Abstract
Purpose
To assess the diagnostic potential of whole-exome sequencing (WES) and elucidate the clinical and genetic characteristics of primary ciliary dyskinesia (PCD) in the Korean population.
Materials and Methods
Forty-seven patients clinically suspected of having PCD were enrolled at a tertiary medical center. WES was performed in all patients, and seven patients received biopsy of cilia and transmission electron microscopy (TEM).
Results
Overall, PCD was diagnosed in 10 (21.3%) patients: eight by WES (8/47, 17%), four by TEM. Among patients diagnosed as PCD based on TEM results, two patients showed consistent results with WES and TEM of PCD (2/4, 50%). In addition, five patients, who were not included in the final PCD diagnosis group, had variants of unknown significance in PCD-related genes (5/47, 10.6%). The most frequent pathogenic (P)/likely pathogenic (LP) variants were detected in DNAH11 (n=4, 21.1%), DRC1 (n=4, 21.1%), and DNAH5 (n=4, 21.1%). Among the detected 17 P/LP variants in PCD-related genes in this study, 8 (47.1%) were identified as novel variants. Regarding the genotype–phenotype correlation in this study, the authors experienced severe PCD cases caused by the LP/P variants in MCIDAS, DRC1, and CCDC39.
Conclusion
Through this study, we were able to confirm the value of WES as one of the diagnostic tools for PCD, which increases with TEM, rather than single gene tests. These results will prove useful to hospitals with limited access to PCD diagnostic testing but with relatively efficient in-house or outsourced access to genetic testing at a pre-symptomatic or early disease stage.
Keywords: Primary ciliary dyskinesia, genetic testing, whole exome sequencing, copy number variants analysis, transmission electron microscopy
Graphical Abstract
INTRODUCTION
Primary ciliary dyskinesia (PCD) is a rare, heterogeneous group of diseases caused by structural or functional abnormalities in the motile cilia of multiple organs. The cause of motile cilia abnormality is a pathogenic (P) variant in PCD-related genes, encoding ciliary proteins that regulate ciliary structural, motility assembly, and transport components.1 Although the global PCD prevalence is reportedly 1:10000, this might be an underestimation owing to undiagnosed patients and the yet-to-be-prevalence from East Asia.2,3
The clinical manifestations of PCD are diverse, including recurrent oto-rhino-pulmonary infections, respiratory distress syndrome in the neonatal period, bronchiectasis from early childhood, situs inversus, and infertility.4,5 Accurate diagnosis of PCD in the early phase is pivotal for patients to prevent disease development and slow its progression.6 A clinical diagnostic prediction tool, such as PICADAR, was reported to identify PCD patient efficiently.7 However, its non-specific manifestations and the difficulties associated with accessing the highly sophisticated diagnostic tests and facilities often delay diagnosis and treatment.1,8,9 These effects are compounded by the requirement for invasive procedures, including nasal or lung biopsies, and patient participation, such as in nasal nitric oxide (nNo) testing, particularly in children.1,9,10
Recent PCD guidelines specify genetic testing as one of the essential methods for PCD diagnosis.1,8,9,10,11,12 In fact, P variants have been identified in more than 40 PCD-related genes with a genetic diagnostic yield estimated to be 30%–70% among suspected or confirmed PCD patients. However, this yield is expected to increase as new genes related to PCD are identified.8,9,12,13,14 Recently, access to genetic testing, either in-house or through outsourcing, has gradually become easier. Indeed, hospitals with limited access to PCD diagnostic tests, such as nNO and high-speed video microscopy (HSVM), can readily access genetic tests to diagnose PCD at a pre-symptomatic or early stage.
The primary purpose of this study is to investigate the diagnostic efficacy of whole-exome sequencing (WES) with or without transmission electron microscopy (TEM) analysis in patients with clinically suspected PCD. In addition, we aim to describe the clinical and genetic characteristics of PCD and elucidate the association between the genotypes and phenotypic manifestations in this patient group.
MATERIALS AND METHODS
Patients
From April 2020 to February 2021, 47 patients who were clinically suspected of having PCD from 46 unrelated families were prospectively enrolled at a single tertiary medical center in Korea. Clinically suspected PCD was defined when at least two of the following clinical criteria of the European Respiratory Society guidelines for diagnosing PCD were fulfilled: 1) persistent wet cough, 2) persistent rhinitis, 3) chronic middle ear disease with or without hearing loss, 4) unexplained neonatal respiratory distress in term infant, 5) situs anomalies, and 6) unexplained bronchiectasis in the chest computerized tomography (CT) scan.15 The patient’s medical history and clinical characteristics, including family history and physical examination findings, were evaluated. Additionally, radiological, pathological (TEM results), and functional testing results, including spirometry and echocardiogram, were collected for each patient.
The study was approved by the Institutional Review Board of the Yonsei University Health System, Severance Hospital (No. 2019-3160-007). All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual patients (or their legal guardian if the patient was younger than 19 years of age).
Genetic analysis
The WES was performed for all enrolled patients with quantified DNA. Genomic DNA from the leukocytes of each patient was extracted using the DNeasy kit (Qiagen, Hilden, Germany), according to manufacturer’s guidelines. Subsequently, libraries were generated using an Illumina TruSeq sample preparation kit. Whole exomes were captured using an Illumina TruSeq Exome enrichment kit and then sequenced on an Illumina HiSeq next-generation sequencer, as previously described.16 DNA was fragmented to 100 bp fragments, end-repaired, ligated to adapters, and hybridized with probes. The sequenced reads were mapped to the human reference genome (GRCh37), and sequencing alignment was performed using the Burrows–Wheeler Aligner software package. For all patients, copy number variations (CNVs) and large deletion/insertion was screened using the ExomeDepth software. The detected sequence variants and CNVs were confirmed using Sanger sequencing or quantitative PCR. If consent was obtained from the patient’s family, segregation tests were performed.
Data analysis and interpretation of the detected variants
DNA variants were prioritized using the following criteria: 1) sequence quality; 2) allele frequency [filtering out the variants of the dbSNP database, Exome Aggregation Consortium (ExAC) and Korean Reference Genome Database (KRGDB; http://coda.nih.go.kr/)]; and 3) presence in HGMD (http://www.hgmd.cf.ac.uk), OMIM (www.omim.org), dbSNP (https://www.ncbi.nlm.nih.gov/snp/), or ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) databases. Variants were identified by in silico prediction algorithms, Polymorphism Phenotyping version 2 (PolyPhen-2), and Sorting Tolerant from Intolerant (SIFT; https://sift.bii.a-star.edu.sg/). After comprehensively analyzing all of the results, we classified detected variants into a five-tier level as P, likely pathogenic (LP), variant of uncertain significance (VUS), likely benign, or benign, according to the American College of Medical Genetics and Genomics (ACMG) guidelines.17
PCD diagnosis
Patients were diagnosed with PCD when they presented with the abovementioned characteristic clinical features, and TEM revealed typical ciliary ultrastructural defects of the respiratory epithelium, or genetic testing results were positive.18 Pathologic criteria of TEM followed the guideline for class 1 or class 2 defects based on the international consensus guideline for reporting TEM results in the diagnosis of PCD.19 We determined a positive result in the genetic testing if bi-allelic P/LP variants in one known autosomal recessive PCD-related genes or a hemizygous P/LP in X-linked PCD-related genes were detected, according to the diagnostic guideline.14
RESULTS
Patient characteristics
Of the 47 patients enrolled in this study (median age: 29.6 years; range: 4–67 years), 23 (48.9%) were pediatric patients, below 19 years old. Among all patients, 22 (46.8%) were male and 25 (53.2%) were female. Eleven patients (23.4%) had a family history of recurrent respiratory disease. Twelve patients (25.5%) underwent nasal (n=8) or lung biopsy (n=4) before and after WES, and seven underwent TEM analysis with biopsy samples. Regarding clinical symptoms, 40 patients (85.1%) had a recurrent wet cough, 32 (68.1%) had recurrent sinusitis, 19 (40.4%) had recurrent otitis media, 21 (44.7%) had idiopathic bronchiectasis, and 2 (4.3%) had situs inversus.
Final diagnostic yield of patients with clinically suspected PCD
The final diagnostic rate was 25.5% (12/47); the diagnosed patients included 10 patients with PCD and one each with cystic fibrosis and 5q35.1q35.2 microdeletion.
Among the 10 PCD confirmed patients (10/47, 21.3%), eight patients were confirmed with positive results of WES (8/47, 17%). Four patients were diagnosed as PCD with positive TEM results. Among them, two patients showed consistent results of PCD with WES and TEM (patients 2 and 6), but two patients were diagnosed as having PCD only with the abnormal findings of TEM analysis exclusively, even though the genetic test showed negative results (patients 9 and 10). Notably, for the patient with confirmed PCD based on the TEM result (patient 9), we detected two VUSs in HYDIN, PCD-related genes, as compound heterozygous variants. However, HYDIN has a pseudogene named HYDIN2, and these two variants cannot completely exclude the possibility of being pseudogene sequences using next generation sequencing.
In addition, we found one LP and eight VUS of PCD-related genes in five patients who were clinically suspected PCD. However, we did not include these five patients in the final diagnostic yield group. Details of patients with the final diagnosis are provided in Supplementary Table 1 (only online).
Clinical characteristics of PCD patients
The mean age of the patients confirmed as having PCD was 25.2 years (range: 4–45 years), of which 5 (50.0%) were pediatric patients, aged under 19 years. Four patients (40.0%) had a family history of PCD-related clinical symptoms.
Among the 10 patients with PCD, 10 (100.0%) had recurrent wet cough, 7 (70.0%) had bronchiectasis, 8 (80.0%) had chronic sinusitis, 4 (40.0%) had chronic otitis media, 2 (20.0%) had situs inversus, and 1 (10.0%) had a history of neonatal respiratory distress. In addition, three patients underwent lung transplantation before the WES test was performed, and one of them received lung transplantation three times. Eight patients (80%) underwent pulmonary function test, and the FEV1/FVC ratio was less than the lower limit of normal (z-score <-1.645) in seven patients.
Detection of genetic variants and genotype–phenotype correlation
We detected 17 P/LP variants of seven PCD-related genes in 10 patients and three P/LP variants of CFTR in two patients. The most frequent P/LP variants were detected in DNAH11 (n=4, 22.2%), DRC1 (n=4, 22.2%), and DNAH5 (n=4, 22.2%). Interestingly, we detected a multi-exon deletion of DRC1 in two unrelated patients. This exon deletion was revealed as a known P and homozygous variant; the parents of both patients were identified as asymptomatic carriers.20
Among the 17 P/LP variants in PCD-related genes, 9 (55.6%) had been previously reported, while 8 (47.1%) were identified as novel variants not yet reported.20,21,22,23 Of them, two were in DHAH11, three in DNAH5, two in CCDC39, and one in DNAH14. Regarding the P variant type, four nonsense variants, six missense variants, four multi-exon deletions, and three frameshift variants were detected. In addition, 15 VUSs of five PCD genes in eight patients and three VUSs of CFTR in two patients were identified.
For genotype–phenotype relationships, the authors experienced severe PCD cases with LP/P variants in MCIDAS, DRC1, and CCDC39. One patient with MCIDAS variants, one with CCDC39, and one with DRC1 variants underwent lung transplantation. Another patient with DRC1 variants was awaiting lung transplantation at the time of genetic testing. Notably, both patients with DNAH5 variants presented situs inversus with respiratory manifestations; these symptoms were not observed in patients with variants in other genes.
Of the four patients with positive TEM results, two also tested positive via genetic testing for PCD. One patient with P variants in DRC1 presented microtubular disarrangement with a central apparatus (CA) defect. The patient with outer dynein arm (ODA) carried DNAH5 P/LP variants. In addition, one patient with CA defect had two HYDIN VUSs.
Patients confirmed to be PCD-positive based on familial genetic testing
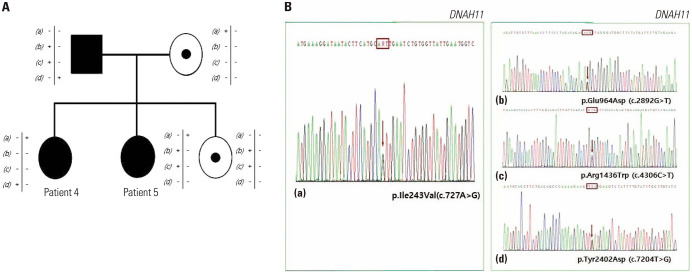
Co-segregation analysis can be a pivotal component in interpreting and determining the pathogenicity of newly identified variants, particularly for PCD-related genes, as there are few known P variants. In this study, one father of two siblings (patients 4 and 5) diagnosed as having PCD was also identified to have PCD from the co-segregation test. Initially, patients 4 and 5 with clinically suspected PCD underwent WES; one shared LP (c.727A>G) and three non-shared VUSs (c.2892G>T, c.2047C>t and c.7204T>G) in DHAN11. The co-segregation test results for the subjects’ father revealed that he had bronchiectasis with emphysematous changes of unknown etiology from his early twenties. According to our Sanger sequencing analysis, he had three VUSs (c.2892G>T, c.4306C>T, and c.7204T>G) that were not shared in either daughter, and his wife carried one LP variant (c.727A>G) in DNAH11. After genetic testing, the analysis of HSVM conducted at another hospital on the sibling’s father presented the hyperkinetic beating patterns of axonemes. This result was also consistent with the PCD findings. Taken together, patients 4 and 5 and their father were diagnosed with PCD, and the two VUSs in their family were re-classified as LP according to the ACMG guidelines (Fig. 1).
Fig. 1. (A) Family pedigrees of patients 4 and 5 in DNAH11. (B) Sanger sequencing results of the maternal allele (a) and paternal alleles (b), (c), (d).
DISCUSSION
Herein, we present the first PCD cohort data, including the diagnostic yield in patients clinically suspected of having PCD and their clinical and genetic characteristics, in Korea. This study obtained a diagnostic yield of 21.3 % (10/47) when diagnosing PCD using WES and/or TEM. If the pathogenicity of VUSs could be re-classified through the accumulation of data, the diagnostic yield can increase to 29.7% (14/47).
The diagnostic yield in this study was comparable to that reported previously. One study reported diagnostic yield as 21.7% (10/46) in patients with clinically suspected PCD (mean age: 16.6 years, range: 1–64 years). They applied Sanger sequencing to screen variant hotspots in DNA11 and DNAH5, followed by targeted exome sequencing with 32 known PCD genes.24 In another study presenting WES results in 13 adult patients with nontuberculous mycobacteria infections and suspected PCD in Korea, 30.8% (4/13) were found to carry biallelic loss-of-function variants in PCD-causing genes.25 Other studies reported higher diagnostic yields, which was postulated to be due to a high proportion of consanguinity.8,9,26,27,28 The low consanguineous marriage rate in Korea and the fact that this study did not include consanguineous families might account, in part, for the relatively low diagnostic yield.
According to the guidelines of diagnosing PCD, any single diagnostic test is insufficient. Therefore, a combination of diagnostic tests, including TEM analysis, genetic testing, HSVM, and nNO, is recommended to accurately diagnose PCD.1,10,15,29 Of note, the possibility of PCD should not be excluded in patients with clinically suspected PCD, even if the genetic test results are negative. However, similar to our hospital, several others might not be equipped with facilities to conduct diagnostic testing for PCD, such as the nNO or HSVM test, and access to genetic testing, either in-house or through outsourcing, is gradually becoming easier. Therefore, the WES results obtained in this study are expected to be helpful in diagnosing PCD patients at an early stage in hospitals with limited access to other diagnostic tests. As a database of PCD-related genes is generated, genetic testing is considered a more important first-tier diagnostic method for patients with clinical symptoms suggestive of PCD.
In terms of gene distribution, DNAH11, DNAH5, and DRC1 were the most prevalent in this study. Although this result tended to agree with those of other studies, certain differences were noted depending on ethnicity (Supplementary Table 2, only online). DNAH5 has been described as the most prevalent gene in Caucasian and East Asian populations; however, it is rare in Arab populations.13,24,27,30,31,32 Meanwhile, RSPH9, which was not found in this study, is reportedly a major gene in Arab populations.8,14,33 The relatively high frequency of DRC1 in PCD patients is a noticeable finding of the present study. Contrary to the low prevalence (<1%) of DRC1 in patients from Western countries, DRC1 exon 1–4 deletions are highly recurrent in East Asian patients, particularly in Japanese and Korean populations.34,35,36
PCD is a heterogeneous disease that can be caused by more than 40 genes involved in the structure and function of each part of primary cilia (Table 1). Therefore, the spectrum and severity of PCD manifestations can be determined based on the causative gene; sufficient genotype-phenotype information can lead to the accurate prediction of disease prognosis.
Table 1. Genes Known to Induce the Development of PCD.
| PCD genes associated with laterality defect | ||
| Defect of ODA | ||
| Heavy chain: DNAH5*, DNAH9†, DNAH11* | ||
| Intermediate chain: DNAI1, DNAI2, NME8 | ||
| Light chain: DNAL1 | ||
| Docking defect of ODA | ||
| ARMC4, CCDC103, CCDC151, CCDC114, MNS1†, TTC25 | ||
| Defect of 96nm axonemal ruler | ||
| CCDC39*‡, CCDC40‡ | ||
| Defects of cytoplasmic preassembly of DA | ||
| DNAAF1, DNAAF2, DNAAF3, DNAAF4, DNAAF5, DNAFF6, LRRC6, ZMYND10, SPAG1, C21ORF59, CFAP300 | ||
| PCD genes without laterality defect | ||
| Defect of radial spoke | ||
| RSPH1†, RSPH3, RSPH4A, RSPH9, DNAJB13 | ||
| Defect of central pair associated protein | ||
| HYDIN, STK36 | ||
| Defect of nexin dynein regulatory complex (N-DRC) | ||
| CCDC65, CCDC164(DRC1)*, GAS8 | ||
| Defect of transcription factor | ||
| MCIDAS *‡ | ||
| Defect of amplification of centrioles | ||
| CCNO ‡ | ||
PCD, primary ciliary dyskinesia; DA, dynein arm; ODA, outer dynein arm.
*Genes associated with confirmed PCD diagnosis in the study; †Genes known to be associated with severe clinical symptoms; ‡Genes known to be associated with relatively mild clinical symptoms.
In terms of WES results and TEM correlations, the results obtained in this study were consistent with previous research findings. MTD or CA abnormalities have been reported to be related to DRC1, and ODA defect alone is a classical finding of DNAH5.36,37,38,39 TEM findings of patient 2 with P variants in DRC1 and patient 6 with P variants in DNAH5 were consistent with those results.
Regarding the genotype–clinical phenotype correlation in this study, the authors experienced severe PCD cases with MCIDAS, DRC1, and CCDC39 variants presenting with relatively severe and rapid lung function impairment, and those who underwent or awaited lung transplantation. The patient with MCIDAS and CCDC39 variants showed severe pulmonary manifestations, which was consistent with earlier reports.1,40 However, regarding DRC1, previous studies reported contradictory genotype-phenotype correlations in DRC1 variants.41,42 For example, Japanese PCD patients with DRC1 showed milder clinical phenotypes compared to the patients in this study, with subtle ciliary alterations. Therefore, in the case of PCD caused by DRC1 variants, further research is needed to confirm a more exact phenotypic pattern. One possibility is that additional factors, such as digenic or environmental cofactors, may have influenced the prognosis in our patient with DRC1 mutation. Although the entire genotype does not exhibit identical phenotypic correlations, this genotype-phenotype tendency might inform the patient’s prognosis, particularly if they were diagnosed in the early phase.
Proper genetic counseling for the affected family is essential in the case of PCD. Additionally, family-based segregation investigation based on the appropriate genetic counseling is important not only to elucidate the pathogenicity of VUSs, but also for the provision of valuable information regarding the diagnosis and treatment of additional patients in the proband family. In our study, two sisters were diagnosed with PCD, and their father was also diagnosed with PCD, according to the segregation study results (Fig. 1). For the family members who have been definitively diagnosed, better treatment options should be possible to improve the overall prognosis. Indeed, the diagnosis of PCD at an early disease stage via genetic analysis should facilitate conservative management, including physiotherapy and active infection control, that can effectively slow the progression of the disease.
The current study has certain limitations. First, it was designed as a single-center study, and the number of patients included was small. Second, the long-term prognosis of patients was not revealed due to the short recruitment and follow-up periods. PCD tends to get worse with age, and it was difficult to accurately compare disease severity according to the causative genes due to the broad age spectrum of the patients. In particular, for the genotype–phenotype correlation analysis, further longitudinal analysis with larger number of patients is required to confirm our findings.
In this study, we confirmed the value of WES as a diagnostic tool for PCD, particularly for hospitals in which accessibility to diagnostic tests for PCD are limited, and genetic testing is available relatively easily. Although the higher diagnostic rate was secured through complementary tests, namely, TEM rather than single gene testing, genetic tests are likely to have more diagnostic potential with additional genetic information in the future. Furthermore, elucidating specific PCD genotypes can help manage patients and predict the progression of patients’ manifestations. In addition, by providing appropriate genetic counseling for diseases, it will be possible to identify family members of patients, even in the asymptomatic period, to potentially prevent the development of genetic diseases.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Kyung Won Kim.
- Data curation: Jiyoung Oh.
- Formal analysis: Jiyoung Oh, Jin-Sung Lee, and Kyung Won Kim.
- Funding acquisition: Jin-Sung Lee.
- Investigation: all authors.
- Methodology: Jiyoung Oh, Sun Och Yoon, and Kyung Won Kim.
- Project administration: Kyung Won Kim and Jiyoung Oh.
- Resources: all authors.
- Supervision: Kyung Won Kim.
- Validation: Kyung Won Kim.
- Visualization: Jiyoung Oh.
- Writing —original draft: Jiyoung Oh.
- Writing—review & editing: Jiyoung Oh and Kyung Won Kim.
- Approval of final manuscript: all authors.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
SUPPLEMENTARY MATERIALS
Clinical and Genetic Profiles of Patients Diagnosed with PCD and Suspected of PCD
Genetic Prevalence of PCD According to Ethnicity or Country
References
- 1.Lucas JS, Davis SD, Omran H, Shoemark A. Primary ciliary dyskinesia in the genomics age. Lancet Respir Med. 2020;8:202–216. doi: 10.1016/S2213-2600(19)30374-1. [DOI] [PubMed] [Google Scholar]
- 2.Kuehni CE, Frischer T, Strippoli MP, Maurer E, Bush A, Nielsen KG, et al. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010;36:1248–1258. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 3.Mirra V, Werner C, Santamaria F. Primary ciliary dyskinesia: an update on clinical aspects, genetics, diagnosis, and future treatment strategies. Front Pediatr. 2017;5:135. doi: 10.3389/fped.2017.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucas JS, Burgess A, Mitchison HM, Moya E, Williamson M, Hogg C. Diagnosis and management of primary ciliary dyskinesia. Arch Dis Child. 2014;99:850–856. doi: 10.1136/archdischild-2013-304831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubbo B, Lucas JS. Clinical care for primary ciliary dyskinesia: current challenges and future directions. Eur Respir Rev. 2017;26:170023. doi: 10.1183/16000617.0023-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioannou P, Kouis P, Kakkoura MG, Kaliva M, Toliopoulou A, Andreou K, et al. Health related quality of life in adult primary ciliary dyskinesia patients in Cyprus: development and validation of the Greek version of the QOL-PCD questionnaire. Health Qual Life Outcomes. 2020;18:105. doi: 10.1186/s12955-020-01360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behan L, Dimitrov BD, Kuehni CE, Hogg C, Carroll M, Evans HJ, et al. PICADAR: a diagnostic predictive tool for primary ciliary dyskinesia. Eur Respir J. 2016;47:1103–1112. doi: 10.1183/13993003.01551-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shamseldin HE, Al Mogarri I, Alqwaiee MM, Alharbi AS, Baqais K, AlSaadi M, et al. An exome-first approach to aid in the diagnosis of primary ciliary dyskinesia. Hum Genet. 2020;139:1273–1283. doi: 10.1007/s00439-020-02170-2. [DOI] [PubMed] [Google Scholar]
- 9.Baz-Redón N, Rovira-Amigo S, Paramonov I, Castillo-Corullón S, Cols Roig M, Antolín M, et al. Implementation of a gene panel for genetic diagnosis of primary ciliary dyskinesia. Arch Bronconeumol (Engl Ed) 2021;57:186–194. doi: 10.1016/j.arbres.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro AJ, Zariwala MA, Ferkol T, Davis SD, Sagel SD, Dell SD, et al. Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr Pulmonol. 2016;51:115–132. doi: 10.1002/ppul.23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro AJ, Davis SD, Polineni D, Manion M, Rosenfeld M, Dell SD, et al. Diagnosis of primary ciliary dyskinesia. An official American Thoracic Society clinical practice guideline. Am J Respir Crit Care Med. 2018;197:e24–e39. doi: 10.1164/rccm.201805-0819ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yiallouros PK, Kouis P, Kyriacou K, Evriviadou A, Anagnostopoulou P, Matthaiou A, et al. Implementation of multigene panel NGS diagnosis in the national primary ciliary dyskinesia cohort of Cyprus: an island with a high disease prevalence. Hum Mutat. 2021;42:e62–e77. doi: 10.1002/humu.24196. [DOI] [PubMed] [Google Scholar]
- 13.Guan Y, Yang H, Yao X, Xu H, Liu H, Tang X, et al. Clinical and genetic spectrum of children with primary ciliary dyskinesia in China. Chest. 2021;159:1768–1781. doi: 10.1016/j.chest.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alzaid M, Al-Mobaireek K, Almannai M, Mukhtar G, Eltahir S, Zafar A, et al. Clinical and molecular characteristics of primary ciliary dyskinesia: a tertiary care centre experience. Int J Pediatr Adolesc Med. 2021;8:258–263. doi: 10.1016/j.ijpam.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas JS, Barbato A, Collins SA, Goutaki M, Behan L, Caudri D, et al. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J. 2017;49:1601090. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omoyinmi E, Melo Gomes S, Standing A, Rowczenio DM, Eleftheriou D, Klein N, et al. Brief report: whole-exome sequencing revealing somatic NLRP3 mosaicism in a patient with chronic infantile neurologic, cutaneous, articular syndrome. Arthritis Rheumatol. 2014;66:197–202. doi: 10.1002/art.38217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall CR, Scherer SW, Zariwala MA, Lau L, Paton TA, Stockley T, et al. Whole-exome sequencing and targeted copy number analysis in primary ciliary dyskinesia. G3 (Bethesda) 2015;5:1775–1781. doi: 10.1534/g3.115.019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoemark A, Boon M, Brochhausen C, Bukowy-Bieryllo Z, De Santi MM, Goggin P, et al. International consensus guideline for reporting transmission electron microscopy results in the diagnosis of primary ciliary dyskinesia (BEAT PCD TEM criteria) Eur Respir J. 2020;55:1900725. doi: 10.1183/13993003.00725-2019. [DOI] [PubMed] [Google Scholar]
- 20.Kim MJ, Kim S, Chae SW, Lee S, Yoon JG, Kim B, et al. Prevalence and founder effect of DRC1 exon 1-4 deletion in Korean patients with primary ciliary dyskinesia. J Hum Genet. 2023;68:369–374. doi: 10.1038/s10038-023-01122-8. [DOI] [PubMed] [Google Scholar]
- 21.Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, et al. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun. 2014;5:4418. doi: 10.1038/ncomms5418. [DOI] [PubMed] [Google Scholar]
- 22.Xia H, Huang X, Deng S, Xu H, Yang Y, Liu X, et al. DNAH11 compound heterozygous variants cause heterotaxy and congenital heart disease. PLoS One. 2021;16:e0252786. doi: 10.1371/journal.pone.0252786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornef N, Olbrich H, Horvath J, Zariwala MA, Fliegauf M, Loges NT, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi K, Kitano M, Kiyotoshi H, Ikegami K, Ogawa S, Ikejiri M, et al. A targeted next-generation sequencing panel reveals novel mutations in Japanese patients with primary ciliary dyskinesia. Auris Nasus Larynx. 2018;45:585–591. doi: 10.1016/j.anl.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Cho EH, Ki CS, Yun SA, Kim SY, Jhun BW, Koh WJ, et al. Genetic analysis of Korean adult patients with nontuberculous mycobacteria suspected of primary ciliary dyskinesia using whole exome sequencing. Yonsei Med J. 2021;62:224–230. doi: 10.3349/ymj.2021.62.3.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emiralioğlu N, Taşkıran EZ, Koşukcu C, Bilgiç E, Atilla P, Kaya B, et al. Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: first results from Turkey. Pediatr Pulmonol. 2020;55:383–393. doi: 10.1002/ppul.24583. [DOI] [PubMed] [Google Scholar]
- 27.Fassad MR, Patel MP, Shoemark A, Cullup T, Hayward J, Dixon M, et al. Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J Med Genet. 2020;57:322–330. doi: 10.1136/jmedgenet-2019-106501. [DOI] [PubMed] [Google Scholar]
- 28.Boaretto F, Snijders D, Salvoro C, Spalletta A, Mostacciuolo ML, Collura M, et al. Diagnosis of primary ciliary dyskinesia by a targeted next-generation sequencing panel: molecular and clinical findings in Italian patients. J Mol Diagn. 2016;18:912–922. doi: 10.1016/j.jmoldx.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antony D, Becker-Heck A, Zariwala MA, Schmidts M, Onoufriadis A, Forouhan M, et al. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum Mutat. 2013;34:462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fassad MR, Shoman WI, Morsy H, Patel MP, Radwan N, Jenkins L, et al. Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia. Clin Genet. 2020;97:509–515. doi: 10.1111/cge.13661. [DOI] [PubMed] [Google Scholar]
- 32.Paff T, Kooi IE, Moutaouakil Y, Riesebos E, Sistermans EA, Daniels HJMA, et al. Diagnostic yield of a targeted gene panel in primary ciliary dyskinesia patients. Hum Mutat. 2018;39:653–665. doi: 10.1002/humu.23403. [DOI] [PubMed] [Google Scholar]
- 33.Demir Eksi D, Yilmaz E, Basaran AE, Erduran G, Nur B, Mihci E, et al. Novel gene variants associated with primary ciliary dyskinesia. Indian J Pediatr. 2022;89:682–691. doi: 10.1007/s12098-022-04098-z. [DOI] [PubMed] [Google Scholar]
- 34.Keicho N, Hijikata M. Genetic predisposition to diffuse panbronchiolitis. Respirology. 2011;16:581–588. doi: 10.1111/j.1440-1843.2011.01946.x. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto K, Hijikata M, Zariwala MA, Nykamp K, Inaba A, Guo TC, et al. Recurring large deletion in DRC1 (CCDC164) identified as causing primary ciliary dyskinesia in two Asian patients. Mol Genet Genomic Med. 2019;7:e838. doi: 10.1002/mgg3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi K, Xu Y, Kitano M, Chiyonobu K, Abo M, Ikegami K, et al. Copy number variation in DRC1 is the major cause of primary ciliary dyskinesia in the Japanese population. Mol Genet Genomic Med. 2020;8:e1137. doi: 10.1002/mgg3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei C, Yang D, Wang R, Ding S, Wang L, Guo T, et al. DRC1 deficiency caused primary ciliary dyskinesia and MMAF in a Chinese patient. J Hum Genet. 2022;67:197–201. doi: 10.1038/s10038-021-00985-z. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro AJ, Leigh MW. Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: genetic defects with normal and non-diagnostic ciliary ultrastructure. Ultrastruct Pathol. 2017;41:373–385. doi: 10.1080/01913123.2017.1362088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kano G, Tsujii H, Takeuchi K, Nakatani K, Ikejiri M, Ogawa S, et al. Whole-exome sequencing identification of novel DNAH5 mutations in a young patient with primary ciliary dyskinesia. Mol Med Rep. 2016;14:5077–5083. doi: 10.3892/mmr.2016.5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amirav I, Wallmeier J, Loges NT, Menchen T, Pennekamp P, Mussaffi H, et al. Systematic analysis of CCNO variants in a defined population: implications for clinical phenotype and differential diagnosis. Hum Mutat. 2016;37:396–405. doi: 10.1002/humu.22957. [DOI] [PubMed] [Google Scholar]
- 41.Benjamin AT, Ganesh R, Chinnappa J, Kinimi I, Lucas J. Primary ciliary dyskinesia due to DRC1/CCDC164 gene mutation. Lung India. 2020;37:179–180. doi: 10.4103/lungindia.lungindia_361_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keicho N, Hijikata M, Morimoto K, Homma S, Taguchi Y, Azuma A, et al. Primary ciliary dyskinesia caused by a large homozygous deletion including exons 1-4 of DRC1 in Japanese patients with recurrent sinopulmonary infection. Mol Genet Genomic Med. 2020;8:e1033. doi: 10.1002/mgg3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and Genetic Profiles of Patients Diagnosed with PCD and Suspected of PCD
Genetic Prevalence of PCD According to Ethnicity or Country
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.