Abstract
This study aimed to map MDRO carriage and potential transmission within and between three Flemish tertiary care hospitals and their neighbouring nursing homes. A cross-sectional MDRO prevalence survey was organized between October 2017 and February 2019. Perianal swabs were cultured for detection of MDRO. Determination of clonal relatedness based on wgMLST allelic profiles was performed. The prevalence of MDRO in Belgian hospitals and NHs is on the rise, compared to previous studies, and transmission in and between institutions is observed. These results re-emphasize the need for a healthcare network-wide infection prevention strategy in which WGS of MDRO strains can be supportive.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10096-023-04699-2.
Keywords: MDRO, Screening, Whole-genome sequencing, wgMLST, Molecular typing, Transmission
Introduction
Increased antimicrobial resistance (AMR) constitutes a major public health concern in healthcare and community settings [1]. At the healthcare facility level, multidrug-resistant organisms (MDRO) can spread by physical contact between patients and healthcare workers or through contaminated surfaces and medical devices. Furthermore, the transfer of patients between hospitals and other healthcare institutions, such as nursing homes (NHs), within and between healthcare networks plays an important role in the spread of MDROs [2]. As data on MDRO carriage in Belgian healthcare institutions are scarce and transmission between institutions has not been studied on a large scale, our study explored ESBL-E, CPE and VRE prevalence and transmission in three tertiary care hospitals and 13 NHs in Flanders, Belgium, using a combination of conventional culture techniques and WGS.
Materials and methods
Study design
A cross-sectional MDRO prevalence survey, the i-4-1-Health project, was organized in the cross-border region of the southern part of the Netherlands and Flanders, Belgium. In this paper, the results obtained between October 2017 and February 2019 from the Flemish hospitals (n = 3) and NHs (n = 13) are presented.
Sampling and microbiological analysis
The microbiological method of the project is described by Kluytmans – van den Bergh et al. [3].
Whole-genome sequencing
Sequencing was performed on an Illumina MiSeq sequencer, using the MiSeq reagent kit v2 generating 250-bp paired-end reads. wgMLST was performed using the BioNumerics software v7.6.3. (bioMérieux, France). Clonal relatedness between isolates was determined based on the similarity of wgMLST allelic profiles. By combining both sequencing and epidemiological data of well-described outbreaks, similarity thresholds for clonal relatedness were determined. Sequencing data should be combined with the metadata to make conclusions for an outbreak, especially in the ‘grey zone’(Table S1). Using a BLAST-based approach requiring at least 95% identity with the reference sequence and at least 95% reference length coverage, genes known to confer resistance were identified from the assembled genomes. Reference sequences and mutations from the Center for Genomic Epidemiology’s ResFinder (database version 2019-08-21) and PointFinder (database version 2019-07-02) databases were used, respectively.
Results
Sample and patient information
In total, 2536 perianal or stoma swabs were obtained from patients in Flemish tertiary care hospitals (n = 1878) and NH residents (n = 658). After the exclusion of six swabs (0.2%) due to poor sampling quality and 125 swabs (6.7%) due to duplicate sampling from previously sampled hospitalized patients, 2405 swabs remained for analysis. Of those, 1748 (73%) were obtained in hospitals and 657 (27%) in NHs (Table 1).
Table 1.
MDRO prevalence in perianal swabs from hospitalized patients (H) and nursing home residents (NH)
| % screened patients or residents | N samples | ESBL-E, n (%) | CPE, n (%) | VRE, n (%) | |
|---|---|---|---|---|---|
| H 1 | 34.6 | 485 | 77 (15.9) | 11 (2.3) | 3 (0.6) |
| H 2 | 79.9 | 602 | 69 (11.5) | 5 (0.8) | 5 (0.8) |
| H 3 | 86.0 | 661 | 53 (8.0) | 2 (0.3) | 8 (1.2) |
| Total H | 59.8 | 1748 | 199 (11.4) | 18 (1.0) | 16 (0.9) |
| NH 1 | 78.8 | 82 | 5 (6.1) | 0 (0.0) | 0 (0.0) |
| NH 2 | 70.4 | 76 | 10 (13.2) | 0 (0.0) | 0 (0.0) |
| NH 3 | 60.2 | 53 | 6 (11.3) | 0 (0.0) | 0 (0.0) |
| NH 4 | 23.6 | 17 | 2 (11.8) | 0 (0.0) | 1 (5.9) |
| NH 5 | 43.2 | 51 | 8 (15.7) | 0 (0.0) | 0 (0.0) |
| NH 6 | 56.2 | 73 | 8 (11.0) | 0 (0.0) | 0 (0.0) |
| NH 7 | 57.5 | 65 | 23 (35.4) | 0 (0.0) | 0 (0.0) |
| NH 8 | 65.5 | 78 | 10 (12.8) | 0 (0.0) | 0 (0.0) |
| NH 9 | 55.2 | 32 | 1 (3.1) | 0 (0.0) | 1 (3.1) |
| NH 10 | 17.2 | 15 | 6 (40.0) | 0 (0.0) | 0 (0.0) |
| NH 11 | 28.8 | 23 | 1 (4.3) | 0 (0.0) | 0 (0.0) |
| NH 12 | 61.4 | 62 | 6 (9.7) | 0 (0.0) | 0 (0.0) |
| NH 13 | 31.6 | 30 | 6 (20.0) | 0 (0.0) | 0 (0.0) |
| Total NH | 51.6 | 657 | 92 (14.0) | 0 (0.0) | 2 (0.3) |
| Total | 57.3 | 2405 | 291 (12.1) | 18 (0.7) | 18 (0.7) |
H hospital, NH nursing home, ESBL-E extended-spectrum β-lactamase-producing Enterobacterales, CPE carbapemase-producing Enterobacterales, VRE vancomycin-resistant Enterococci
Prevalence of ESBL-E, CPE and VRE in hospitals and NHs
The percentage of screened patients or residents is based on the number of patients of residents present on a ward. The genotypic confirmed ESBL-E, CPE and VRE isolates are shown in Table 1. Of the MRDOs, ESBL-E were cultured most often (11.4% in hospitals and 14.0% in NHs) compared to a lower prevalence of VRE (0.9% in hospitals and 0.3% in NHs) and CPE (1.0% in hospitals and undetected in NHs). Significant differences in prevalence of MDRO between the individual hospitals (p < 0.001) and between NHs (p < 0.001) were detected, while no significant variation in prevalence between the hospital and NH sector was observed.
Phenotype-genotype correlation for MDRO detection
In all VRE (15 E. faecium and 3 E. avium strains with an MIC for vancomycin of > 4 mg/l), a resistance gene could be detected (14 vanA genes and 4 vanB genes). However, for ESBL-E and CPE, phenotypic resistance could not always be confirmed by the presence of a resistance gene or mutation, resulting in lower genetic confirmation rates, especially for CPE (Table 2). The presence of other β-lactamase genes and porins are usually responsible for an ESBL-like phenotype.
Table 2.
Confirmation rates of phenotypical ESBL-E and CPE strains by the presence of resistance genes
| ESBL -E | CPE | |||||
|---|---|---|---|---|---|---|
| P (n) | G (n) | C (%) | P (n) | G (n) | C (%) | |
| Escherichia coli | 249 | 233 | 93.6 | 11 | 3 | 27.3 |
| Klebsiella pneumoniae | 69 | 66 | 95.7 | 15 | 9 | 60.0 |
| Enterobacter cloacae complex | 19 | 17 | 89.5 | 6 | 1 | 16.7 |
| Citrobacter spp. | 14 | 5 | 35.7 | 7 | 7 | 100 |
| Proteus spp. | 10 | 1 | 10.0 | 2 | 0 | 0.0 |
| Klebsiella spp. | 7 | 3 | 42.9 | 2 | 0 | 0.0 |
| Klebsiella aerogenes | 6 | 3 | 50.0 | 3 | 2 | 66.7 |
| Hafnia alvei | 2 | 0 | 0.0 | 0 | 0 | - |
| Morganella morganii | 0 | 0 | - | 2 | 1 | 50.0 |
| Serratia marcescens | 0 | 0 | - | 1 | 0 | 0.0 |
| Total | 376 | 328 | 87.2 | 49 | 23 | 46.9 |
ESBL-E extended-spectrum β-lactamase-producing Enterobacterales, CPE carbapenemase-producing Enterobacterales, P phenotypically confirmed, G genotypically confirmed, C genotypic confirmation rate
ESBL-E were detected in 291 of the 2405 swabs, corresponding with 328 ESBL-E strains (predominantly E. coli and K. pneumoniae) as 37 swabs contained multiple ESBL-E strains. Over half (51.5%) of the ESBL production capability could be attributed to the presence of the blaCTX-M-15 gene.
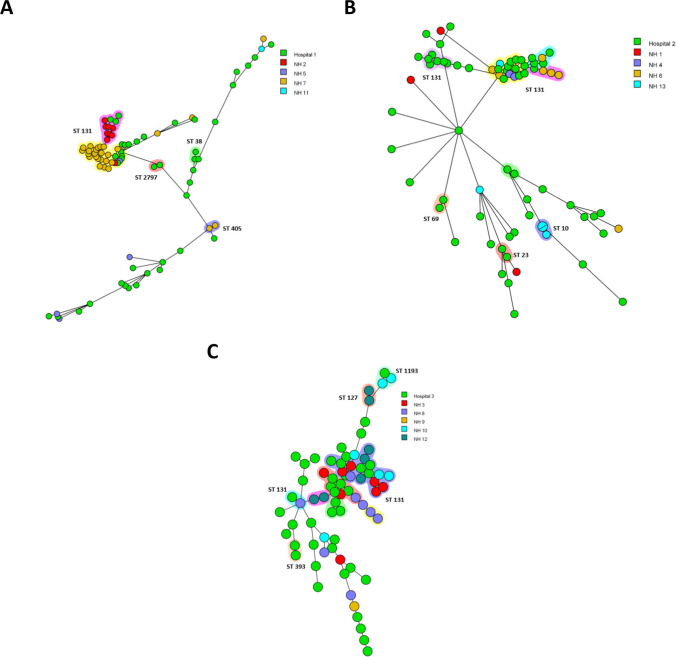
To study local and regional clonal relatedness of ESBL-E. coli, separate spanning trees were created for each hospital and its surrounding NHs (Fig. 1A–C). NH 3 is located in a range of < 10 km; NHs 4, 5 and 11 in a range of 11–25 km; NHs 2, 6, 8, 10, 12 and 13 in a range of 26–50 km and NHs 1, 7 and 9 in a range of 51–75 km from the tertiary care institutions.
Fig. 1.
Three minimum spanning trees based on wgMLST analysis of 233 ESBL-E. coli isolates per geographical region around the three hospitals (hospital 1 (A), hospital 2 (B) and hospital 3 (C) and their surrounding NHs). Isolates are represented by circles connected by branches proportional to the allelic distance, colours of the circle represent the origin location, the shading represent clonal clusters and ST number represent sequence types
The first region enclosed 84 ESBL-E. coli isolates from hospital 1 and four NHs (Fig. 1A). Five clonal clusters could be visualized, of which two are large ones with over 10 ST131 ESBL-E. coli isolates. The three smaller clusters comprise two isolates each and are within hospital 1 (two clusters of ST2797 and ST38) and NH 7 (a cluster of ST405).
In the second region, the isolates obtained from the hospitalized patients predominate in five of the seven clusters (Fig. 1B). Four clonal clusters (clusters of ST131, ST69, ST23 and ST10) contain isolates obtained from hospital 2 or NH13. The large cluster of ST131 comprises isolates of hospital 2, NH4, NH6 and NH13.
In the third region around hospital 3, a high level of clonality and clustering among the collected ESBL-E.coli could be observed (Fig. 1C). Five of nine clonal clusters consist of isolates obtained within hospital 3 (cluster of ST393 and ST131), NH8 (cluster of ST131) and NH12 (cluster of ST127 and ST131). NH3 (seven isolates in total in the two clusters) and hospital 3 (ten isolates in total in the two clusters) have many related isolates and are located in a range of < 10 km. In all regions, there were clusters of isolates in the hospital and the attached nursing homes indicative of transmission between healthcare institutes.
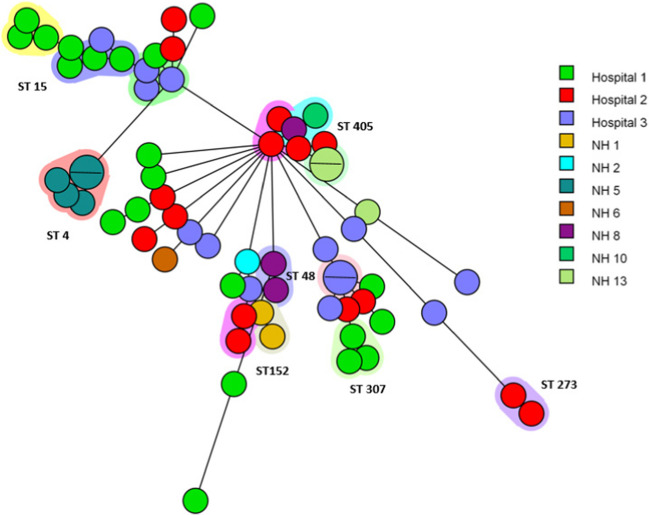
A minimum spanning tree of all ESBL-K. pneumoniae isolates (n = 66) is visualized in Fig. 2. One cluster contains two isolates (ST405) from NH 8 (one) and NH 10 (one) which differ by 3 alleles and have a similarity of 99.9%. NH 8 and 10 are from the same region (within 11–25 km).
Fig. 2.
A minimum spanning tree based on wgMLST analysis of 66 ESBL-producing Klebsiella pneumoniae isolates. Isolates are represented by circles connected by branches proportional to the allelic distance, colours of the circle represent the location, the shading represent clonal clusters, branches and numbers represent allelic differences between isolates and ST number represent sequence types
Carbapenemase-producing Enterobacterales
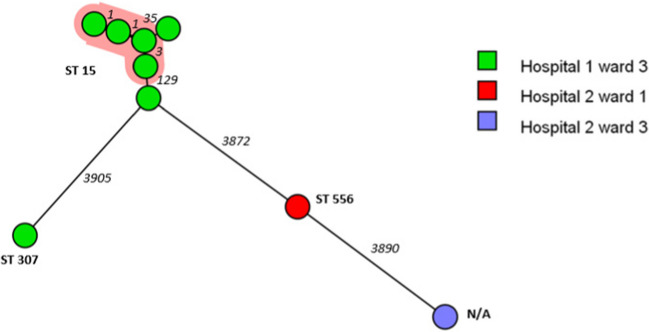
Twenty-three (1.3%) CPE isolates were detected; all of them originated from hospitalized patients. These CPE isolates consisted of six different species of which K. pneumoniae (n = 9) and Citrobacter spp. (n = 7) were predominant. The presence of the blaOXA-48 gene explained the carbapenem resistance in all but one isolate, a Citrobacter freundii strain carrying a blaVIM-4 resistance gene. All the blaOXA-48 positive isolates contained the ca. 64kb IncL/M plasmid. Analysis of clonal relatedness of these CPE revealed a cluster of 4 ST15 K. pneumoniae CPE in ward 3 of hospital 1 (Fig. 3). In total, 10 blaOXA-48 producing bacteria of three different species, K. pneumoniae, Citrobacter spp. and E. cloacae complex were detected in this specific ward of hospital 1.
Fig. 3.
A minimum spanning tree based on wgMLST analysis of 9 OXA-48-positive Klebsiella pneumoniae in two hospitals. Isolates are represented by circles connected by branches proportional to the allelic distance, colours of the circle represent the origin location, the shading represent clonal clusters, branches and numbers represent allelic differences between isolates and ST numbers represent sequence types
Vancomycin-resistant Enterococci
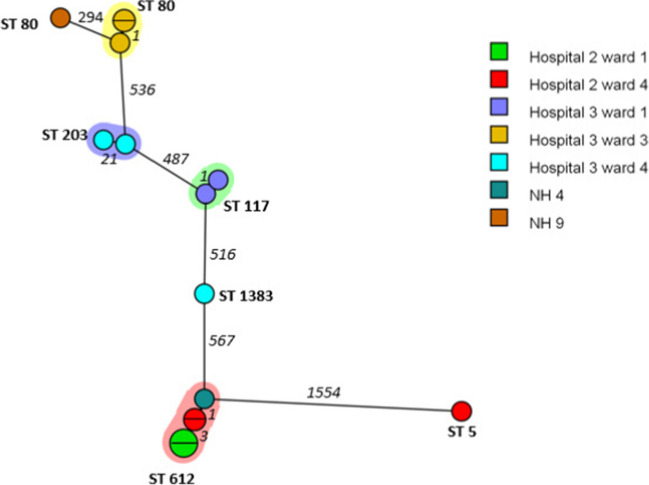
Eighteen (0.7%) VRE isolates were detected in the hospitals and NHs. In total, 14 isolates were vanA-positive and four isolates contained vanB. Hospital 3 had three individual clusters of two isolates (ST80, ST203, ST117) on three different wards (Fig. 4). The cluster of five isolates with ST612 consisted of strains of hospital 2 from two different wards and NH 4. Hospital 2 and NH 4 are located within a range of 11–25 km.
Fig. 4.
A minimum spanning tree based on wgMLST analysis of 15 vancomycin resistant Enterococcus faecium isolates. Isolates are represented by circles connected by branches proportional to the allelic distance, colours of the circles represent the origin location, the shading represent clonal clusters, branches and numbers represent allelic differences between isolates and ST number represent sequence types
Discussion
The prevalence of ESBL-E was found to be 11.4% in participating hospitals and 14.0% in NHs. Notably, the NH data indicates a rising trend in ESBL-E carriage, with rates increasing from 6.2% in 2011 to 11.3% in 2015 [4]. This upward trajectory may also be observed in Belgian hospitals, as evidenced by a noteworthy increase in the occurrence of ESBL-E. coli isolates in clinical samples, escalating from 3.9% in 2005 to 10.1% in 2018 [5].
Among the detected ESBL genes, blaCTX-M-15 was the most prevalent, followed by other blaCTX-M-type genes. Rodriguez-Villalobos et al. reported an increased presence of blaCTX-M-type genes in clinical isolates in Belgium in 2008 (54%) compared to 2006 (23%) with 51.5% of the ESBL-E. coli isolates and 31% of the ESBL-K. pneumoniae isolates harbouring the blaCTX-M-15 gene in 2008 [6]. In this study, the percentage of E. coli containing the blaCTX-M-15 gene was also 51.5%. However, a remarkable increase could be noted in the K. pneumoniae isolates as blaCTX-M-15 was now present in 90.9% of ESBL-K. pneumoniae strains. The predominant ST type among the blaCTX-M-15 gene harbouring E. coli strains is ST131. This pandemic clone is pathogenic with an ability to cause frequent infections in the community, especially in the elderly, and with the remarkable feature of a low level of core genome recombinations [7], as also visible in the minimum spanning tree.
Unlike ESBL-E, CPE’s were not detected in the studied NHs, which is in line with previous Belgian studies conducted in NHs in 2011 [8] and 2015 [4]. In contrast to NHs, CPE could be detected in the hospitals with a mean prevalence of 1% and predominance of K. pneumoniae harbouring blaOXA-48 in line with previous findings [9]. The blaOXA-48 gene containing CPE isolates in our study (E. coli, K. pneumoniae, E. cloacae complex, Citrobacter spp., K. aerogenes and M. morgannii) all harboured the ca. 64kb IncL/M plasmid. The blaOXA-48 gene on the 64kb IncL/M plasmid is known to be widely distributed in Europe, especially in the Mediterranean area [10]. Therefore, focusing on the clonal spread of one bacterial species may be insufficient to control a plasmid-mediated outbreak [10], as illustrated by the different CPE species (K. pneumoniae, Citrobacter spp. and E. cloacae complex) isolated in ward 3 of hospital 1.
In total, 18 VRE isolates were detected with MIC values ranging from 12 to 256 mg/l. The detection of only two VRE in two different NHs is in accordance with the low prevalence described before [4], [11].
To conclude, our study shows, compared to previous studies, that the prevalence of MDRO in Flemish hospitals and NHs has increased over recent years and transmission of ESBL-E, CPE and/or VRE within hospital wards and in and between institutions could be observed. These results re-emphasize the importance of developing an inter-institutional collaboration for infection prevention in the healthcare network.
Supplementary information
Acknowledgements
We are grateful to the infection control practitioners in the participating hospitals, the collaborators in the participating NHs and the collaborators in the participating laboratories for their contribution to the collection of the epidemiological, microbiological and whole-genome sequence data. We are grateful to Dr. A. Huss (Utrecht University, Utrecht, the Netherlands) for generating geodetic data.
i-4-1-Health Study Group
Lieke van Alphen (Maastricht University Medical Center+, Maastricht, the Netherlands), Nicole van den Braak (Avans University of Applied Sciences, Breda, the Netherlands), Caroline Broucke (Agency for Care and Health, Brussels, Belgium), Anton Buiting (Elisabeth-TweeSteden Ziekenhuis, Tilburg, the Netherlands), Liselotte Coorevits (Ghent University Hospital, Ghent, Belgium), Sara Dequeker (Agency for Care and Health, Brussels, Belgium and Sciensano, Brussels, Belgium), Jeroen Dewulf (Ghent University, Ghent, Belgium), Wouter Dhaeze (Agency for Care and Health, Brussels, Belgium), Bram Diederen (ZorgSaam Hospital, Terneuzen, the Netherlands), Helen Ewalts (Regional Public Health Service Hart voor Brabant, Tilburg, the Netherlands), Herman Goossens (University of Antwerp, Antwerpen, Belgium and Antwerp University Hospital, Antwerp, Belgium), Inge Gyssens (Hasselt University, Hasselt, Belgium), Casper den Heijer (Regional Public Health Service Zuid-Limburg, Heerlen, the Netherlands), Christian Hoebe (Maastricht University Medical Center+, Maastricht, the Netherlands and Regional Public Health Service Zuid-Limburg, Heerlen, the Netherlands), Casper Jamin (Maastricht University Medical Center+, Maastricht, the Netherlands), Patricia Jansingh (Regional Public Health Service Limburg Noord, Venlo, the Netherlands), Jan Kluytmans (Amphia Hospital, Breda, the Netherlands and University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands), Marjolein Kluytmans–van den Bergh (Amphia Hospital, Breda, the Netherlands and University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands), Stefanie van Koeveringe (Antwerp University Hospital, Antwerp, Belgium), Sien de Koster (University of Antwerp, Antwerp, Belgium), Christine Lammens (University of Antwerp, Antwerp, Belgium), Isabel Leroux-Roels (Ghent University Hospital, Ghent, Belgium), Hanna Masson (Agency for Care and Health, Brussel, Belgium), Ellen Nieuwkoop (Elisabeth-TweeSteden Ziekenhuis, Tilburg, the Netherlands), Anita van Oosten (Admiraal de Ruyter Hospital, Goes, the Netherlands), Natascha Perales Selva (Antwerp University Hospital, Antwerp, Belgium), Merel Postma (Ghent University, Ghent, Belgium), Stijn Raven (Regional Public Health Service West-Brabant, Breda, the Netherlands), Veroniek Saegeman (University Hospitals Leuven, Leuven, Belgium), Paul Savelkoul (Maastricht University Medical Center+, Maastricht, the Netherlands), Annette Schuermans (University Hospitals Leuven, Leuven, Belgium), Nathalie Sleeckx (Experimental Poultry Centre, Geel, Belgium), Arjan Stegeman (Utrecht University, Utrecht, the Netherlands), Tijs Tobias (Utrecht University, Utrecht, the Netherlands), Paulien Tolsma (Regional Public Health Service Brabant Zuid-Oost, Eindhoven, the Netherlands), Jacobien Veenemans (Admiraal De Ruyter Hospital, Goes, the Netherlands), Dewi van der Vegt (PAMM Laboratory for Pathology and Medical Microbiology, Veldhoven, the Netherlands), Martine Verelst (University Hospitals Leuven, Leuven, Belgium), Carlo Verhulst (Amphia Hospital, Breda, the Netherlands), Pascal De Waegemaeker (Ghent University Hospital, Ghent, Belgium), Veronica Weterings (Amphia Hospital, Breda, the Netherlands), Clementine Wijkmans (Regional Public Health Service Hart voor Brabant, Tilburg, the Netherlands), Patricia Willemse-Smits (Elkerliek Hospital, Helmond, the Netherlands), Ina Willemsen (Amphia Hospital, Breda, the Netherlands).
Author contributions
Conception and design of the study: A.S, M.F.Q.K.-V.D.B, J.A.J.W.K, H.G., W.D., I.L. Acquisition of data: M.S.V.K., N.P.S., W.D., I.L., A.S. Analysis and/or interpretation of data: M.S.V.K., D.D.C., M.F.Q.K.-V.D.B. Drafting of the manuscript: M.S.V.K., D.D.C., V.M. Revising the manuscript: M.S.V.K., A.S, V.M., S.D.K, N.P.S, H.J., D.D.C, K.D.B, K.M, M.F.Q.K.-V.D.B, J.A.J.W.K, H.G., W.D., I.L. Approval of the manuscript: M.S.V.K., A.S, V.M., S.D.K., N.P.S, H.J., D.D.C, K.D.B, K.M, M.F.Q.K.-V.D.B, J.A.J.W.K, H.G., W.D., I.L. and the i-4-1-Health Study Group.
Funding
The i-4-1-Health project was financed by the Interreg V Flanders The Netherlands program, the crossborder cooperation program with financial support from the European Regional Development Fund (ERDF) (0215). Additional financial support was received from the Dutch Ministry of Health, Welfare and Sport (325911), the Dutch Ministry of Economic Affairs (DGNR-RRE/14191181), the Province of Noord-Brabant (PROJ-00715/PROJ-01018/PROJ-00758), the Belgian Department of Agriculture and Fisheries (no reference), the Province of Antwerp (1564470690117/1564470610014) and the Province of EastFlanders (E01/subsidie/VLNL/i-4-1-Health). The authors are free to publish the results from the project without interference from the funding bodies. Selective and non-selective agar plates, ETEST® strips and VITEK® 2 AST cards were provided by bioMérieux (Marcy l’Etoile, France); FecalSwabs® and tryptic soy broths were provided by Copan Italy (Brescia, Italy). The authors are free to publish the results from the project without interference from bioMérieux or Copan Italy.
Data availability
The data that support the findings of this study are available from the corresponding author, Stefanie van Kleef- van Koeveringe, upon reasonable request.
Declarations
Ethics approval statement
The study protocol was reviewed and approved by the Ethics Committee of the University Hospital Antwerp (Antwerp, Belgium) (Belgian Registration Number B300210733784), the Ethics Committee of the University Hospital Ghent (Ghent, Belgium) (Belgian Registration Number B670201733428) and the Ethics Committee of the University Hospitals Leuven (Leuven, Belgium) (S59580 BD1 & S61353). The study was judged to be beyond the scope of the Law on Experiments on Humans, dated May 7th, 2004. Written or verbal informed consent for data collection and taking a perianal swab or faecal sample for microbiological culture was obtained from all participants or their legal representatives.
Competing interests
The authors declare no competing interests.
Footnotes
Wouter Dhaeze and Isabel Leroux-Roels share equal last authorship.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.European Centre for Disease Prevention and Control . Surveillance of antimicrobial resistance in Europe annual report of the European antimicrobial resistance surveillance network (EARS-Net) 2017 [Internet] ECDC: Surveillance Report; 2018. [Google Scholar]
- 2.Ciccolini M, Donker T, Köck R, Mielke M, Hendrix R, Jurke A et al (2013) Infection prevention in a connected world: the case for a regional approach. Int J Med Microbiol [DOI] [PubMed]
- 3.Kluytmans-van den Bergh M, Lammens C, Perales Selva N, Verhulst C, Buiting A, Leroux-Roels I, et al. Microbiological methods to detect intestinal carriage of highly-resistant microorganisms (HRMO) in humans and livestock in the i-4-1-Health Dutch-Belgian cross-border project. 2019. [Google Scholar]
- 4.Latour K, Huang T-D, Jans B, Berhin C, Bogaerts P, Noel A, et al. Prevalence of multidrug-resistant organisms in nursing homes in Belgium in 2015. PLoS One. 2019;14(3):e0214327. doi: 10.1371/journal.pone.0214327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latour K (2018) Surveillance of antimicrobial resistant bacteria in Belgian hospitals, Brussel
- 6.Rodriguez-Villalobos H, Bogaerts P, Berhin C, Bauraing C, Deplano A, Montesinos I et al (2011) Trends in production of extended-spectrum β-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J Antimicrob Chemother [DOI] [PubMed]
- 7.Nicolas-Chanoine MH, Bertrand X, Madec JY (2014) Escherichia coli st131, an intriguing clonal group. Clin Microbiol Rev [DOI] [PMC free article] [PubMed]
- 8.Saegeman VSM, Ntoumpanaki M, Niclaes L, Schuermans A (2014) Carbapenemase producing Enterobacteriaceae in Belgian nursing homes: myth or real danger? Acta Clin Belg [DOI] [PubMed]
- 9.Huang TD, Bogaerts P, Berhin C, Hoebeke M, Bauraing C, Glupczynski Y (2017) Increasing proportion of carbapenemase-producing Enterobacteriaceae and emergence of a MCR-1 producer through a multicentric study among hospital-based and private laboratories in Belgium from September to November 2015. Eurosurveillance [DOI] [PMC free article] [PubMed]
- 10.Hidalgo L, de Been M, Rogers MRC, Schürch AC, Scharringa J, van der Zee A et al (2019) Sequence-based epidemiology of an OXA-48 plasmid during a hospital outbreak. Antimicrob Agents Chemother [DOI] [PMC free article] [PubMed]
- 11.Jans B, Schoevaerdts D, Huang TD, Berhin C, Latour K, Bogaerts P et al (2013) Epidemiology of multidrug-resistant microorganisms among nursing home residents in Belgium. PLoS One [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, Stefanie van Kleef- van Koeveringe, upon reasonable request.