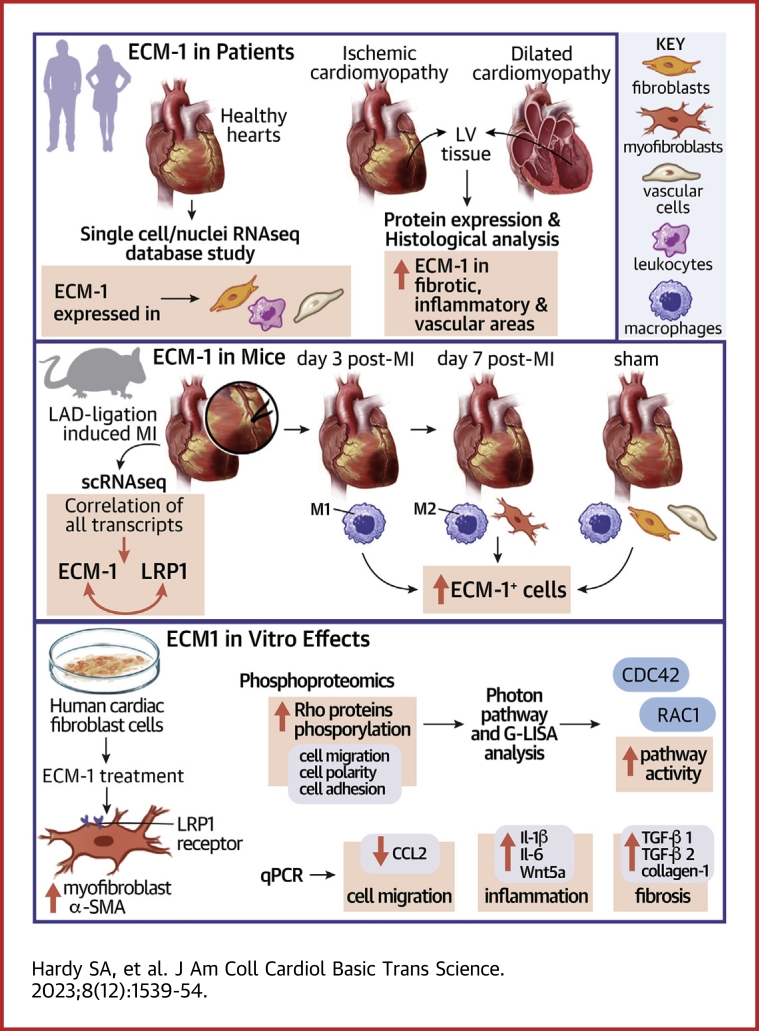
Visual Abstract
Key Words: extracellular matrix, fibroblasts, fibrosis, heart, inflammation, myocardial infarction
Highlights
-
•
ECM-1 is up-regulated in ischemic and dilated failing human hearts and predominantly localized to fibrotic, inflamed, and perivascular areas.
-
•
After MI, ECM-1 cellular sources shift from M1 macrophages early, to myofibroblasts and M2 macrophages later.
-
•
ECM-1 expression is associated with LRP1, inflammation, cell communication, cell migration, and matrix remodeling–associated signaling pathways according to scRNAseq analyses.
-
•
ECM-1 binds cell surface receptor LRP1 and stimulates profibrotic fibroblast-to-myofibroblast transition. LRP1 inhibition is sufficient to block this effect. ECM-1 regulates key profibrotic and inflammatory pathways, activates Rac1,2,3 GTPase, and inhibits human cardiac fibroblast migration.
-
•
ECM-1 may represent a novel link mediating inflammation-induced fibrotic signaling in the heart.
Summary
Irreversible fibrosis is a hallmark of myocardial infarction (MI) and heart failure. Extracellular matrix protein-1 (ECM-1) is up-regulated in these hearts, localized to fibrotic, inflammatory, and perivascular areas. ECM-1 originates predominantly from fibroblasts, macrophages, and pericytes/vascular cells in uninjured human and mouse hearts, and from M1 and M2 macrophages and myofibroblasts after MI. ECM-1 stimulates fibroblast-to-myofibroblast transition, up-regulates key fibrotic and inflammatory pathways, and inhibits cardiac fibroblast migration. ECM-1 binds HuCFb cell surface receptor LRP1, and LRP1 inhibition blocks ECM-1 from stimulating fibroblast-to-myofibroblast transition, confirming a novel ECM-1-LRP1 fibrotic signaling axis. ECM-1 may represent a novel mechanism facilitating inflammation-fibrosis crosstalk.
Myocardial infarction (MI) remains one of the largest causes of death and heart failure worldwide. MI results in mass myocyte death, which promotes intense inflammatory responses, extracellular matrix (ECM) remodeling, and cardiac fibrosis.1, 2, 3 Because the adult mammalian heart has limited regenerative capacity,4,5 necrosis and inflammation are followed by profibrotic ECM remodeling and scarring. This process produces a functional scar in an attempt to maintain the integrity of the heart and prevent cardiac rupture, but it impairs cardiac structure and function, and promotes the progression of heart failure (HF).3,6 The molecular events fine-tuning ECM remodeling and fibrosis in cardiac wound healing remain incompletely understood, so we currently have limited treatment options to prevent or reverse post-MI inflammation and scarring.
There is a recent appreciation for the critical role of inflammation-fibrosis crosstalk, whereby fibroblasts and leukocytes cooperate at numerous points to orchestrate wound healing.7, 8, 9 Some leukocytes, like macrophages, have a particularly intimate relationship with fibroblasts in wound healing after MI, and have been demonstrated to directly influence fibroblast cells and carry out paracrine inflammatory-fibrosis crosstalk via secretion of cytokines, chemokines, and other factors.8,10 Macrophages have even been shown to carry out ECM remodeling and even contribute to fibrotic tissue deposition themselves.11, 12, 13, 14 This novel axis of inflammation-fibrosis crosstalk may be why unidirectional therapies, which target fibrosis or inflammation alone, have failed to substantially reduce the mortality associated with fibrotic cardiac diseases.7 As such, inflammation-fibrosis crosstalk is a key factor in the investigation of novel therapeutic strategies to battle the mortality of fibrotic cardiac disease.
Extracellular matrix protein (ECM)-1 is an 85-kDa glycoprotein encoded by the ECM-1 gene located on human chromosome 1q21.15 ECM-1 interacts with many ECM proteins16 and acts as a structural basement membrane protein, as well as regulating inflammatory cell function.17, 18, 19 We recently identified that ECM-1 is expressed by monocyte/macrophage-like cells in mouse bone marrow, and that it stimulates collagen-I production in human cardiac fibroblasts (HuCFbs).1 However, the precise mechanisms involved, as well as cellular expression profiles of ECM-1 in the heart, remain unknown. Therefore, we investigated the spatiotemporal cellular origin of ECM-1 in healthy and diseased human and mouse hearts, and ECM-1-dependent HuCFb signaling mechanisms.
Methods
Human studies, ethics, and patient characteristics
The use of human biomaterials was approved by the Ethics Committee of the Medical University of Graz (20-277 ex 08/09, 26-355 ex 13/14, and 28-508 ex 15/16) and conformed to all pertaining regulations and the principles of the Declaration of Helsinki. Left ventricular cardiac samples were collected from patients with ischemic cardiomyopathy (ICM) and dilated cardiomyopathy (DCM), and nonfailing (NF) control subjects (n = 8/group) for immunoblotting at the time of cardiac transplantation or explantation. All ICM and DCM patients were male, and the NF group included 2 women. The average ages were 63.3 ± 1.8, 58.9 ± 3.0, and 64.4 ± 4.2 years, respectively, in the ICM, DCM, and NF groups. The average ejection fractions were 27.5% ± 2.6%, 19.4% ± 1.9%, and 61.9% ± 1.6%, respectively. Left ventricular cardiac samples used for immunohistochemistry (IHC) were collected from heart disease patients at routine autopsy (n = 12; 75% male, age 66 ± 4.16 years; ICM n = 8, DCM n = 1, NF n = 4). All ICM hearts were from patients with a clinical history of HF and concordant pathologic and histologic findings. The Ethical Committee approved the study and waived the requirement for informed consent when human tissue originated from deceased persons; all other persons provided written informed consent.
Analysis of single-cell and single-nuclear RNA-sequencing data
Published single-cell (sc) RNA-sequencing (RNAseq) data of murine cardiac interstitial cells of ventricles from sham and MI hearts from Farbehi et al20 are available from ArrayExpress under accession codes E-MTAB-7376. Similarly, published scRNAseq data of murine cardiac interstitial cells of sham and transverse aortic constriction (TAC) hearts from Alexanian et al21 are available in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE155882.
Briefly, for data collected by Farbehi et al,20 8- to 12-week-old male C57Bl/6J mice were anesthetized and intubated, their hearts exposed, and the left anterior descending coronary artery ligated permanently. Hearts were harvested at 3 or 7 days after surgery. Sham-operated mice were subject to the same procedure without ligation and cells isolated from sham or MI hearts were profiled for scRNAseq using the 10× Genomics Chromium system. For analysis parameters and details see the Supplemental Methods.
Processed and annotated human heart single nuclei (sn)/scRNA-seq data from Litviňuková et al22 were loaded in the Seurat R package for further analysis. A normalized gene expression score per cell base for ECM-1 was extracted with the use of the AddModuleScore function.
Cell culture
All cell cultures were carried out with a commercially sourced adult human cardiac fibroblast (HuCFb) cell line (Sigma Aldrich; cat no. 306-05A) sourced from normal human adult heart ventricle (52-year-old White male), and performed with the recommended reagents per the manufacturer’s protocols, except for the “starving media” formulation of DMEM/F-12 GlutaMAX (ThermoFisher; cat no. 31331028) supplemented with 0.5% fetal bovine serum (FBS) (ThermoFisher; cat no. 10099141). Briefly, HuCFb cells were cultured to 70% confluence, the growth media replaced with starving media for 16 hours, and then treated with recombinant proteins in starving media at the following concentrations: ECM-1 (20 ng/mL; R&D systems; catno. 3937-EC), and TNF-α (20 ng/mL; Gibco; cat no. PHC3011). Protein and mRNA were then extracted from cells (Supplemental Methods). Alternatively, MTT Cell Proliferation Kit I (Roche; cat no. 11 465 007 001) and wound healing assays were performed on live HuCFb cells in culture (Supplemental Methods). Methods for Rac1,2,3 and RhoA G-LISA assays and immunofluorescence/immunocytochemistry are specified in the Supplemental Methods. Protein lysates from cultured HuCFb cells were also used for phosphoproteomics and for His-tag–dependent pull-down of ECM-1 protein-protein binding partners followed by mass spectrometry (in-gel digestion workflow). For details, see the Supplemental Methods. All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE23 partner repository with the dataset identifier PXD027626.
Tissue and sample preparation and analysis
Human samples were snap frozen in liquid nitrogen on explantation and stored at −80 °C. mRNA and protein extraction and quantification methods are specified in the Supplemental Methods. Protein and mRNA extracted from human samples and HuCFb cells were subject to immunoblotting and/or quantitative polymerase chain reaction (qPCR) analysis, as previously described.24,25 All mRNA expression data are expressed as delta-delta threshold cycle (ΔΔCt), relative to control mRNA expression, and normalized to tpt1 housekeeping gene. All immunoblot protein expression data were normalized to either GAPDH, β-tubulin (55 kDa), or β-actin (42 kDa) as the loading control for relative band density analysis, but in the case of phosphoproteins and active β-catenin, data were normalized as a ratio of the active or phosphorylated form/total (nonphosphorylated). For specific methods of protein isolation and quantification, RNA isolation and quantification, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblot, qPCR, immunohistochemistry, in situ hybridization (RNAscope), and our list of qPCR primer sequences, see the Supplemental Methods.
Statistical analysis
Data are represented as mean ± SD. All data sets were assessed for normality of distribution (parametric distribution) with the use of Shapiro-Wilk tests. Parametric data were analyzed with the use of t-tests for 2 groups and analysis of variance (ANOVA) with post hoc Šídák’s multiple comparisons test for more than 2 groups, or repeated-measures 2-way ANOVA (or repeated measures mixed-effects analysis to account for missing values) with post hoc Šídák’s multiple comparisons test, as detailed in the figures. Pearson’s correlation and simple linear regression analysis were used to test the strength of linear relationships between 2 variables. Fisher’s exact test was used to assess association/enrichment of categoric data. Statistical analysis was conducted with the use of GraphPad Prism (version 9.1.0) unless otherwise stated. All tests were conducted with an assigned significance level of P < 0.05. Statistical analyses of scRNAseq and proteomics experiments are provided in the Supplemental Tables.
Results
ECM-1 expression in healthy and failing human hearts
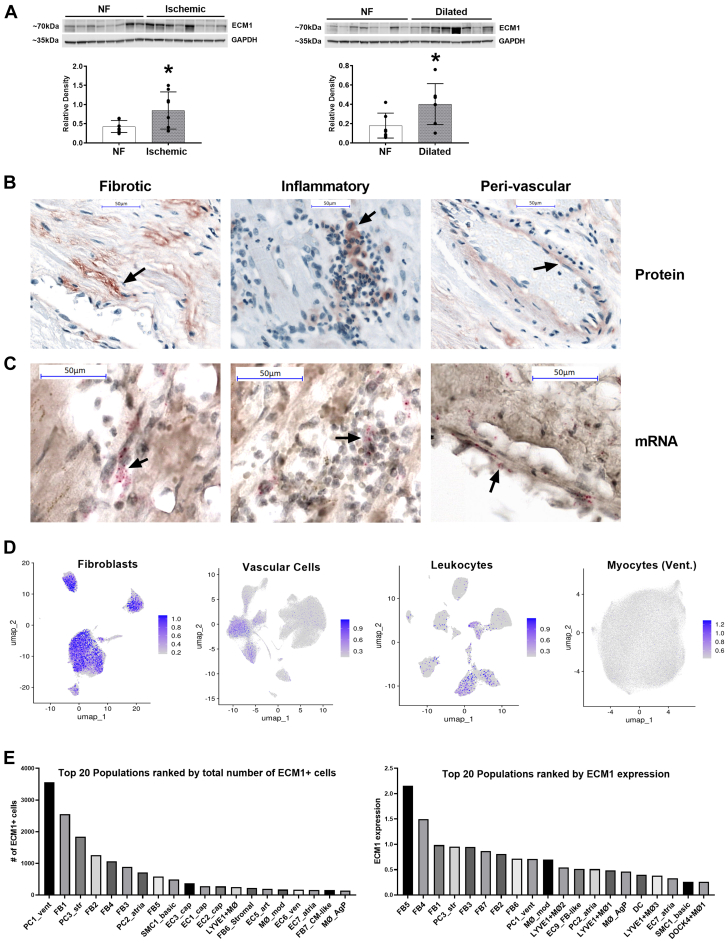
We have previously shown ECM-1 expression in murine hearts after MI.1 We hypothesized that this would be the same in human hearts and quantified the expression levels of ECM-1 in human NF hearts relative to ICM and DCM HF patients. ECM-1 protein expression was significantly up-regulated in heart tissue from both ICM and DCM patients (Figure 1A). To visualize the localization of ECM-1 expression, we conducted ECM-1 IHC and in situ hybridization experiments. ECM-1 expression was predominantly interstitial, localized to fibrotic, inflammatory, and perivascular areas in ICM patients (Figure 1B) and predominantly fibrotic and perivascular in DCM patients (Supplemental Figure 2A), where less inflammation was evident. In situ hybridization of ICM patients confirmed that ECM-1 was locally transcribed in these areas (Figure 1C). Next, we analyzed ECM-1 expression in sc- and snRNAseq in healthy human heart donors in the data set generated by Litviňuková et al.22 Cells expressing ECM-1 in the human heart were predominantly fibroblasts, leukocytes, and vascular cells (Figure 1D, Supplemental Figure 1). A small number of ECM-1+ leukocytes were identified in these data owing to low leukocyte cell numbers in healthy hearts relative to diseased hearts (Supplemental Table 1). The cell type with the highest ECM-1 expression level was “FB5_ECM-organising” (23.15% ECM-1+), which are ventricular fibroblasts enriched for ECM production, remodeling, and degradation genes (Figure 1E); see Supplemental Table 2 for detailed cell characteristics and Supplemental Tables 3 and 4 for detailed data. The leukocyte subpopulation with the highest number of ECM-1+ cells was LYVE1+ tissue-resident macrophages (LYVE1+MØ1), which are enriched in clathrin and cathepsin genes associated with cardiovascular remodeling.26 Interestingly, PC1_vent had the overall highest number of ECM-1+ cells in the data set and was the most abundant cell type, with 50,095 cells, nearly double the second most abundant cell type, FB1_VT-canonical (26,632 cells). PC1_vent are pericytes enriched for adhesion molecules NCAM2 and CD38 and the microvascular morphogenesis and endothelial cell crosstalk gene CSPG4.
Figure 1.
ECM-1 Is Upregulated in Human Failing Hearts and Is Localized to Fibrotic Tissue, Leukocytes, and Endothelia
(A) Western blotting shows that extracellular matrix protein (ECM)-1 protein is up-regulated in ischemic and dilated human heart failure patients, presented as mean ± SD, analyzed by means of Student’s t-test (n = 8/group). ∗P < 0.05. (B) Immunohistochemistry of ECM-1 protein expression in the human heart (NF: n = 4; heart failure: n = 8) shows that ECM-1 expression is predominantly interstitial, localized to fibrotic, inflammatory, and perivascular areas (×40 magnification; scale bars = 50 μm). (C) mRNA in situ hybridization (n = 3) confirms transcription of ECM-1 in these areas (×40 magnification zoomed in; scale bars = 50 μm). (D) ECM-1-specific analysis of single-cell/single-nuclear RNA sequencing dataset from Litviňuková et al22 identifies ECM-1 expression by fibroblasts, leukocytes, and vascular cells in the healthy human heart (gray = all cells; blue overlay = ECM-1+ cells). (E) The top 20 ECM-1-expressing cell subpopulations ranked by total number of ECM-1+ cells and by ECM-1 expression level (expressed as a Log2 average). NF = nonfailing.
Together, these data reveal spatiocellular ECM-1 expression in healthy human hearts that is amplified in failing human hearts. Fibroblast and leukocyte (macrophage) subsets as well as pericytes are the predominant cellular ECM-1 sources.
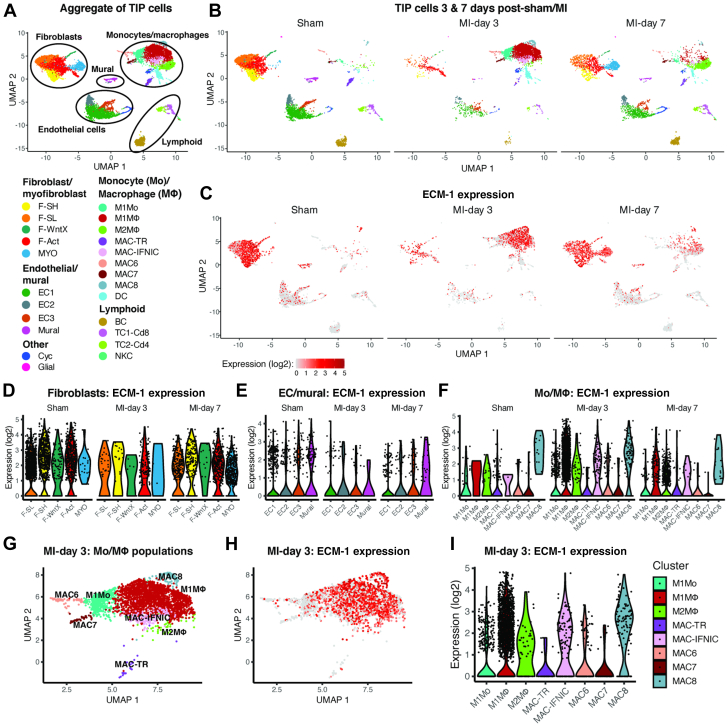
Dynamic ECM-1 expression patterns after MI
To investigate the origin of ECM-1 in the heart and its dynamic cellular expression changes with cardiac disease, we analyzed ECM-1 expression in the mouse scRNAseq data set of interstitial cells after MI generated by Farbehi et al.20 Consistent with our analysis in human heart tissue (Figure 1), ECM-1 was expressed most abundantly by fibroblasts and monocytes/macrophages (Mo/MΦ) (Figures 2A to 2C). Furthermore, we found a dynamic shift of ECM-1 cellular expression between fibroblasts and Mo/MΦs over the time course of wound healing after MI. In the uninjured sham-operated heart, the number of ECM-1+ cells was highest in unactivated F-SL and F-SH fibroblasts (which express genes either for enriched cell adhesion [F-SH] or cell secretion and cell signaling processes [F-SL]). At day 3 after MI, ECM-1 expression was highest in M1MΦs, with expression also in M1Mos, activated fibroblasts (F-Acts), MAC8s, and interferon-inducible cell (IFNIC) MΦs. At day 7 after MI, ECM-1 expression significantly increased in F-Acts, myofibroblasts, and M2MΦs, which are involved in inflammation resolution and repair (Figures 2D to 2F); see Supplemental Table 5 for detailed cell characteristics and Supplemental Table 6 for detailed data. ECM-1 was also expressed by some vascular mural cells, which encompass pericytes and endothelial cells, however ECM-1+ cell numbers decreased after MI. Little ECM-1 expression was observed in lymphoid cell populations at all time points. Of note, although leukocyte numbers were present at small numbers in sham-operated hearts, tissue-resident MΦs (MAC-TR) had the highest ECM-1+ cell number of all leukocyte subpopulations, which was consistent with our data in the human heart (Figure 1).
Figure 2.
Expression of ECM-1 in Cardiac Interstitial Cells Before and After MI
(A) UMAP plot showing cell populations in single-cell RNA sequencing of cardiac interstitial cells from sham-operated and MI hearts combined, and (B) faceted according to condition. (C) ECM-1 expression on UMAP coordinates according to condition. (D, E, F) Violin plots of ECM-1 expression according to condition for cell subpopulations in (D) fibroblasts, (E) endothelial (EC) and mural cells, and (F) monocyte (Mo) and macrophage (MΦ) populations. (G) UMAP plot of Mo/MΦ cell populations at day 3 after MI. (H, I) ECM-1 expression in day-3 post-MI Mo/MΦ cells on (H) UMAP coordinates and (I) in violin plots.Abbreviations as in Figure 1.
To contrast our findings in ischemic hearts to ECM-1 expression in nonischemic HF, we used the mouse scRNAseq dataset of interstitial cells after TAC (an animal model of non-ischemic cardiomyopathy) generated by Alexanian et al.21 Similarly to MI, ECM-1 was expressed most abundantly by fibroblasts and MΦs, and ECM-1+ fibroblast and MΦ cell numbers increased after TAC (Supplemental Figures 2B to 2F).
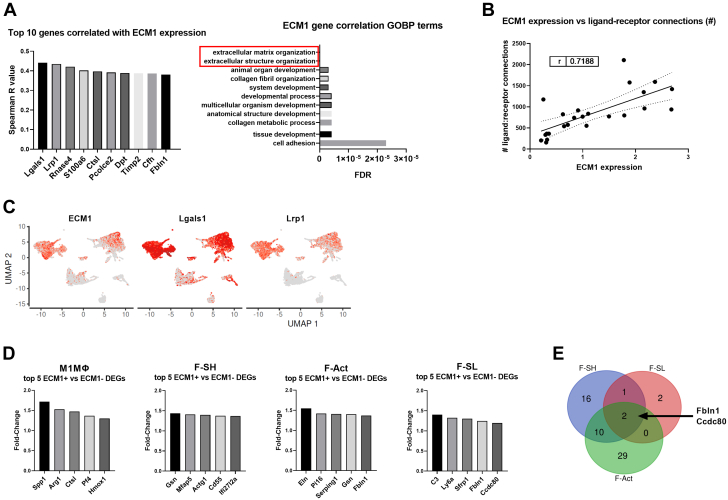
To gain unbiased insights into the function of ECM-1 in the heart after MI, we next correlated ECM-1 expression levels with all genes in the Farbehi et al20 dataset. We also assessed differentially expressed genes (DEGs) between ECM-1+ and ECM-1− cells (per specific cell population). ECM-1 expression most positively correlated with inflammatory and tissue remodeling genes Galectin-1 (Lgals1) and low-density lipoprotein receptor–related protein 1 (Lrp1).27,28 Of interest, LRP1 is a large cell surface receptor expressed by both MΦs and fibroblasts, that plays a key role in regulating inflammatory and tissue remodeling responses after MI.29 Gene ontology biological process (GOBP) enrichment analysis of correlated genes (eg, LRP1, Tissue inhibitor of metalloproteinases 2 [Timp2], Fibulin-1 [Fbln1], and Collagen type I alpha 1 chain [Col1a1]) showed that ECM-1 expression was most significantly associated with ECM and collagen organization, cell adhesion, and developmental processes (Figures 3A and 3C, Supplemental Tables 7 and 8).
Figure 3.
ECM-1 Expression Strongly Correlated With the Number of Outbound Ligand-Receptor Interactions Between Cells, Extracellular Matrix Organization, and Cell Adhesion Genes
(A) Per-cell Spearman correlation analysis of ECM-1 expression against all genes in the mouse single-cell RNA sequencing data set20; all P <0.001. The top 10 positively correlated genes (x-axis) are shown as column graphs of Spearman’s r-value (y-axis). The top ECM-1-correlated genes (with Spearman’s r > 0.30) were subject to gene ontology biological process (GOBP) enrichment analysis via the GOnet/DICE online tool. The top 10 GOBPs are presented as a bar chart ranked by false discovery rate (FDR) P value. (B) The average ECM-1 expression level (Log2) of each cell subpopulation is positively correlated with the number of significant outbound ligand-receptor connections (r = 0.719; P < 0.001), analyzed by Pearson’s correlation and simple linear regression. (C) ECM-1, Lgals1, and Lrp1 expression overlayed on the UMAP plot of cardiac interstitial cells from sham and MI (as shown in Figure 2A), to compare the cellular expression profile of the top 2 ECM-1 correlated genes. (D) Differentially expressed genes (DEGs) between ECM-1+ and ECM-1− cells (per cell subpopulation) within the major ECM-1-expressing cell subpopulations were assessed via MAST. Only M1MΦ, F-Act, F-SH, and F-SL returned lists of significant DEGs, and the top 5 up-regulated genes in ECM-1+ cells (x-axis) are shown in column graphs as fold change in gene expression (y-axis). (E) All up-regulated DEGs for M1MΦ, F-Act, F-SH, and F-SL were subject to Venn diagram analysis. No genes were commonly up-regulated in all 4 cell types; Fbln1 and Ccdc2 were commonly up-regulated in ECM-1+ F-Act, F-SH, and F-SL cells. Abbreviations as in Figures 1 and 2.
DEG analysis and GOBP enrichment showed that genes up-regulated in ECM-1+ F-Acts and F-SH fibroblasts were over-represented for extracellular structure and ECM organization (Figure 3D, Supplemental Tables 9 and 10). Furthermore, the 2 cellular adhesion proteins Ccdc80 and Fbln1 were commonly up-regulated in ECM-1+ F-Acts and F-SH, and F-SL fibroblasts (Figure 3E). Up-regulated DEGs in ECM-1+ M1MΦs showed over-representation of chemotactic and migration-associated processes (Figure 3D, Supplemental Table 11). Finally, via new analysis of our published cell-cell communication data,20 we identified a strong correlation between ECM-1 expression levels and the number of significant outbound ligand-receptor connections (Figure 3B), suggesting a functional association between ECM-1 expression and intercellular signaling crosstalk.
Together, these data reveal that ECM-1 is predominantly expressed by fibroblasts and MΦs in both ischemic and nonischemic cardiomyopathies. The cellular expression patterns of ECM-1 shift between fibroblasts and MΦs after MI, supporting a key role for ECM-1 in fibroblast-MΦ crosstalk in wound healing. ECM-1 expression is correlated with Lgals1 and Lrp1 expression, and functionally associated with ECM and collagen organization/remodeling, cardiac inflammation, cell-cell/cell-matrix adhesion, cell migration, and intercellular signaling crosstalk.
ECM-1 stimulates inflammatory and fibrotic signaling pathways and myofibroblast transition in human cardiac fibroblasts
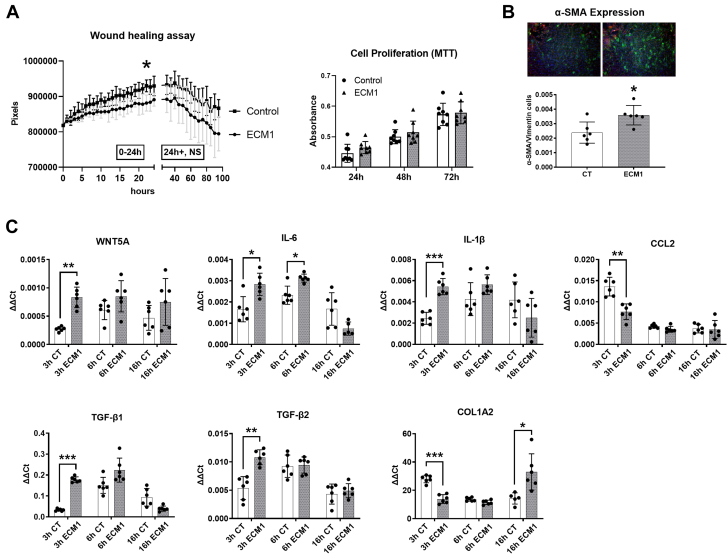
Next, to understand the effect of ECM-1 in wound healing and fibrosis after MI we aimed to test ECM-1’s impact on fibroblast functionality. Wound healing assays showed that ECM-1 significantly reduced the rate of HuCFb migration in vitro over 24 hours, relative to control cells treated with media alone (Figure 4A). ECM-1 had no direct effect on cell proliferation (MTT assay). ECM-1 significantly stimulated fibroblast-to-myofibroblast transition after 48 hours of treatment relative to control (Figure 4B).
Figure 4.
ECM-1 Inhibits Human Cardiac Fibroblast Cell Migration in Culture, and Stimulates Fibroblast-to-Myofibroblast Transition and Inflammatory and Fibrotic Signaling Pathways in HuCFbs
(A) ECM-1 treatment (20 ng/mL) significantly reduced the migration rate of HuCFb cells over 24 hours relative to control subjects (n = 4/group), as assessed via both repeated-measures 2-way analysis of variance (ANOVA) with post hoc Šídák’s multiple comparisons test (P = 0.044) and inclination of the linear equation of growth rate via Student’s t-test (P = 0.027). ECM-1 had no effect on cell proliferation assessed via MTT assay after 24, 48, or 72 hours of treatment, as assessed via repeated-measures 2-way ANOVA with post hoc Šídák’s multiple comparisons test. (B) Immunofluorescence shows ECM-1 stimulates fibroblast-to-myofibroblast transition in HuCFbs after 48 hours of treatment, as analyzed by Student’s t-test (P = 0.014; n = 6; ×2.5 magnification; green = α-smooth muscle actin [SMA]; red = vimentin; blue = DAPI/nuclei). (C) HuCFbs were treated with ECM-1 (20 ng/mL) or medium alone for 3, 6, or 16 hours and assessed via quantitative polymerase chain reaction for mRNA expression of Wnt5a, interleukin (IL)-1β2, IL-6, CCL2, transforming growth factor (TGF)-β1, TGF-β2, and collagen type I alpha 2 chain (Col1a2). Each gene was assessed via repeated-measures 2-way ANOVA (IL-6 and Col1a2 were assessed via repeated-measures mixed-effects analysis to account for missing values) with post hoc Šídák’s multiple comparisons test; expression of all genes are presented as the delta-delta threshold cycle (ΔΔCt) relative to Tpt1 housekeeping gene expression, n = 6/group. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. CT = control; other abbreviations as in Figures 1 and 2.
We then used qPCR and immunoblotting to investigate the influence of ECM-1 over inflammatory and fibrotic pathways, chemotactic pathways (CCL2 [MCP-1]), and Wnt5a, which modulates noncanonic Wnt signaling, Rho protein signaling, cell/microtubule architecture organization, cell adhesion, and cell chemotaxis. ECM-1 significantly up-regulated mRNA expression of interleukin (IL)-1β, IL-6, and Wnt5a at 3 hours after treatment, and down-regulated CCL2 mRNA expression, relative to control (Figure 4C). ECM-1 significantly up-regulated TGF-β1 and TGF-β2 expression at 3 hours after treatment, followed by an up-regulation of Col1a2 expression at 16 hours.
Taken together, these data demonstrate that ECM-1 inhibits HuCFb cell migration, stimulates fibroblast-to-myofibroblast transition, and up-regulates inflammatory and fibrotic genes.
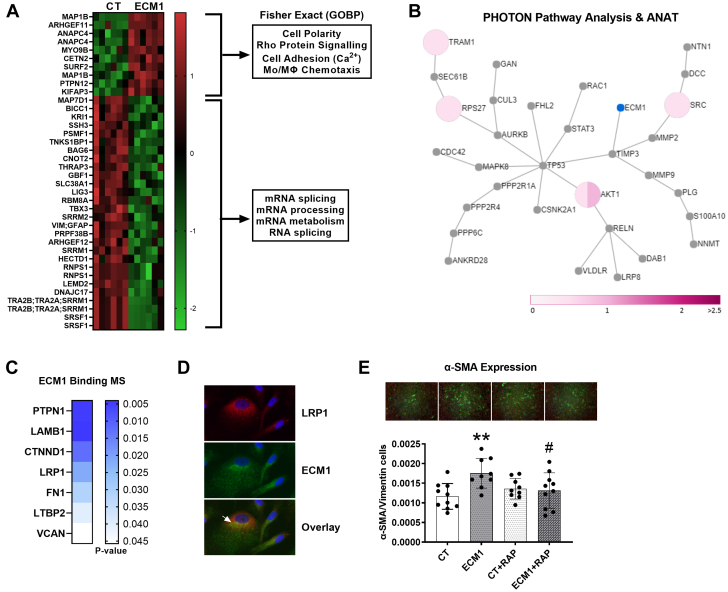
ECM-1-dependent HuCFb cell signaling
Next, we aimed to investigate potential signaling cascades and elucidate the mechanisms that underpin these profibrotic, proinflammatory, and chemotactic effects of ECM-1 on HuCFbs. With the use of unbiased phosphoproteomics mass spectrometry (serine/threonine), we identified 38 differentially phosphorylated proteins in ECM-1-treated HuCFbs relative to control cells treated with media alone (Figure 5A); for full data see Supplemental Table 12. MAP1B was the most highly phosphorylated protein relative to control samples (27.3-fold, q-value <0.0001). Interestingly, most proteins (28 proteins) were dephosphorylated in the ECM-1 treatment group, with only 8 proteins (10 including multiple P-sites) increasing in phosphorylation relative to control: MAP1B, ARHGEF11, ANAPC4, MYO9B, CETN2, SURF2, PTPN12, and KIFAP3. Of note, ARHGEF11, MYO9B, and PTPN12 have significant influence over Rho protein/Rho GTPase signal transduction. Analysis of GOBPs with the use of Fisher’s exact test complemented this, showing over-representation of calcium-dependent cell-cell adhesion, Rho protein signal transduction, cell polarity, microtubule-based transport, and Mo/MΦ chemotaxis processes (Supplemental Table 13). Gene ontology cellular compartment locations associated with microtubule-rich cellular projections and nonmotile primary cilia also were over-represented. PHOTON pathway analysis of proteins increased in phosphorylation status with ECM-1 treatment identified 18 proteins with significant signal functionality scores (≥6), including RAC1, CDC42, LRP8, S100a10, and RELN; this indicates involvement in active mediation of ECM-1-dependent signal transduction. ANAT reconstruction of this ECM-1 network also showed ECM-1 signaling through TIMP3 and TP53 to influence pathways including Rho GTPase mediators RAC1, CDC42, and LRP8 receptor (Figure 5B). G-LISA assay confirmed this, showing increased RAC1,2,3 activation in response to ECM-1 treatment (Supplemental Figure 3).
Figure 5.
Proteomics Reveals ECM-1-HuCFb Cell Membrane Binding Partners and Downstream Signaling Mechanisms: ECM-1 Binds LRP1 Cell Surface Receptor, Which Is Required for ECM-1-Dependent Stimulation Of Profibrotic Fibroblast-to-Myofibroblast Transition
(A) HuCFb cells were treated with ECM-1 (20 ng/mL) for 10 minutes and subject to phosphoproteomics (Ser/Thr) mass spectrometry (titanium dioxide workflow; n = 6/group). Thirty-eight proteins with significantly different phosphorylation levels were identified between CT (medium alone) and ECM-1-treated cells, as assessed via Student’s t-test with permutation-based FDR; data are presented as the Log2 difference (CT − ECM-1) in phosphorylation level; green = down-regulated; red = up-regulated. Note that proteins measured multiple times represent different phosphosites within those proteins. Fisher’s exact enrichment testing was conducted on significantly up-regulated and down-regulated phosphorylation sites in ECM-1-treated samples (separately), and 4 of the most over-represented GOBPs are shown. (B) Our list of differential phosphorylation sites in ECM-1-treated HuCFbs was then subjected to PHOTON pathway analysis (2-sided) with reconstruction ANAT (ECM-1 as the response source) to visualize the ECM-1-dependent signaling network. The ANAT network for ECM-1 signaling in the “greater” PHOTON analysis direction (signaling score ≥6) is shown with ECM-1 highlighted in blue. All proteins shown in the network represent involvement in active mediation of ECM-1-dependent signal transduction. Pink pie charts represent fold changes of measured phosphorylation sites (see scale bar), with multiple phosphorylation sites represented by a pie chart divided by the number of phosphorylation sites measured. (C) ECM-1 binding partners were pulled down from a purified HuCFb membrane protein lysate, with the use of recombinant human ECM-1 protein as bait; 58 potential ECM-1-binding proteins were identified and 7 proteins of particular interest are presented: PTPN1, LAMB1, CTNND1, LRP1, FN1, LTBP2, and VCAN. (D) Immunofluorescence of HuCFb cells in culture showed that ECM-1 colocalizes with LRP1 (×40 magnification zoomed in); arrowhead shows an example area of interest (red = low-density lipoprotein receptor–related protein 1 [LRP1]; green = ECM-1; blue = DAPI/nuclei). (E) Immunofluorescence shows ECM-1 stimulates fibroblast-to-myofibroblast transition in HuCFbs at 48 hours of treatment (P = 0.003), and RAP (LRPAP1)–dependent LRP1 inhibition blocks this effect (ECM-1 vs ECM-1+RAP: P = 0.038; CT vs ECM-1+RAP; nonsignificant), presented as mean ± SD, analyzed by 1-way ANOVA with post hoc Šídák’s multiple comparisons test (n ≥ 9; ×2.5 magnification; green = α-SMA; red = vimentin; blue = DAPI/nuclei); #P < 0.05 comparing ECM-1+RAP and ECM-1 treatment groups. ∗P < 0.05; ∗∗P < 0.01. Abbreviations as in Figure 1, Figure 2, Figure 3, Figure 4.
To complement phosphoproteomics, ECM-1 binding partners were pulled down from a purified HuCFb membrane protein lysate. This was done using recombinant human ECM-1 protein which was pre-bound (“ECM-1” sample) to His-Tag Isolation Dynabeads (“control” sample; Dynabeads had no pre-bound ECM-1). With this approach, we identified 58 potential ECM-1-binding proteins. Proteins of interest were objectively selected from this list, first based on their subcellular localization as membrane and/or extracellular matrix (via the UniProt Knowledgebase30), with priority given to cell surface receptors or proteins that were without bias implicated in or correlated with ECM-1 signaling or expression in our sc/snRNAseq (Figures 2 and 3), eg, LRP1, or phosphoproteomics data (Figure 5A). Further literature review of selected proteins was conducted to prioritize targets based on involvement in fibrosis, inflammation, and wound healing after MI. The 7 identified proteins were PTPN1, LAMB1, CTNND1, LRP1, FN1, LTBP2, and VCAN (Figure 5C). CTNND1 was of interest because it positively controls Rac1 activity and Rho protein signal transduction.31,32 Indeed, ECM-1 co-localized with and phosphorylated CTNND1 in HuCFb cells (Supplemental Figure 3). However, of particular relevance to fibrotic signaling after MI is ECM-1-LRP1 cell surface receptor binding29; consistently with our mass spectrometry data, ECM-1 mRNA expression was also highly correlated with LRP1 expression in our scRNAseq analysis (Figure 3C). Immunofluorescence supported ECM-1-LRP1 binding, showing that ECM-1 and LRP1 co-localize in HuCFb cells (Figure 5D). Finally, to determine if ECM-1-LRP1 binding is required to achieve profibrotic effects, we co-treated HuCFb cells with ECM-1 and the potent LRP1 inhibitor protein LRPAP1 (also known as RAP) for 48 hours and assessed the extent of fibroblast-to-myofibroblast transition. LRP1 inhibition was sufficient to block ECM-1-dependent stimulation of fibroblast-to-myofibroblast transition (Figure 5E). These data confirm a novel profibrotic ECM-1-LRP1 cell signaling axis in HuCFb cells.
Together, our unbiased mass spectrometry–based experiments demonstrate that ECM-1 binds various membrane-associated proteins, including the cell surface receptor LRP1, and induces phosphorylation of a variety of proteins associated with Rho protein signaling, cell-cell adhesion, cell polarity, microtubule dynamics, and chemotaxis/motility in HuCFbs. RAP-dependent LRP1 inhibition demonstrates that ECM-1 signals, at least in part, by binding the LRP1 cell surface receptor to achieve myofibroblast transformation in HuCFb cells.
Discussion
Cardiac wound healing and tissue remodeling are complex dynamic processes. Wound healing with MI involves mass recruitment of leukocytes to areas of tissue damage and intrinsically relies on tightly coordinated cell-cell signaling, interaction with the ECM, and dynamic collaboration between leukocytes (particularly MΦs) and fibroblasts.3,33 Here, we confirmed our previous mouse data1 and show that ECM-1 expression is up-regulated in the human heart in both ischemic and non-ischemic HF, has profibrotic effects and plays a pivotal role in facilitating inflammatory-to-fibrotic signaling crosstalk.
ECM-1 expression patterns in the healthy and diseased heart
We investigated spatiotemporal expression patterns and showed that fibroblasts, macrophages, and pericytes/vascular mural cells express ECM-1 in fibrotic, inflammatory, and perivascular areas in both mice and humans. We showed that the cellular origin of up-regulated ECM-1 in the inflammatory phase at day 3 after MI is mostly recruited M1MΦs. Later, ECM-1 expression is sustained by M2MΦs involved in inflammation resolution and wound repair, and profibrotic fibroblast subsets such as myofibroblasts, F-Acts, F-SLs, and F-SHs. Thus, ECM-1 plays a dynamic role in both inflammation and fibrosis in post-MI wound healing. Similarly, in nonischemic cardiomyopathy, ECM-1 was expressed predominantly by fibroblasts. This was reflected by IHC showing ECM-1 protein predominantly localized to fibrotic and perivascular areas in human DCM patients. Of note, DCM/TAC is not associated with the same level of acute inflammatory cell infiltrate as MI, which limits the comparability of ECM-1 expression in inflammatory cell infiltrates across models.
We first reported that ECM-1 expression is in leukocytes after MI and in Mo/MΦ-like cells in mouse bone marrow.1 Since then, ECM-1 has been shown to control M1MΦ polarization in inflammatory bowel disease,19 demonstrating the importance for ECM-1 in MΦ-dependent inflammation. Herein, we have used more powerful and unbiased technologies to refine our previous findings and demonstrate the specific cells of origin in the heart throughout MI, namely MΦs and fibroblasts. Of note, a distinct neutrophil population was not defined in our data, which may be due in part to the fact that neutrophil cell numbers in the heart peak at day 1 after MI and may be largely cleared by day 3 after MI.3,34 However, the MAC6 population encompassed cells that showed up-regulation of neutrophil (Ly6g), granulocyte (S100a9 and Csf3r), and eosinophil (Siglecf) markers, but none showed a relevant level of ECM-1 positivity. It is noteworthy to address that in our previous work1 we did not detect ECM-1 protein expression in fibroblasts cultured in vitro. This may be due to technical reasons related to the sensitivity of antibody testing, low levels of protein translation in cultured cells, and ECM-1 protein secretion into media. Here we used the more sensitive technique of RNAseq and were able to detect significant ECM-1 mRNA expression.
Gene correlation analyses (and GOBP enrichment) of our previously published mouse scRNAseq dataset,20 indicated that the role of ECM-1 cellular expression in the heart after MI was in ECM and collagen organization (or remodeling), cell-cell/cell-matrix adhesion, cell migration, and cardiac inflammation; we confirmed that ECM-1 influences HuCFb cell migration in vitro via wound healing assay and a corresponding down-regulation of CCL2 (MCP-1) mRNA expression. We found a strong positive correlation between ECM-1 expression and the number of outbound ligand-receptor connection in cells, suggesting a functional association between ECM-1 expression and intercellular signaling crosstalk. We also found that the 2 cellular adhesion proteins Ccdc80 and Fbln1 were commonly up-regulated in ECM-1+ F-Acts, F-SHs, and F-SLs, further supporting that ECM-1 expression is associated with cell-cell/cell-ECM adhesion processes. Indeed, Fbln1 and these other processes also have prominent roles in pulmonary fibrosis, which may be important areas of future investigations for ECM-1.35
Our proposed functions of ECM-1 are strongly supported by a recent study that showed up-regulation of genes associated with cell adhesion, ECM organization, and cell migration in ECM-1 small interfering RNA–silenced dermal fibroblasts.36 Several other previous studies are also consistent with our results, suggesting that ECM-1 is a structural basement membrane protein that binds many ECM proteins (Fbln1, Fibronectin, Fibulin-3, Laminin-332, insulin-like growth factor, epidermal growth factor, and matrix metalloproteinase-9), and is essential for the maintenance of ECM and overall structural integrity of the skin.16 Consistently, ECM-1 gene mutation is a major cause of the skin disorder lipoid proteinosis.37
Contrary to our proposed functions for ECM-1 in the heart, a study of the liver suggested that ECM-1 has antifibrotic properties via sequestering TGF-β in its latent form in the ECM.38 That study showed that ECM-1 expression decreases with increasing severity of liver disease in both mice and human patients. The reason for the discrepancy possibly lies in differences in injury models (chronic liver disease vs acute MI injury) or in inflammatory and fibrotic responses between the liver as a regenerative organ and the adult heart being nonregenerative.5 Regardless, considering our previous study1 alongside both our unbiased (cellular expression profile of ECM-1 in the mouse and human heart) and targeted approaches here, it appears that ECM-1 does not have antifibrotic effects in the heart; and consistent with that, ECM-1 has also been identified as a biomarker for fibrosis in the development of fibrostenotic Crohn’s disease.39
ECM-1 stimulates fibroblast-to-myofibroblast transition, inhibits cell migration, and up-regulates key fibrotic and inflammatory pathways in human cardiac fibroblasts
Next, we aimed to understand the effect of ECM-1 in wound healing and fibrosis after MI and test ECM-1’s impact on fibroblast functionality. We found that ECM-1 stimulated fibroblast-to-myofibroblast transition. Indeed, ECM-1 up-regulated key profibrotic genes TGF-β1, TGF-β2 (3 hours), and Col1a2 (16 hours), and proinflammatory genes IL-1β and IL-6; as well as noncanonic Wnt gene Wnt5a. These data are directly in line with our previous study1 and strongly support a profibrotic role for ECM-1 in wound healing after MI. Further functional data showed that ECM-1 inhibited the rate of HuCFb migration in wound healing assays and led to a down-regulation of CCL2 (MCP-1) mRNA expression. However, although ECM-1 clearly influences cell migration in vitro, these effects may be micro-environment specific and different in vivo.40 This may explain why our results contradict previous studies in breast cancer cells showing that ECM-1 increases cell migration and invasion in this setting.41
Together, these findings suggest that ECM-1 facilitates inflammatory-to-fibrotic signaling by stimulating fibroblast-to-myofibroblast transition and up-regulation of classic profibrotic and proinflammatory pathways.
ECM-1-dependent human cardiac fibroblast cell signaling mechanisms
Phosphoproteomics identified that ECM-1 affects pathways associated with Rho protein signaling, cell-cell adhesion, cell polarity, microtubule dynamics, and cell chemotaxis in HuCFbs. Rho GTPases are small molecular switches that control an array of signal transduction pathways, such as cell polarity, microtubule/actin cytoskeleton dynamics, and membrane transport pathways.42 Of interest, we identified 4 differentially phosphorylated proteins involved in Rho protein signaling: ARHGEF1143,44 and ARHGEF12,45 which are Rho guanine nucleotide exchange factors associated with adherens/tight junctions, cell motility, and Rho protein signaling; the Rho GTPase activator MYO9B, involved in cell motility; and PTPN12, a core controller of Rho GTPase activity, fibroblast cell motility, actin cytoskeleton remodeling, and cell adhesion.46, 47, 48 Fisher’s exact test and PHOTON pathway analysis were both supportive of this, showing over-representation of Rho protein signal transduction and that the Rho GTPase regulator proteins CDC42 and RAC1 are involved in active mediation of ECM-1-dependent HuCFb signal transduction, respectively.42 We later confirmed this directly, showing that ECM-1 treatment up-regulates Rac1,2,3 GTPase activity in HuCFbs. Consistent with these results, Gómez-Contreras et al49 showed ECM-1 is essential in the regulation of cell architecture and microtubule organization in triple-negative breast cancer cells, and that small interfering RNA against ECM-1 significantly distorted cellular architecture. They suggested that the calcium-binding protein s100a4 and/or Rho-family GTPases are the main effectors of these ECM-1-dependent cytoskeletal changes. Another recent study suggested a similar influence of ECM-1 over AKT/FAK/Rho/cytoskeleton signaling in cancer cells.50
Our protein-protein binding mass spectrometry data provided insight to upstream cell surface/membrane binding partners of ECM-1, which may initiate these signaling cascades. Of key interest was our finding of ECM-1-LRP1 binding. LRP1 is a large (∼600 kDa) cell surface receptor expressed by both MΦs and fibroblasts (and many other cell types) that is known to interact with various ligands, including ECM glycoproteins and growth factors such as TGF-β1.51 LRP1 plays several key roles in regulating inflammatory and tissue remodeling responses after MI29 and has recently been under investigation as a promising therapeutic target to improve wound healing after MI.28,30 Interestingly, studies have suggested that LRP1 acts as a co-receptor and integrator of TGF-β signaling.52,53 Fittingly, we show that LRPAP1 (RAP)–dependent inhibition of LRP1 blocks the ability of ECM-1 to stimulate fibroblast-to-myofibroblast transition, strongly suggesting that ECM-1-LRP1 binding is required to achieve this profibrotic effect. These findings provide the first evidence for a novel ECM-1-LRP1–dependent profibrotic signaling axis in HuCFb cells, which may prove a promising target for therapeutic intervention in future studies. Because the spatiotemporal cellular expression patters of LRP1 and ECM-1 are similar after MI (Figure 3C), it is enticing to speculate that ECM-1 originating from MΦs and fibroblasts represents a novel form of ECM-1-LRP1–dependent inflammation-fibrosis crosstalk. However, future studies using in vitro co-culture (MΦs and cardiac fibroblasts) and in vivo and genetic models will be needed to provide evidence for this.
Also of note was ECM-1-CTNND1 binding (also known as p120 catenin). This, alongside our phosphoproteomics data, implicates ECM-1 in Rho protein (Rac) signal transduction and cell adhesion. CTNND1 is a key regulator of cell-cell adhesion, cell migration, and cytoskeletal dynamics via controlling Rho family GTPase activity33,54,55 and is known to positively control Rac1 activity.31,55, 56, 57 Indeed, ECM-1 and CTNND1 protein co-localized in HuCFbs, and ECM-1 treatment of HuCFbs leads to phosphorylation of CTNND1 at ser252 and ser268, 2 sites known to regulate CTNND1 activity.57,58 Interestingly, PTPN12 identified in our phosphoproteomics specifically targets and dephosphorylates CTNND1, thereby controlling CTNND1’s Rho GTPase activity and effects on cell motility.32,47 Espejo et al32 showed that PTPN12 knockdown increased cell motility via altering CTNND1 phosphorylation and cellular distribution. Thus, it is enticing to speculate that ECM-1 influences HuCFb cell migration through a CTNND1/PTPN12/CCL2 axis. Future studies should investigate this ECM-1-CTNND1 binding and PTPN12-Rho/Rac1 protein signaling with the use of genetic knockout/silencing models.
Together, our findings suggest that ECM-1 facilitates inflammatory-to-fibrotic signaling via a novel ECM-1-LRP1–dependent mechanism and stimulation of traditional profibrotic and proinflammatory pathways and Rho/Rac1 protein signaling, thereby regulating cell adhesion, cell migration, and cell cytoskeletal/morphogenic properties. Thus, ECM-1 may represent a new mechanism in facilitating inflammation-to-fibrosis crosstalk in the post-MI heart through a macrophage-fibroblast ECM-1-LRP1 cell signaling axis.
Study limitations
The methods of this work are preclinical and were designed to provide the first description of the spatiotemporal cellular origins of ECM-1 in healthy and diseased human and mouse hearts, as well as ECM-1-dependent HuCFb signaling mechanisms. The data presented here provide a fundamental cellular and molecular knowledge base for future work to determine the potential of therapeutically targeting ECM-1 to ameliorate wound healing with fibrotic cardiac diseases. We note that the study is limited by a lack of experiments using in vivo and in vitro ECM-1 genetic knockout/overexpression models. This was beyond the scope of the present study, and we note this as a critical next step for future research. Future studies should use an ECM-1 genetic knockout animal model, perhaps alongside viral overexpression/rescue of ECM-1 (eg, adeno-associated virus), in experiments of surgically induced MI (left anterior descending coronary artery ligation). Future studies should also investigate the upstream factors that drive or mediate a shift in ECM-1 expression between cell subtypes (eg, fibroblasts or M1MΦs) with fibrotic cardiac disease.
Conclusions
We elucidated the spatiotemporal cell-specific expression profile of ECM-1 in healthy and diseased human and mouse hearts for the first time and found that MΦs and fibroblasts dynamically express ECM-1. ECM-1 stimulates the expression of inflammatory and fibrotic genes, stimulates fibroblast-to-myofibroblast transition, and induces phosphorylation and activity of Rho/Rac1 signaling proteins in HuCFbs. ECM-1 binds cell surface receptor LRP1, and LRP1 inhibition blocks ECM-1 from stimulating fibroblast-to-myofibroblast transition, strongly suggesting a novel profibrotic ECM-1-LRP1 cell signaling axis in HuCFbs. Thus, ECM-1 may represent a novel mechanism facilitating inflammation-to-fibrosis crosstalk in wound healing after MI.
Funding Support and Author Disclosures
This work was supported by the Austrian Society of Cardiology, ERA-CVD, and Austrian Science Fund AIR-MI consortium (I 4168-B to Dr Rainer); John Hunter Charitable Trust (G1800510 to Dr Boyle); Hunter Medical Research Institute and Emlyn and Jennie Thomas Postgraduate Medical Research Scholarship (G2100164 and G1800696 to Dr Boyle); Australian Commonwealth funded Research Training Program stipend (to Drs Hardy and Mabotuwana); Medical University of Graz Doctoral School of Translational Molecular and Cellular Biosciences [to Drs Hardy and Mabotuwana] and Doctoral School of Molecular Medicine [to Dr Rech]; Austrian Society of Cardiology (to Dr Hardy); National Health and Medical Research Council (NHMRC) (2000615 and 1074386 to Dr Harvey, 1079187 and 1175134 to Dr Hansbro, and 1156898 and 20000483 to Dr Starkey); NHMRC Senior Principal Research Fellowship (1118576 to Dr Harvey); Stem Cells Australia (SR110001002 to Dr Harvey); the Victor Chang Cardiac Research Institute, Australia [to Dr Harvey]; the University of Technology Sydney, Australia [to Dr Hansbro]; Monash University, Australia (to Dr Starkey); Australian Research Council (DE170100226 to Dr Starkey); Foundation Leducq (15CVD03 and 13CVD01 to Dr Harvey); Austrian Science fund (KLI645, W1226, and F73 to Dr Birner-Gruenberger); Interdisciplinary Centre for Clinical Research, University Hospital Würzburg (E-353 to Dr Cochain, E-354 to Dr Campos Ramos); the German Research Foundation [471705758 and 458539578 to Dr Cochain, DFG SFB1525 project no. 453989101 to Drs Cochain and Campos Ramos, and 411619907 to Dr Campos Ramos]; and the European Research Area Network-Cardiovascular Diseases/German Federal Ministry of Education and Research (01KL1902 to Dr Campos Ramos). All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Fibrosis is a key factor in heart disease that impedes cardiac function and prognosis. Yet, no therapies specifically target profibrotic signaling in the heart, and once established, fibrotic remodeling is largely irreversible. Inflammation-fibrosis crosstalk, whereby ECM remodeling and fibrotic tissue deposition is tightly connected to inflammation and vice versa, is now thought to be critical in orchestrating wound healing and cardiac scarring. It is suggested that this is why unidirectional therapies targeting fibrosis or inflammation alone have failed to substantially reduce the mortality associated with cardiac diseases. Here, we represent ECM-1 as a potential novel mediator of inflammation-induced fibrotic signaling in the heart and confirm its regulation in human heart failure.
TRANSLATIONAL OUTLOOK: ECM-1 may serve as an attractive future treatment target to prevent excessive and detrimental fibrosis. Because fibrosis and scarring is a universal response to injury in many organs, this has potential implications beyond heart disease.
Acknowledgments
The authors thank Viktoria Trummer-Herbst, Andreas Somma, and Sylvia Schauer, Medical University of Graz, Austria, for their expert technical assistance.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures, tables, and methods, please see the online version of this paper.
Contributor Information
Andrew J. Boyle, Email: andrew.boyle@newcastle.edu.au.
Peter P. Rainer, Email: peter.rainer@medunigraz.at.
Appendix
References
- 1.Hardy S.A., Mabotuwana N.S., Murtha L.A., et al. Novel role of extracellular matrix protein 1 (ECM1) in cardiac aging and myocardial infarction. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sansbury B.E., DeMartino A.M., Xie Z., et al. Metabolomic analysis of pressure-overloaded and infarcted mouse hearts. Circ Heart Fail. 2014;7:634–642. doi: 10.1161/CIRCHEARTFAILURE.114.001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forte E., Furtado M.B., Rosenthal N. The interstitium in cardiac repair: role of the immune–stromal cell interplay. Nat Rev Cardiol. 2018;15:601–616. doi: 10.1038/s41569-018-0077-x. [DOI] [PubMed] [Google Scholar]
- 4.Kretzschmar K., Post Y., Bannier-Hélaouët M., et al. Profiling proliferative cells and their progeny in damaged murine hearts. Proc Natl Acad Sci U S A. 2018;115 doi: 10.1073/pnas.1805829115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadek H., Olson E.N. Toward the goal of human heart regeneration. Cell Stem Cell. 2020;26:7–16. doi: 10.1016/j.stem.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W., Frangogiannis N.G. Fibroblasts in post-infarction inflammation and cardiac repair. Biochim Biophys Acta. 2013;1833:945–953. doi: 10.1016/j.bbamcr.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Linthout S., Miteva K., Tschope C. Crosstalk between fibroblasts and inflammatory cells. Cardiovasc Res. 2014;102:258–269. doi: 10.1093/cvr/cvu062. [DOI] [PubMed] [Google Scholar]
- 8.Okyere A.D., Tilley D.G. Leukocyte-dependent regulation of cardiac fibrosis. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas T.P., Grisanti L.A. The dynamic interplay between cardiac inflammation and fibrosis. Front Physiol. 2020;11:1133. doi: 10.3389/fphys.2020.529075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung M., Ma Y., Iyer R.P., et al. IL-10 improves cardiac remodeling after myocardial infarction by stimulating M2 macrophage polarization and fibroblast activation. Basic Res Cardiol. 2017;112:33. doi: 10.1007/s00395-017-0622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hume R.D., Chong J.J.H. The cardiac injury immune response as a target for regenerative and cellular therapies. Clin Ther. 2020;42:1923–1943. doi: 10.1016/j.clinthera.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Suthahar N., Meijers W.C., Silljé H.H.W., de Boer R.A. From inflammation to fibrosis—molecular and cellular mechanisms of myocardial tissue remodelling and perspectives on differential treatment opportunities. Curr Heart Fail Rep. 2017;14:235–250. doi: 10.1007/s11897-017-0343-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simões F.C., Cahill T.J., Kenyon A., et al. Macrophages directly contribute collagen to scar formation during zebrafish heart regeneration and mouse heart repair. Nat Commun. 2020;11:600. doi: 10.1038/s41467-019-14263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouton A.J., DeLeon-Pennell K.Y., Rivera Gonzalez O.J., et al. Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res Cardiol. 2018;113:26. doi: 10.1007/s00395-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits P., Ni J., Feng P., et al. The human extracellular matrix gene 1 (ECM1): genomic structure, cDNA cloning, expression pattern, and chromosomal localization. Genomics. 1997;45:487–495. doi: 10.1006/geno.1997.4918. [DOI] [PubMed] [Google Scholar]
- 16.Oyama N., Merregaert J. In: Textbook of Aging Skin. Farage M.A., Miller K.W., Maibach H.I., editors. Springer; Berlin, Heidelberg: 2017. The extracellular matrix protein 1 (ECM1) in molecular-based skin biology; pp. 91–110. [Google Scholar]
- 17.Li Z., Zhang Y., Liu Z., et al. ECM1 controls TH2 cell egress from lymph nodes through re-expression of S1P. Nat Immunol. 2011;12:178–185. doi: 10.1038/ni.1983. [DOI] [PubMed] [Google Scholar]
- 18.He L., Gu W., Wang M., et al. Extracellular matrix protein 1 promotes follicular helper T cell differentiation and antibody production. Proc Natl Acad Sci U S A. 2018;115:8621–8626. doi: 10.1073/pnas.1801196115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Li X., Luo Z., et al. ECM1 is an essential factor for the determination of M1 macrophage polarization in IBD in response to LPS stimulation. Proc Natl Acad Sci U S A. 2020;117:3083. doi: 10.1073/pnas.1912774117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farbehi N., Patrick R., Dorison A., et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife. 2019;8 doi: 10.7554/eLife.43882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexanian M., Przytycki P.F., Micheletti R., et al. A transcriptional switch governs fibroblast activation in heart disease. Nature. 2021;595:438–443. doi: 10.1038/s41586-021-03674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litviňuková M., Talavera-López C., Maatz H., et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Riverol Y., Csordas A., Bai J., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim R.Y., Pinkerton J.W., Essilfie A.T., et al. Role for NLRP3 inflammasome-mediated, IL-1β-dependent responses in severe, steroid-resistant asthma. Am J Resp Crit Care Med. 2017;196:283–297. doi: 10.1164/rccm.201609-1830OC. [DOI] [PubMed] [Google Scholar]
- 25.Ljubojević-Holzer S., Kraler S., Djalinac N., et al. Loss of autophagy protein ATG5 impairs cardiac capacity in mice and humans through diminishing mitochondrial abundance and disrupting Ca2+ cycling. Cardiovasc Res. 2022;118:1492–1505. doi: 10.1093/cvr/cvab112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim H.Y., Lim S.Y., Tan C.K., et al. Hyaluronan receptor LYVE-1-expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen. Immunity. 2018;49:326–341.e7. doi: 10.1016/j.immuni.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Seropian I.M., González G.E., Maller S.M., Berrocal D.H., Abbate A., Rabinovich G.A. Galectin-1 as an emerging mediator of cardiovascular inflammation: mechanisms and therapeutic opportunities. Mediators of Inflamm. 2018;2018 doi: 10.1155/2018/8696543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potere N., Del Buono M.G., Mauro A.G., Abbate A., Toldo S. Low density lipoprotein receptor-related protein-1 in cardiac inflammation and infarct healing. Front Cardiovasc Med. 2019;6:51. doi: 10.3389/fcvm.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toldo S., Austin D., Mauro A.G., et al. Low-density lipoprotein receptor–related protein-1 is a therapeutic target in acute myocardial infarction. J Am Coll Cardiol Basic Trans Science. 2017;2:561–574. doi: 10.1016/j.jacbts.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The UniProt C. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizoguchi T., Ikeda S., Watanabe S., Sugawara M., Itoh M. Mib1 contributes to persistent directional cell migration by regulating the Ctnnd1-Rac1 pathway. Proc Natl Acad Sci U S A. 2017;114:E9280–e9289. doi: 10.1073/pnas.1712560114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espejo R., Jeng Y., Paulucci-Holthauzen A., et al. PTP-PEST targets a novel tyrosine site in p120 catenin to control epithelial cell motility and Rho GTPase activity. J Cell Sci. 2014;127:497–508. doi: 10.1242/jcs.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epifano C., Megias D., Perez-Moreno M. p120-catenin differentially regulates cell migration by Rho-dependent intracellular and secreted signals. EMBO Rep. 2014;15:592–600. doi: 10.1002/embr.201337868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frodermann V., Nahrendorf M. Neutrophil–macrophage cross-talk in acute myocardial infarction. Eur Heart J. 2017;38:198–200. doi: 10.1093/eurheartj/ehw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G., Cooley M.A., Jarnicki A.G., et al. Fibulin-1c regulates transforming growth factor-β activation in pulmonary tissue fibrosis. JCI Insight. 2019;5 doi: 10.1172/jci.insight.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Utsunomiya N., Utsunomiya A., Chino T., Hasegawa M., Oyama N. Gene silencing of extracellular matrix protein 1 (ECM1) results in phenotypic alterations of dermal fibroblasts reminiscent of clinical features of lichen sclerosus. J Dermatol Sci. 2020;100:99–109. doi: 10.1016/j.jdermsci.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Hamada T., McLean W.H., Ramsay M., et al. Lipoid proteinosis maps to 1q21 and is caused by mutations in the extracellular matrix protein 1 gene (ECM1) Hum Mol Genet. 2002;11:833–840. doi: 10.1093/hmg/11.7.833. [DOI] [PubMed] [Google Scholar]
- 38.Fan W., Liu T., Chen W., et al. ECM1 prevents activation of transforming growth factor beta, hepatic stellate cells, and fibrogenesis in mice. Gastroenterology. 2019;157:1352–1367.e13. doi: 10.1053/j.gastro.2019.07.036. [DOI] [PubMed] [Google Scholar]
- 39.Wu J., Lubman D.M., Kugathasan S., et al. Serum protein biomarkers of fibrosis aid in risk stratification of future stricturing complications in pediatric Crohn’s disease. Am J Gastroenterol. 2019;114:777–785. doi: 10.14309/ajg.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venhuizen J.-H., Jacobs F.J.C., Span P.N., Zegers M.M. P120 and E-cadherin: double-edged swords in tumor metastasis. Semin Cancer Biol. 2020;60:107–120. doi: 10.1016/j.semcancer.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 41.Steinhaeuser S.S., Morera E., Budkova Z., et al. ECM1 secreted by HER2-overexpressing breast cancer cells promotes formation of a vascular niche accelerating cancer cell migration and invasion. Lab Invest. 2020;100:928–944. doi: 10.1038/s41374-020-0415-6. [DOI] [PubMed] [Google Scholar]
- 42.Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 43.Itoh M., Tsukita S., Yamazaki Y., Sugimoto H. Rho GTP exchange factor ARHGEF11 regulates the integrity of epithelial junctions by connecting ZO-1 and RhoA–Myosin II signaling. Proc Natl Acad Sci U S A. 2012;109:9905. doi: 10.1073/pnas.1115063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuki Y., Takaki M., Sakamoto S., et al. Human Rho guanine nucleotide exchange factor 11 (ARHGEF11) regulates dendritic morphogenesis. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lessey-Morillon E.C., Osborne L.D., Monaghan-Benson E., et al. The RhoA guanine nucleotide exchange factor, LARG, mediates ICAM-1-dependent mechanotransduction in endothelial cells to stimulate transendothelial migration. J Immunol. 2014;192:3390–3398. doi: 10.4049/jimmunol.1302525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansour M., Nievergall E., Gegenbauer K., et al. PTP-PEST controls EphA3 activation and ephrin-induced cytoskeletal remodelling. J Cell Sci. 2016;129:277–289. doi: 10.1242/jcs.174490. [DOI] [PubMed] [Google Scholar]
- 47.Lee K.-J., Kim Y., Kim M.S., et al. CD99-PTPN12 axis suppresses actin cytoskeleton-mediated dimerization of epidermal growth factor receptor. Cancers. 2020;12:2895. doi: 10.3390/cancers12102895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young K.A., Biggins L., Sharpe H.J. Protein tyrosine phosphatases in cell adhesion. Biochem J. 2021;478:1061–1083. doi: 10.1042/BCJ20200511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gómez-Contreras P., Ramiro-Díaz J.M., Sierra A., et al. Extracellular matrix 1 (ECM1) regulates the actin cytoskeletal architecture of aggressive breast cancer cells in part via S100A4 and Rho-family GTPases. Clin Exp Metastasis. 2017;34:37–49. doi: 10.1007/s10585-016-9827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin H., Wang J., Li H., et al. Extracellular matrix protein-1 secretory isoform promotes ovarian cancer through increasing alternative mRNA splicing and stemness. Nat Commun. 2021;12:4230. doi: 10.1038/s41467-021-24315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calvier L., Boucher P., Herz J., Hansmann G. LRP1 deficiency in vascular SMC leads to pulmonary arterial hypertension that is reversed by PPARγ activation. Circ Res. 2019;124:1778–1785. doi: 10.1161/CIRCRESAHA.119.315088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boucher P., Li W.P., Matz R.L., et al. LRP1 functions as an atheroprotective integrator of TGFβ and PDFG signals in the vascular wall: implications for Marfan syndrome. PLoS One. 2007;2:e448. doi: 10.1371/journal.pone.0000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xian X., Ding Y., Dieckmann M., et al. LRP1 integrates murine macrophage cholesterol homeostasis and inflammatory responses in atherosclerosis. Elife. 2017;6 doi: 10.7554/eLife.29292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anastasiadis P.Z., Moon S.Y., Thoreson M.A., et al. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- 55.Wildenberg G.A., Dohn M.R., Carnahan R.H., et al. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 56.Hou J.C., Shigematsu S., Crawford H.C., Anastasiadis P.Z., Pessin J.E. Dual regulation of Rho and Rac by p120 catenin controls adipocyte plasma membrane trafficking. J Biol Chem. 2006;281:23307–23312. doi: 10.1074/jbc.M603127200. [DOI] [PubMed] [Google Scholar]
- 57.Hong J.Y., Oh I.-H., McCrea P.D. Phosphorylation and isoform use in p120-catenin during development and tumorigenesis. Biochim Biophys Acta Mol Cell Res. 2016;1863:102–114. doi: 10.1016/j.bbamcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendonsa A.M., Bandyopadhyay C., Gumbiner B.M. p120-catenin phosphorylation status alters E-cadherin mediated cell adhesion and ability of tumor cells to metastasize. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.