracellularly without cleavage of a signal peptide. J Bacteriol. 1985;163:88–93. doi: 10.1128/jb.163.1.88-93.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Herrero L A, Olabarria G, Berenguer J. Surface proteins and a novel transcription factor regulate the expression of the S-layer gene in Thermus thermophilus HB8. Mol Microbiol. 1997;24:61–72. doi: 10.1046/j.1365-2958.1997.3191683.x. [DOI] [PubMed] [Google Scholar]
- 30.Forsyth M H, Atherton J C, Blaser M J, Cover T L. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088–3094. doi: 10.1128/iai.66.7.3088-3094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita M, Morooka T, Fujimoto S, Moriya T, Amako K. Southern blotting analyses of strains of Campylobacter fetus using the conserved region of sapA. Arch Microbiol. 1995;164:444–447. doi: 10.1007/BF02529743. [DOI] [PubMed] [Google Scholar]
- 32.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 33.Ghigo J M, Wandersman C. A carboxy-terminal four amino acid motif is required for secretion of the metalloprotease PrtG through the Erwinia chrysanthemi protease secretion pathway. J Biol Chem. 1994;269:1–7. [PubMed] [Google Scholar]
- 34.Gilson L, Mahanty H K, Kolter R. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 1990;9:3875–3884. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerrant R L, Lahita R G, Winn W C, Roberts R B. Campylobacteriosis in man: pathogenic mechanisms and review of 91 bloodstream infections. Am J Med. 1978;65:584–592. doi: 10.1016/0002-9343(78)90845-8. [DOI] [PubMed] [Google Scholar]
- 36.Guzzo J, Pages J-M, Duong F, Lazdunski A, Murgier M. Pseudomonas aeruginosa alkaline protease: evidence for secretion genes and study of secretion mechanism. J Bacteriol. 1991;173:5290–5297. doi: 10.1128/jb.173.17.5290-5297.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 38.Kawai E, Akatsuka H, Idei A, Shibatani T, Omori K. Serratia marcescens S-layer protein is secreted extracellularly via an ATP-binding cassette exporter, the Lip system. Mol Microbiol. 1998;27:941–952. doi: 10.1046/j.1365-2958.1998.00739.x. [DOI] [PubMed] [Google Scholar]
- 39.Kenny B, Taylor S, Holland I B. Identification of individual amino acids required for secretion within the haemolysin (HlyA) C-terminal targeting region. Mol Microbiol. 1992;6:1477–1489. doi: 10.1111/j.1365-2958.1992.tb00868.x. [DOI] [PubMed] [Google Scholar]
- 40.Koronakis E, Hughes C, Milislav I, Koronakis V. Protein exporter function and in vitro ATPase activity are correlated in ABC-domain mutants of HlyB. Mol Microbiol. 1995;16:87–96. doi: 10.1111/j.1365-2958.1995.tb02394.x. [DOI] [PubMed] [Google Scholar]
- 41.Koronakis V, Hughes C. Synthesis, maturation and export of the E. coli hemolysin. Med Microbiol Immunol. 1996;185:65–71. doi: 10.1007/s004300050016. [DOI] [PubMed] [Google Scholar]
- 42.Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 43.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Létoffé S, Delepelaire P, Wandersman C. Cloning and expression in Escherichia coli of the Serratia marcescens metalloprotease gene: secretion of the protease from E. coli in the presence of the Erwinia chrysanthemi protease secretion functions. J Bacteriol. 1991;173:2160–2166. doi: 10.1128/jb.173.7.2160-2166.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Létoffé S, Delepelaire P, Wandersman C. Protein secretion in Gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 1996;15:5804–5811. [PMC free article] [PubMed] [Google Scholar]
- 47.étoffé S, Wandersman C. Secretion of CyaA-PrtB and HlyA-PrtB fusion proteins in Escherichia coli: involvement of the glycine-rich repeat domain of Erwinia chrysanthemi protease B. J Bacteriol. 1992;174:4920–4927. doi: 10.1128/jb.174.15.4920-4927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy E C, Doyle D, Burda K, Corbeil L B, Winter A J. Superficial antigens of Campylobacter (Vibrio) fetus: characterization of an antiphagocytic component. Infect Immun. 1975;11:517–525. doi: 10.1128/iai.11.3.517-525.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei Z, Blaser M J. Pathogenesis of Campylobacter fetus infections: role of surface array proteins in virulence in a mouse model. J Clin Invest. 1990;85:1036–1043. doi: 10.1172/JCI114533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pei Z, Ellison R T I, Lewis R, Blaser M J. Purification and characterization of a family of high molecular weight surface-array proteins from Campylobacter fetus. J Biol Chem. 1988;263:6416–6420. [PubMed] [Google Scholar]
- 51.Salmond G P C, Reeves P J. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:8–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- 53.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 54.Sleytr U B, Messner P, Pum D, Sara M. Crystalline bacterial surface layers. Mol Microbiol. 1993;10:911–916. doi: 10.1111/j.1365-2958.1993.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 55.Smibert R M. The genus Campylobacter. Annu Rev Microbiol. 1978;32:673–709. doi: 10.1146/annurev.mi.32.100178.003325. [DOI] [PubMed] [Google Scholar]
- 56.Stanley P, Koronakis V, Hughes C. Mutational analysis supports a role for multiple structural features in the C-terminal secretion signal of Escherichia coli haemolysin. Mol Microbiol. 1991;5:2391–2403. doi: 10.1111/j.1365-2958.1991.tb02085.x. [DOI] [PubMed] [Google Scholar]
- 57.Suh Y, Benedik M J. Production of active Serratia marcescens metalloprotease from Escherichia coli by α-hemolysin HlyB and HlyD. J Bacteriol. 1992;174:2361–2366. doi: 10.1128/jb.174.7.2361-2366.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson S A, Sparling P F. The RTX cytotoxin-related FrpA protein of Neisseria meningitidis is secreted extracellularly by meningococci and by HlyBD+Escherichia coli. Infect Immun. 1993;61:2906–2911. doi: 10.1128/iai.61.7.2906-2911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tummuru M K R, Blaser M J. Characterization of the Campylobacter fetus sapA promoter: evidence that the sapA promoter is deleted in spontaneous mutant strains. J Bacteriol. 1992;174:5916–5922. doi: 10.1128/jb.174.18.5916-5922.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tummuru M K R, Blaser M J. Rearrangement of sapA homologs with conserved and variable regions in Campylobacter fetus. Proc Natl Acad Sci USA. 1993;90:7265–7269. doi: 10.1073/pnas.90.15.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wandersman C. Secretion across the bacterial outer membrane. Trends Genet. 1992;8:317–321. doi: 10.1016/0168-9525(92)90264-5. [DOI] [PubMed] [Google Scholar]
- 62.Wang B, Kraig E, Kolodrubetz D. A new member of the S-layer protein family: characterization of the crs gene from Campylobacter rectus. Infect Immun. 1998;66:1521–1526. doi: 10.1128/iai.66.4.1521-1526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson H R, Archer C D, Liu J, Turnbough C L., Jr Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol. 1992;174:514–524. doi: 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolff N, Ghigo J-M, Delepelaire P, Wandersman C, Delepierre M. C-terminal secretion signal of an Erwinia chrysanthemi protease secreted by a signal peptide-independent pathway: proton NMR and CD conformational studies in membrane-mimetic environments. Biochemistry. 1994;33:6792–6801. doi: 10.1021/bi00188a007. [DOI] [PubMed] [Google Scholar]