Highlights
-
•
Positive cases and deaths are still happening after 3 years with new COVID-19 strains.
-
•
Vaccinations and booster shots are currently being administered all over the world.
-
•
Pharmaceutical companies are working to develop effective vaccines against COVID-19.
-
•
One health rule, immunity-boosting, hygiene, masks, and immunization may control COVID-19..
Keywords: SARS-CoV-2, Variants & subvariants, Vaccines, Booster doses, Immunity
Abstract
The severe acute respiratory syndrome coronavirus disease 2 instigated by coronavirus disease of 2019 (COVID-19) has delivered an unfathomable obstruction that has touched all sectors worldwide. Despite new vaccine technologies and mass administration of booster doses, the virus persists, and unknown the ending of the pandemic for new variants and sub-variants. Moreover, whether leaning on home medications or using plant extracts is sufficient often to combat the virus has generated tremendous interest in the scientific fraternity. Different databases including PubMed, Scopus, Web of Science, and Google Scholar used to find published articles linked with related topics. Currently, COVID-19 third and fourth shots of vaccines are progressively administered worldwide, where some countries trail others by a significant margin. Many proteins related to viral activity have changed, possibly boosting the virus infectivity and making antibodies ineffective. This study will reminisce the viral genome, associated pathways for viral protein functions, variants, and their mutations. The current, comprehensive review will also provide information on vaccine technologies developed by several biotech companies and the efficacy of their doses, costs including boosters on a mass level. As no vaccine is working to protect fully against all the variants, the new proactive vaccine research needs to be conducted based on all variants, their sub-lineage, and mutations.
1. Introduction
Coronavirus disease of 2019 (COVID-19) has been spreading since late 2019 with a massive challenge to the world's healthcare system for generating active vaccines [1]. This outbreak was proclaimed a pandemic on March 11, 2020, by the World Health Organization (WHO), for its severity [2], [3]. WHO confirmed around 768 million positive reports that had been recorded up to March 22, 2023, including 6.9 million fatalities [4], [5].
Positive SARS-CoV-2 patients displayed a notable feature of medical manifestations that appeared 2–14 days after infection [4], [5], [6], [7], [8]. The common COVID-19 symptoms are fever, cough, chill, difficulties in breathing, fatigue, body aches, and headache are most common [5], [6], [9]. Several techniques are available to detect SARS-CoV-2, where the gold standard and mostly used technique is reverse transcription polymerase chain reaction (RT-PCR) to identify genetic markers [10], [11]. Serological methods, molecular techniques, whole genome sequencing (WGS), and enzyme-linked immunosorbent assay (ELISA) are commonly used methods for this virus [12], [13], [14]. At least 10 significant variants SARS-CoV-2 have emerged because of genetic mutation, a frequent occurrence among RNA viruses [15]. Alpha, Beta, Gamma, Delta, Epsilon, Zeta, Eta, Theta, Iota, Kappa, Lambda, Omicron, and neo-COV-2 have become a grave threat to public health [8]. The WHO has classified these variants into variants of interest (VOIs), variants under monitoring (VUMs), and variants of concern (VOCs). The Delta and Omicron variants and their sub-lineages pose an even more than other variants [8]. WHO declared variant B.1.1.529 as a VOC, which was named Omicron on November 26 2021 [6], [7]. These variants have caused widespread concern and alarm, resulting in the imposition of stricter lockdown measures and travel restrictions globally [10], [16]. The discovery of these increasingly severe and extremely communicable varieties is a significant setback in the ongoing battle against the COVID-19 catastrophe [17], [18].
Although immunization against COVID-19 reduces its positivity and mortality, SARS-CoV-2 with its novel and more potent forms, continues to appear, resulting in a sluggish response in vaccine campaigns and immunization [19]. Several COVID-19 vaccines from Moderna, Sinopharm, Oxford-AstraZeneca, Pfizer-BioNTech, and Sputnik Janssen have been approved for use against coronavirus infection [20]. This review presents COVID-19 genomic structural features, mutations, vaccination strategy, and mode of action, including their pros and cons. The effectiveness of booster shots of vaccines against different variants, coverage of global vaccines, and difficulties and costs behind COVID-19 vaccines are also included in this article. Finally, a therapeutically active new vaccine against SARS-CoV-2 development necessity is added.
2. SARS-CoV-2 genomic construction
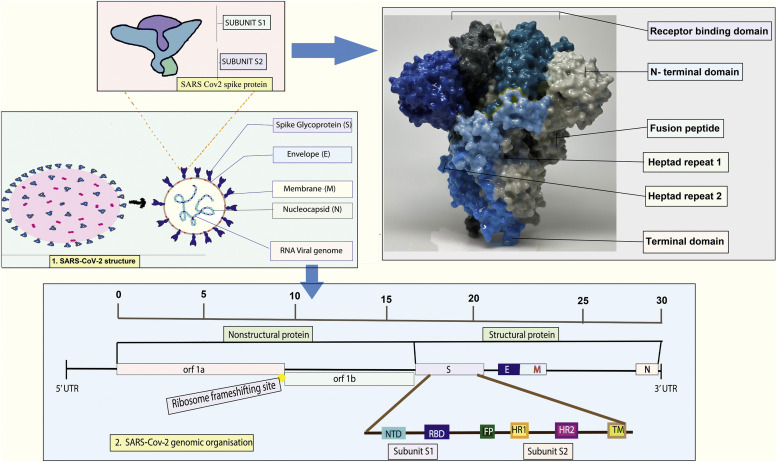
SARS-CoV-2 is a representative of Nidovirales order, Coronaviridae family, the Betacoronavirus genus, and the severe acute respiratory syndrome-related coronavirus species. The causal factor of COVID-19 is a positive-strand RNA virus encapsulated in a lipid bilayer with a genome size of approximately 30 kilobases [21]. The virus acquires its enveloped lipid bilayer by budding from the host cells through an orchestrated mechanism involving the spike protein interaction with the ACE2 receptor on the host cell membrane. The genomic spectrum of SARS-CoV-2 has several functional and regulatory segments and proteins covering structural and nonstructural proteins (nsps) (Fig. 1) [4].
Fig. 1.
A detailed depiction of the genomic structure of SARS-CoV-2, with a focus on the structural and nonstructural protein segments and their smaller subunits. The diagram provides a comprehensive understanding of this virus's molecular organization and complexity, particularly with respect to its protein-coding regions [27], [28].
Fig. 1 represents SARS-CoV-2 structure which contains mainly spike glycoprotein with 2 subunits (receptor binding domain, N- terminal, fusion peptide, heptad repeat 1 and 2), envelope, membrane, nucleocapsid, and viral genome. Both structural and nonstructural proteins are found in the genomic organization of this virus. The formation and assembly of the virion are facilitated by the structural proteins, which consist of the spike (S) glycoprotein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein. Nucleocapsid protein (N) is physically connected to the nucleic acid, which is located in the vacuum of the Golgi body and endoplasmic reticulum (ER) [22]. The most physically complex protein, known as the membrane protein (M protein), affects how the virus envelope is shaped [23]. All other structural proteins can bind to this protein. The envelope, or E protein, is the tiniest component of SARS-CoV-2 but is crucial to viral development and maturity [22]. SARS CoV-2 is deficient in the hemagglutinin esterase gene, present in the lineage of human coronavirus (hCoV) HKU1, betacoronavirus.
SARS-CoV-2 viral entry into human cells is facilitated with the help of S glycoprotein, a trimeric transmembrane protein. This is accomplished by interaction with the ACE2 receptor on the host cell's surface, which mediates viral entrance. The measured weight of the S protein is 600-kDa, which has 66 N-linked glycans that explain its heavy glycosylation [17]. The elements are known as spike proteins translated from the viral RNA template by the infected host, and the spike is integrated into the lipid bilayer around the virus core [24]. Two main portions of the spike proteins of SARS-CoV-2 are transported to the plasma membrane, and the cellular furin proteases cut the spike at the furin cleavage site (FCS) between the S1 and S2 regions. The severed fragments joined forces to produce a semi-mature spike on the cell surface [25].
The nonstructural proteins (nsps) are tangled in viral replication and transcription, including nsp1 to nsp16. Nsp1 is a multifunctional protein that interferes with host gene expression and immune response. Nsp2 is an enzyme-like protease that splits the SARS-CoV-2 polyprotein into functional nsps [26]. Nsp3 protein acts as a multi-domain involved in various aspects of viral replication, including viral RNA synthesis and modification and host immune evasion. Nsp4 is a transmembrane protein involved in viral replication complex assembly. Nsp5 functions as a protease that enzymatically cleaves both viral and host proteins, rendering it as a viable therapeutic target. Nsp6 is a transmembrane protein involved in viral replication complex assembly and host cell membrane regulation. Nsp7 and nsp8 are cofactors forming a complex in viral RNA synthesis. Nsp9 is a single-stranded RNA-binding protein involved in viral RNA synthesis [7]. Nsp10 is a cofactor for nsp16, which plays a role in viral RNA methylation. Nsp12 is an RNA-dependent RNA polymerase responsible for viral RNA synthesis. Nsp13 is a helicase that unwinds double-stranded RNA during viral replication [9]. Nsp14 is an exoribonuclease that proofreads viral RNA during replication. Nsp15 acts as an endoribonuclease that cleaves viral RNA to prevent recognition by host immune sensors. Nsp16 is a methyltransferase that modifies viral RNA to evade host immune recognition (Table 1).
Table 1.
Comprehensive overview of the critical functions of nonstructural proteins encoded by the SARS-CoV-2 genome, highlighting their interplay and role in the viral life cycle.
| Nonstructural protein | Function |
|---|---|
| NSP1 or leader protein | Attaches to the host cell's 40S ribosome, causing a halt in protein synthesis, and stimulates the degradation of host cell mRNA [29] |
| NSP2 | Contributes to the disruption of the host cell's homeostasis. |
| NSP3 (papain like proteinase) | Facilitates viral activity by producing NSP1, NSP2, and NSP3 [30] |
| NSP4 | Alteration of cell membranes in SARS-CoV-2 [31] |
| NSP5 (3C-like proteinase) | Splits the NSP polyprotein into 11 distinct segments through cleavage [32] |
| NSP6 (putative transmembrane domain) | generate autophagosomes [33] |
| NSP7 | Forms a dimer with NSP8 and NSP12, resulting in the RNA polymerase function of NSP8 [34] |
| NSP8 | Stimulates NSP12 [35] |
| NSP9 | Crucial for the replication of the virus [35] |
| NSP10 | Boosts the function of NSP14 and NSP16 [36] |
| NSP11 | Unknown |
| NSP12 (RNA dependent RNA polymerase) | Replicates the viral RNA [37] |
| NSP13 (Helicase) | Separates the duplex RNA [38] |
| NSP14 (3′–5′ endonuclease, N7-methyltransferase) | 5′-cap RNA (3′–5′ exonuclease, guanine N7-methyltransferase) [39] Proofreading of viral genome |
| NSP15 (endoRNAse) | Degrade RNA to evade host defense [40] |
| NSP16 (2′-O-ribose-methyltransferase) | The 5′-cap shields mRNA from degradation, promotes its translation, and blocks the regulation of innate immunity [41] |
The functions of nonstructural proteins of SARS-CoV-2 are tabulated in Table 1. Various fragments of the SARS-CoV-2 proteins have been studied as potential targets for vaccine development and therapeutic intervention. The S protein receptor-binding domain (RBD) is a major target for neutralizing antibodies and has been used in vaccine development. The N protein has also been studied as a potential vaccine candidate. Nsp5 has been targeted for drug development, and inhibitors of this protease have been shown to have antiviral activity. Nsp12 is another target for drug development, and remdesivir, a nucleotide analogue, has been approved to treat COVID-19 [42].
3. Functions of the spike protein in SARS-CoV-2 pathogenesis
A crucial step in membrane fusion is the attachment of the S protein to the ACE2 [43]. The plasma membrane containing transmembrane protease serine 2 (TMPRSS2) is responsible for digesting S1 and S2 at the S29 location next to the FCS for optimum cellular infection. Thus, to effectively promote the viral membrane-host cell membrane fusion for virus entry, the RBD (in S1) of Spike can link with the ACE2 of the host cell surface receptor [15]. A furin-like protease must break down S1/S2 to activate S, which then changes its shape before following fusion [44]. The 3 RBDs of the S glycoprotein can be seen in different earlier stages of fusions with confirmations in either direction [26], [43]. The RBDs can only assume an up conformation to reveal the receptor binding sites. Recombinantly produced S proteins of the virus have been found in the states such as “RBD down,” “one RBD up,” and “two-RBD up” [45]. S pursues a regular class I fusion protein activation route: major alterations to its structure, including removing its S1 subunit and adding the fusion peptide (FP) to the target cell membrane [44]. S changes into a needle-shaped post-fusion form after membrane fusion, with 3 helixes coaxially entwined. The Wuhan-Hu-1 strain, having full-length S protein with 1273 residues, contains a signal peptide (N-terminal), S1(RBD), and S2 (fusion fragment). As opposed to this, the S1 was further split into the RBD, C-terminal domains (CTD1 and CTD2), and N-terminal domain (NTD) [46]. On the other hand, S2 includes a fusion-peptide proximal region (FPPR), heptad repeats (HR1 and HR2), fusion peptide (FP), cytoplasmic tail (CT), transmembrane segment (TM), central helix (CH), connector domain (CD) [44].
The SARS-CoV-2 virus utilizes the homotrimer spike glycoprotein on its surface to recognize the ACE-2 receptor. Following this recognition, the virus-host cellular membrane fusion is necessary for the viral nucleocapsid to enter the host cells. The S glycoprotein-ACE2 receptor attachment initiates the viral accession into the host organism's cells (lung cells concerning type 2 pneumocytes) [44]. The S protein binding domain (331–524 residue), situated along the receptors of the virus, can stably bind to both human and bat ACE2 [44]. The fusion process involving the host cell and viral membrane occurs after the entrance and attachment steps [44]. Once fusion has occurred, the receptor-linked S proteins are activated by a type II transmembrane serine protease (TMPRSS2) from the host cell [44]. Subsequently, the virus can enter the cells because this causes conformational alterations. The 2 proteins, ACE2 and TMPRSS2, are the initial regulators for the trance of the virus.
The mRNA released by the virus further undergoes translation equipping its genomic materials. The viral genome is assisted by a large number (approximately 14) of open reading frames (ORF), each of which encodes a different set of structural and nonstructural proteins essential to the virus's continued existence and pathogenicity. During its transformation phase, the mechanism allows the production of gene fragments encoding nonstructural mega proteins such as ORF1a and ORF1b and their further assembly into super-imposed mega proteins, pp1a, and pp1ab. Polyprotein processing involves the replacement of polyproteins by protease enzymes such as papain-like proteases (PLpro) and the serine-type Mpro (chymotrypsin-like protease (3CLpro) protease), which are encoded by nsp3 and nsp5, respectively. After then, nonstructural proteins (nsps) 1–11 and 1–16 are produced by the cleavage of pp1a and pp1ab, respectively. The nsps are crucial for diverse viral and host cell-related functions [44].
Following their biosynthesis, a complex network of nonstructural proteins (nsps) coalesces into replicase-transcriptase complexes (RTCs) within double-membrane vesicles (DMVs). These RTCs are primarily assembled by subunits that contain RNA-dependent RNA polymerase (RdRp) and helicases, key enzymes involved in viral RNA replication and transcription. The RdRp domain is found in CoVnsp 12 and AV nsp 9, respectively. Transcription to positive-sense mRNAs is predominantly assisted by RdRp [44]. The transformation of sub-genomic RNA into intermediary products coupled with the negative sense genes of progeny genome are formulated as the endogenous genome associated with the viral entry is turned by the complex [44]. The intermediary chamber facilitates the translation of sub-genomic proteins into structural and accessory proteins like M, S, and E proteins (ERGIC). It is situated between the Golgi apparatus and the endoplasmic reticulum. Meanwhile, the copied genome program may now access the ERGIC to link the N protein to the nucleocapsid structure directly. Small wallet-shaped vesicles, called exosomes, are constructed from the nucleocapsids in this compartment, along with a variety of other structural proteins.
Changes observed in the glycosylation sites and mutations in human SARS-CoV-2 spike proteins have found presence based on a study assessing the reactivity and infectivity concerning the neutralizing antibodies and sera obtained from individuals recovered from infection. They are relevant to the current situation [44]. The spike glycoprotein has reportedly undergone relatively few mutations. The D614G mutation is known to increase the effectiveness of infection and is found to be considerably more common [10], [11]. Two-dimensional maps of the spike protein mutation locations from several Indian isolates related to the virus have been produced [44], [47]. Almost every area of the protein has some form of mutation. The D614G mutation is found in the S1D domain (594–674), which is present in 7859 of the 7915 mutations in this area of the spike protein [44].
4. COVID-19 vaccine technology
The SARS-CoV-2 genome was made available on January 11, 2020. This triggered rapid growth in international research and development (R&D) projects to initiate vaccination programs to eliminate the epidemic. Due to the impact COVID-19 posed on economic resources and human affairs, innovative approaches are speeding up the assessment of potential platforms for next-generation vaccine technologies [3]. Consequently, the initial candidate of the COVID-19 vaccine for human trials was authorized at an astonishing speed on March 16, 2020. There are 115 vaccine candidates currently in different stages of development in the R&D field concerning COVID-19 as of April 8, 2020, 78 of which have been confirmed as active, and 37 of which have not. In the experimental or preclinical stages, 73 of the 78 initiatives have been confirmed as active. Several cutting-edge vaccine candidates, such as Ad5-nCoV from CanSino Biologicals, mRNA-1273 from Moderna, LV-SMENP-DC, INO-4800 from Inovio, and a pathogen-specific APC from Shenzhen Geno-Immune Medical Institute, have already entered clinical studies. Many more vaccine producers have made plans to begin human trials in 2020. Seeing how many different technological platforms are being evaluated for the COVID-19 vaccine is breathtaking. Accordingly, a range of vaccine technologies are under investigation, including viral vectors (both replicating and non-replicating), inactivated viruses, live attenuated viruses, recombinant proteins, nucleic acids (RNA and DNA), peptides, virus-like particles, and recombinant proteins (Fig. 2). According to the WHO data total of 11 COVID-19 vaccines are permitted to use including Serum Institute COVOVAX (Novavax formulation)- approved in 6 countries, 7 trials in 3 countries; Novavax (Nuvaxovid)- approved in 40 countries, 22 trials in 14 countries; Moderna (Spikevax)- approved in 88 countries 70 trials in 24 countries; Pfizer/BioNTech (Comirnaty)- approved in 149 countries 100 trials in 31 countries; CanSino (Convidecia)- approved in 10 countries 14 trials in 6 countries; Janssen (Johnson & Johnson) Jcovden- approved in 113 countries 26 trials in 25 countries; Oxford/AstraZeneca (Vaxzevria)- approved in 149 countries 73 trials in 34 countries; Serum Institute of India Covishield (Oxford/AstraZeneca formulation)- approved in 49 countries 6 trials in 1 country; Bharat Biotech Covaxin- approved in 14 countries 16 trials in 2 countries; Sinopharm (Beijing) Covilo- approved in 93 countries 39 trials in 18 countries; Sinovac CoronaVac- approved in 56 countries 42 trials in 10 countries. Among these vaccines, Sinopharm, Bharat Biotech Covaxin, and Sinovac are inactivated; CanSino, Janssen, Oxford, Covishield are non-replicating Viral vector; Moderna and Pfizer are RNA vaccines; and COVOVAX and Novavax are protein subunit vaccine (https://covid19.trackvaccines.org/agency/who/).
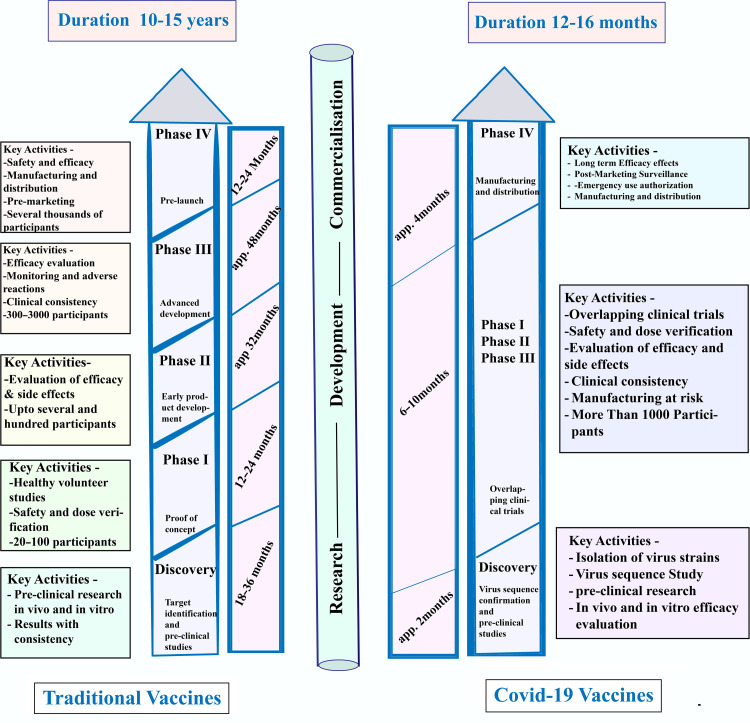
Fig. 2.
A comparative view of the vaccine development process in 2 distinct platforms, traditional vaccines, and COVID-19 vaccines, highlights the similarities and differences between the 2 platforms and emphasizes the unique challenges and advancements associated with COVID-19 vaccine development [48].
This illustration serves as a valuable reference tool for researchers and healthcare professionals seeking to understand the underlying mechanisms and innovations in vaccine development in the context of COVID-19. Fig. 2 describes a comparative overview of traditional and COVID-19 vaccine development. The traditional vaccine needs 10–15 years to develop whereas COVID-19 developed within 1 year. Clinical trial phases in the traditional vaccine require 3∼4 years with help of 3000 participants, which was 6–10 months for COVID-19 using 1000 participants.
4.1. Inactivated vaccines
Attenuated vaccines are based on dead microorganisms and are a highly traditional technological platform that has produced many vaccinations [10], [11], [49], [50]. The constructs of the technology present superior resiliency compared to live attenuated vaccines; however, they are still limited by the short immunological memory, which necessitates the use of bigger doses of vaccine or the combination of the immobilized microbe with an adjuvant. Numerous other SARS-CoV-2 antigens are also targeted by the immune reaction that is produced, in addition to the Spike protein. Although the generated reaction is typically less strong than that caused by viruses that have been attenuated, the vaccine is easier to use, less costly, and considerably safer [47].
BBIBP-CorV, also called the Sinopharm Beijing vaccine, is a COVID-19 vaccine that uses inactivated SARS-CoV-2. This vaccine utilizes multiple strains of the virus, including 19nCoV-CDC-Tan-Strain03 (CQ01), 19nCoV-CDC-Tan-HB02 (HB02), and 19nCoV-CDC-Tan-Strain04 (QD01), acquired from bronchoalveolar lavage (BAL) samples of individuals with COVID-19 during the vaccine development process [47]. Researchers selected the HB02 variant to develop the inactivated vaccine due to its high virus yield and replication levels, and it has proven effective against SARSCoV-2 [47]. The HB02 variant was subsequently isolated and utilized as a viral stock. Vero cells were used to increase the viral stock. After 10 generations of adaption and passing, the initial germ for the inactivated vaccine was obtained with a sequence homology of 99.95%. The collected virus sample was combined with β-propiolactone to inhibit the formation of SARS-CoV-2. Vial contagiousness was eliminated, and formulation stability went hand in hand with the viral inactivation process. Sinopharm's inactivated SARS-CoV-2 vaccine tested positive in phase I/II clinical trials and was shown to be both safe and well-tolerated. Additionally, healthy vaccination recipients showed immunogenicity.
Sinovac, commonly referred to as CoronaVac, is a type of WIV vaccine developed and produced in China [51]. In this context, kidney cells from African green monkeys were used to grow the SARS-CoV-2 vaccine's CN02 strain. After then, β-propiolactone was employed to harvest and inactivate SARS-CoV-2. The material was then concentrated and cleaned. It was eventually deposited on aluminum hydroxide. By employing Water, phosphate-buffered saline (PBS) and sodium chloride, the dilution of the aluminum hydroxide complex prior to disinfection was executed [51]. Based on the data obtained during the phase I/II clinical research volunteers with good health status had been shown tolerating perfectly against the vaccine and could only slightly generate immunogenicity [51].
COVAXIN, also referred to as BBV152, is an Indian-made COVID-19 vaccine that uses inactivated SARS-CoV-2 and is produced by Bharat Biotech [52]. By utilizing β-propiolactone to inactivate the NIV-2020-770 strain, COVAXIN is created. The spike proteins of the strain have the Asp614Gly mutation [52]. According to phase I clinical trial findings, COVAXIN was safe and well tolerated by participants and had the ability to produce immunogenicity and boost the immune response (most notably the T-cell response) following vaccination [52]. Utilizing several chemical approaches, the SARS-CoV-2 is rendered inactive. These potential vaccinations are all intramuscularly administered.
Seven vaccine candidates with differently induced SARS-CoV-2 inactivation processes are now undergoing clinical testing; 4 of them have previously received restricted approval. When they are available, reports from Phase II studies point to the vaccine's safety and high antibody titer. The 7 medical studies are directed by:
-
•
Interim findings for the CoronaVac vaccine, constructed in China by Sinovac Biotech, are anticipated in late November. CoronaVac, meanwhile, has already received permission for restricted usage among the public.
-
•
Chinese Academy of Sciences, China.
-
•
Two of Sinopharm's unique initiatives have received approval for restricted usage in the general population.
-
•
Another vaccine has been authorized for restricted use in general population by Wuhan Institute of Biological Products, China.
-
•
This vaccine, Covaxin, is now undergoing a late-stage Phase III study at Bharat Biotech in India.
-
•
RIBSP, Kazakhstan.
4.2. Viral vector vaccines
Viral vectors have the ability to transport DNA encoding the Spike protein into cells, which is based on the remarkable ability of viruses to infect and distribute mRNA throughout human cells [53]. The use of adenovirus vectors holds great potential in developing COVID-19 vaccines by utilizing this mechanism. Adenoviruses are non-enveloped dsDNA viruses that can cause mild diseases that affect the respiratory tract and eyes and other human illnesses that typically resolve on their own [53]. High-tech vaccine platforms, which rely on adenovirus vectors, are currently gaining attention as reliable transporters of nanotechnology for gene delivery [16]. The SARS-CoV-2 S proteins can replace the E1 and E3 viral genes in order to improve vaccine design and eliminate unwanted antigens [53]. The major advantage of employing adenovirus vectors for drug, gene, and vaccine delivery is that they cannot integrate into the human genome, thereby ensuring safety following injection.
The virus with DNA insertion may endanger its potential to proliferate. Since a preestablished immunity to the virus vector may reduce the effectiveness of the vaccination, primate viruses (from chimpanzee, gorilla, etc.) are usually utilized as vectors. In other instances, the DNA is placed into replicated viral vectors; since these viruses may spread to some extent, they might cause a more potent immune response [53]. The Spike protein, its variations, or the fragmented pieces are also the target antigens for these vaccine studies, if not the only ones. These virus-interceded vaccinations are frequently delivered through the intra-muscular route. However, many intriguing projects try to inhale the vaccine into the nasal for delivery. If successful, the proposed vaccine might stimulate mucosal immunity that can neutralize the virus, preventing it from entering the human body [53].
The AstraZeneca/Oxford vaccine, which is marketed as Vaxzevria, is a recombinant adenovirus vaccine that employs the S glycoprotein to fight against SARS-CoV-2. To produce the SARS-CoV-2 spike protein (S-protein), the vaccine utilizes a chimpanzee adenovirus and a shuttle plasmid that contains the SARS-CoV-2 amino acid sequence and tissue plasminogen activator (tPA) leader at the 5′ end. One of the primary advantages of this vaccine is its affordability in low- and middle-income countries and regions. Additionally, it can be stored at temperatures ranging between 2 and 8 °C, making it a practical option for global distribution [53].
Adenovirus-vectored vaccines include the Johnson & Johnson vaccine, sometimes referred to as Janssen. The adenovirus 26 DNA was modified to include the SARS-CoV-2 spike protein gene (Ad26.COV2-S). By delivering this altered adenovirus immunization, the viral DNA may enter the cell and be released. The spike protein will then be generated using the DNA of the virus. Subsequently, the immune system will get activated, and spike protein-induced antibodies will be generated. As a result, immunization may prevent SARS-CoV-2 infection following virus exposure [53]. After immunization, infection will not be happening because the adenoviral proliferation is precluded, responsible for SARS-CoV-2 transmission. This vaccine may be distributed internationally and requires a higher storage temperature (2∼8 °C) than previously licensed vaccines like Pfizer/BioNTech and Moderna, which need an extremely cold storage environment for the endurance of the SARS-CoV-2 DNA molecules [54]. Sputnik V, marketed as Gam-COVID-Vac, is a type of vaccine that utilizes a heterologous recombinant adenovirus (rAd26 and rAd5) vector [54]. Adenoviral vector-delivered antigens gain the ability to elicit both humoral and cell-mediated immune responses after administration of the initial dose. However, a long-lasting immune response would occur once the second dose of these vaccinations was administered [54]. This vaccines' immunogenicity against the vector components would be their main disadvantage. The prime-boost heterologous vaccination technique can lessen and even get rid of this issue by utilizing 2 distinguishable vectors concerning the vaccination (1st and 2nd dose) purpose. Sputnik V is a double vector vaccine that uses Ad5 and rAd26 to deliver spike proteins encoded by the SARS-CoV-2 virus (rAd26-S and rAd5-S, respectively) [54]. Both heterologous recombinant adenoviruses are administered intravenously at intervals of 21 days. Following the positive findings of the phase I/II clinical studies, this vaccine was given early approval in Russia [54]. According to the findings of phase III clinical studies, Gamaleya's Sputnik V vaccine was 91.6% effective overall on day 21 following the administration of the first dosage (the day of the second injection).
According to the COVID-19 Vaccine Development Landscape, Adenovirus 5 (Ad5)-vectored vaccines, such as the CanSino vaccine (Ad5-nCoV), deliver DNA to cells encoding the spike proteins of SARS-CoV-2. A phase I clinical trial conducted in China using the CanSino vaccine, a type of Ad5-vectored vaccine, demonstrated the vaccine's ability to stimulate both adaptive and innate immunity within 14 and 28 days following a single-dose administration. The study utilized a dose-escalation design and was open-label and non-randomized in nature. These findings provide promising evidence for the vaccine's potential to elicit an immune response against SARS-CoV-2 [54]. Various vaccination initiatives utilizing viral vectors are already well advanced in the clinical testing process, with 4 of them undergoing a Phase III study or having limited authorization. The vaccine's DNA is inserted into cells, and the CanSino vaccine, also known as CanSinoBIO, uses Ad5-vectors for SARS-CoV-2 spike protein encoding.
4.3. RNA vaccines
Even though messenger RNA (mRNA) has not yet resulted in a vaccine that has received its approval for mass use, several initiatives are using it to construct SARS-CoV-2 vaccines. RNA, administration employs a variety of methods. The mRNA vaccination induces the cells to generate antigen proteins once it has been incorporated momentarily. Liposomes transport the mRNA in most of these vaccination efforts. Additionally, regarding anti-SARS-CoV-2 mRNA vaccines, the Spike protein, its variations, or its fragments serve as the primary or sole representation of the target antigen translated by the mRNA. These vaccination formulations must be stored between −30 and −80 °C [55].
The first vaccination formulations to receive EMA conditional approval and Emergency Use Authorization (EUA) from the FDA were Moderna and Pfizer/BioNTech. Moderna (US) developed mRNA-1273, which expresses the full-length S protein and is given in 2 doses through intramuscular injection. Encouraging clinical trial outcomes led to the vaccine's approval for widespread use in humans. Similarly, LNP encapsulated mRNA vaccines were created by Pfizer and BioNTech (BNT162a1, b1, b2, c2). Unlike BNT162b1, which aims to encode the full-length S protein in the prefusion state, BNT162b2 encodes the SARS-CoV2 RBD. These 2 vaccinations can enhance neutralizing antibodies, according to the findings of the clinical experiment that compared them. The company opted to move forward with this formulation in hopes of receiving approval because BNT162b2 mRNA vaccination produced less severe adverse effects [56].
The BNT162b1 mRNA vaccine has demonstrated promising safety results and substantial immune cell responses in its phase I trial, although the BNT162b2 mRNA vaccine has received regulatory approval worldwide. This mRNA vaccine encodes a trimerized, secreted version of the SARS-CoV2 spike (S) glycoprotein RBD, increasing CD4+ and CD8+ T-cell-mediated responses generating IFN-γ after 2 doses. In a randomized, placebo-controlled, double-blind phase 1 study, the SARS-CoV-2 BNT162b1 mRNA vaccine was shown to be safe and immunogenic in both younger and older Chinese individuals [57]. Other companies are testing mRNA-based vaccines, such as the CureVac firm's CVnCoV vaccine, which codes for the S protein and is administered intramuscularly in 2 doses, encapsulated in LNP [58]. While the previous mRNA vaccines from Moderna and BioNTech/Pfizer were thought to be more expensive and unstable, the consequences of CureVac's mRNA vaccine were less effective due to various reasons, such as mRNA conformation, the use of regular uridine instead of modified nucleotides of pseudouridine to reduce inflammatory reactions, and differences in storage temperature and non-coding sections. A Phase 1 evaluation is being conducted among human volunteers employing mRNA lipid-based nanoparticle vaccine for safety and immunogenicity by Acturus/Duke-NUS and Imperial College, London, respectively, looking towards the vaccines ARCT021 (Lunar-COV19) and LNP-nCoVsaRNA, which are designed to be administered intramuscularly and replicate mRNA (Fig. 3).
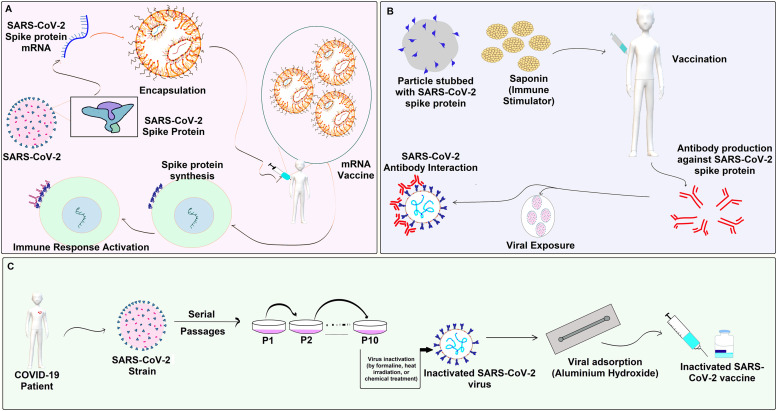
Fig. 3.
Comprehensive overview of the design and development of COVID-19 vaccines, showcasing 3 different approaches to vaccine development: (A) a specific vaccine targeting mRNA, (B) the Novavax COVID-19 vaccine, and (C) a general overview of the overall design and development of COVID-19 vaccines [62].
4.4. DNA vaccines
The platforms based on DNA and mRNA allow for extremely rapid and flexible modification of the coded antigen [59]. DNA vaccines are commonly used in veterinary medicine, however, they are yet to receive approval for human use. These vaccines provide stability and higher production in large amounts by microorganisms. A very brief local electrical pulse may help DNA plasmids injected into muscle or skin infiltrate human cells (electroporation). Plasmid DNA, once introduced, prompts the cell to generate the desired protein for a limited time. Killer T cell activation in conjunction with stimulated antibody production, is the principal effort in ensuring protective immunity when the DNA vaccination strategy is employed [59]. IgG responses are triggered against the RBD and S proteins. A higher titer of neutralizing antibodies was also observed to develop. At least momentarily, the IL-12 immunization protocol enhanced the neutralizing antibodies. Singular prefusion spike protein encoded copy or in conjunction with plasmid IL-12 transfection, based on the electroporation of plasmid DNA is conducted in-silico to produce a novel coronavirus vaccine (CORVax) [60]. On the 20th of August 2021, Zydus Cadila has been granted Emergency Use Authorization (EUA) by the Drug Controller General of India (DCGI) for ZyCoV-D, which is recognized as the world's first Plasmid DNA Vaccine developed to combat COVID-19 [59], [60]. The administration of the vaccine ZyCoV-D is limited to the utilization of the PharmaJet Tropis® Needle-free Injection System. The ZyCoV-D vaccine developed in India uses circular DNA as a protective measure against SARS-CoV-2 infection. The vaccine formulation consists of a 2 mg quantity of a DNA plasmid known as pVAX-1, which encodes the Wuhan Hu-1 spike antigen of the SARS-CoV-2 virus, along with an IgE signal peptide [59].
4.5. Subunit vaccines
Vaccines containing antigenic proteins that are either chemically synthesized peptides or produced through recombinant DNA technology are used to induce a durable immune response for therapeutic or preventative purposes [61]. However, an adjuvant is necessary to augment the vaccine's immunological responses due to the subunit vaccination's limited immunogenicity. An adjuvant may increase the production of immunomodulatory cytokines or increase the half-life of the antigenic substance, triggering an immune response in the body, often by binding to specific antibodies or immune cells. When used in conjunction with a vaccine, adjuvants help alleviate some of the problems associated with protein subunit vaccines [39]. The SARS-CoV-2 S protein is the most efficient foreign entity in generating antibodies that have the ability to render the infection inactive [63]. Two subunits make up the S protein. Compared with the S2 subunit possessing HR 1 & 2, FP, the S1 subunit includes NTD, RBD, and RBM domains [64]. Viruses are able to enter cells via endocytosis owing to the S-Protein-assisted attachment to the hACE2 receptor. Subunit vaccination is primarily directed toward the S-Protein and its antigenic parts as a result. The pre-fusion and post-fusion states are the 2 conformational states in which the dynamic protein known as the S glycoprotein can exist. As a way to secure the epitopes necessary to relay the reactions concerning the antibodies, the antigen cannot but hang on to continued superficial chemistry and protein profiling of the primary pre-fusion spike protein [39]. Utilizing techniques that employ the cloaked RBM as an antigen can enhance the production of neutralizing antibodies and improve the overall effectiveness of the vaccination.
4.6. Booster of COVID-19 vaccines
The immune system undergoes subsequent steps by producing immune cells and associated chemicals and observes reduced immune cell counts when the vaccine is administered. Despite that, a minute amount of T and B cells remain in the bloodstream, safeguarding against potential infections [34]. A booster dose of the vaccine has multiple functions. It replicates the defense mechanism against the foreign particles when multiplying the number of antibody-producing B cells [39]. Even though the B cell number might go down once the infection is in control, the reservoir of memory B cells will combat ensuing infections. The procedure describing the transportation of “Engaged” B cells (the stimulated phase when in contact with the vaccine) to the lymph node is termed “Affinity Maturation” and it is further accelerated by administering the booster doses. Affinity maturation results in the B cells inheriting potential mutations, which may lead them to increased efficacy in their fight against probable infections [39]. Since administering the 2nd dose of the vaccine, several months of the non-intervened state ensures the depression of active antibodies in the body. The 3rd dose of the vaccine, constructed by Oxford-AstraZeneca, Pfizer-BioNTech, Moderna, and Sinovac contributed to increasing neutralizing antibody counts. In a current study in the UK, alternative booster regimens will be tested, including those employing a different vaccine from the initial shots. These “mix and match” tactics may lead to specific antiviral responses coupled with the destruction of infected cells Due to the increase in T cells and antibodies.
A variant of interest (VOI) is defined by a lineage with suspected mutations that result in increased transmissibility, the blockade against therapeutic measurement or vaccination, increased infectivity or instances of infections, and prevalence that has not spread and localized in a particular area. Several VOIs were circulating during the height of the pandemic. In Nigeria, the B.1.525 (eta) and B.1.1.318 variants coexisted with the Alpha variant (Table 2). These 2 variants were shown to infect experimental monkeys and human cell lines aggressively. They both have E484K substitution in the spike RBD. Currently, no VOIs are in existence. A study conducted recently has found suspicious deletions in the antigenic regions of SARS-CoV-2, particularly in the N-terminal region, and concluded that future variants may originate based on this feature.
Table 2.
A comprehensive list of the variants being monitored (VBM), including their origin, global distribution, vaccine efficacy level, and transmission phenomenon, serves as a critical reference tool for public health officials and researchers to track and screen the emergence and feast of new SARS-CoV-2 variants [65].
| WHO labels | Clade | First identified lineage | Top pangoline lineage | Origin | First infection | Vaccine activity against this | Transmission phenomenon |
|---|---|---|---|---|---|---|---|
| Alpha | 20I | B.1.1.7 | B.1.1.7 | Kent, England | November 20 | According to statements made by Pfizer, Moderna, and Johnson & Johnson, their respective vaccines have demonstrated effectiveness in protecting against severe disease and hospitalization associated with the Alpha variant. | 30%–50% more transmissible in comparison with the original SARS-CoV-2 strain |
| Beta | 20H | B.1.351 | B.1.351 | South Africa | July 20 | The Beta variant has been reported to exhibit lower susceptibility to the vaccines developed by AstraZeneca-Oxford, Pfizer-BioNTech, Moderna, and Johnson & Johnson. | The initial coronavirus strain was found to 50% less contagious in comparison with Beta variant. |
| Delta | 21A, 21I, 21J | B.1.617.2 | B.1.617.2 | India | October 5 20 | The 3 vaccines available in the United States have been deemed highly efficacious in protecting against severe illness, hospitalization, and mortality resulting from the Delta variant. However, despite their effectiveness, breakthrough infections of Delta have occurred in some individuals who were fully vaccinated. Moreover, vaccinated individuals who contracted the infection were still capable of transmitting the virus to others, although the period of their infectiousness was likely to be shorter. It is worth noting that no vaccine provides 100% protection against any variant of the virus. | The transmissibility of the Delta variant has been found to be significantly higher, estimated to be around 80–90% more contagious than the Alpha variant. |
| Delta Plus | 21A, 21I, 21J | AY.4.2 | AY.4.2 | UK | December 20 | Partially effective | 10%–20% more transmissible than Delta. |
| Omicron | 21K-L, 22A-F, 23A | BA.1 | BA.5, BF.7, XBB, BN.1, BF.11, BQ.1, BQ1.1 | Botswana and South Africa | November 21 | Experts are still learning about their effectiveness against the latest Omicron subvariants | The most transmissible variant of concern |
On the contrary, a variant of concern (VOC) is described as a lineage that is married with increased transmissibility, disease severity, evidence of elevated infections, substantial protectivity against therapeutic intervention (antibodies from previous infections), and diagnostic detections. The Omicron subvariant XBB 1.5 (Kraken) shows increased transmissibility in the US. The variant may be more transmissible than its ancestor XBB. Omicron BA.2.10.1 and BA.2.75 are the recombinant parents of XBB, and XBB 1.5 is a direct descendant of XBB. However, 2 booster campaigns are persistently effective against the variant XBB 1.5 for at least 3 months, and the susceptibility to severity is low compared to other persisting sub-lineages such as BQ.1. Variants being monitored are defined as variants that have been taken care of by advanced therapeutic measurements and diagnostic protocols. A variant of high consequence (VOHC) is denoted by failure in diagnostic tests, substantially higher prevalence of infections in vaccinated persons, reduced vaccine efficacy, severe clinical onsets, or disease progression. The declaration of any VOHC relevant to COVID-19 has not yet materialized.
5. Vaccine efficacy against variants
Examining a vaccine's effectiveness in research, such as how effectively it prevents symptomatic disease, is one way to determine its efficacy [16]. With 692 million COVID-related illnesses along with a mortality count of 6.90 million as of August 07, 2023, the decisiveness of COVID-19 is undeniable when considering the world perspective The global spread of variants, particularly the Omicron variant, has aggravated more severe infections and transmission [34].
The COVID-19 vaccination campaign is viewed as the most critical measure to combat the pandemic because it helps to minimize the complications associated with COVID-19, thus making it suitable for societies to reopen and promoting economic recovery. As of June 4, 2022, 129 COVID-19 vaccination programs had undergone clinical trials, and 32 had already received approval for mass application. In many nations, widespread immunization campaigns and booster vaccination schedules are in place. Numerous randomized controlled trials (RCT) or observational studies have provided strong evidence for vaccinations' immunogenicity, safety, and effectiveness. The majority of COVID-19 vaccines, nevertheless, were created for the initial strains. However, concerns about the efficacy of the vaccine arose with the emergence of several VOCs, including Alpha, Beta, Gamma, Delta, and Omicron. According to leading research, the VOCs are more infectious and better at evading the immune system than the original strains, suggesting a decline in VE. A number of Randomized Clinical Trials (RCT) studies were conducted to measure VE against morbid outcomes brought on by VOCs. The RCT studies are carried out in an extremely challenging environment, with rigorous limitations on the sample volume and trial settings. The study environment was distinct to the actual research. Public health initiatives, personal self-protective behaviors, access to healthcare, vaccination reluctance, and a variety of study groups have a cumulative impact on how well VE performs in the real world.
Evaluating evidence of public health choices signifies the success of COVID-19 vaccinations in the practical world. The results that arose from several investigations are debatable, despite some studies authenticating the real-world efficiency of COVID-19 vaccinations against the VOCs. Comprehensive evaluation urgently requires meta-analysis based on empirical data. In this work, we sought to comprehensively assess the VE in the context of the actual world against each clinical result brought on by the VOCs.
6. Effectiveness of booster shot upon different variants
The dose registered after the initial vaccination series can be defined as the booster dose. That can be any dose after the first dose or 2 doses of any vaccine. According to several prior investigations, existing variations like Delta and Omicron display robust immune responses to mitigating antibodies produced by initial immunization or prior infestations [58]. However, with a period, the perks of protection fade [58]. With such a decline in immunity, fully immunized individuals do have illnesses that develop suddenly [58]. For instance, with multiyear research within the course of 6 months, its potency throughout preventive care was reduced by a preliminary 90%–60% [58]. As a result, the subject of interest has shifted to the longevity of the resistance of vaccine induction as well as the requirement for recurrent boosting vaccinations.
Boosting shots provide better defense versus the COVID-19 virus. In comparison to the second dose of a specific vaccine (BNT162b2), a trio dosage produced 95.3 percent effectiveness towards COVID-19, according to a stage 3 clinical study (NCT04955626). Towards the end of 2021, once the Delta variant became the most popular type, several nations began to give COVID-19 boosts. However, the Omicron variant offered a different problem as the emerging outbreaks dramatically surged at the climax of 2021, and enhancers efficiently protected over severe sickness and hospitalization brought on by the Delta variant. Thankfully, patients who received boosting of BNT162b2, mRNA-1273 [34], or generic mRNA vaccinations demonstrated a significant rise in serum anti-spike antibody responses and the neutralized titers towards the Omicron variant.
Additionally, it's been demonstrated that CoronaVac and mRNA vaccine enhancers enhance memory B cells specialized for anti-receptor attachment domains and quickly develop responses against various variations, including Omicron. Optimized people can still experience Delta and Omicron outburst infestations, but the viral counts appear to be reduced, and the signs are less severe [34].
Especially in marginalized and elevated populations, repeat immunizations are particularly crucial. For instance, investigations revealed that the addition of BNT162b2 or mRNA-1273 enhancers is required to protect from Delta and Omicron in cancer therapy effectively, organ transplant candidates, and the elderly [34]. Israel began providing an additional booster in January 2022 to aged and immunosuppressed people. In March 2022, subsequent mRNA boosts were also made available in the US to the elderly and those with underlying medical issues. According to future research in Israel, adults over 60 who received 2 BNT162b2 booster doses saw fewer hospitalization and fatalities due to COVID-19 than those who received just one.
This relevance of renewal vaccines, demonstrated in several trials to improve antibody responses and susceptibility towards Delta and Omicron, has indeed been highlighted by introducing the more contagious Delta and Omicron strains. For people receiving genetically identical BNT162b2 supplements, this same efficiency against inflammation to Delta and Omicron has been predicted to be 88∼93 percent and 76 percent, respectively. In comparison, the efficiency against clinical complications induced by Delta and Omicron was anticipated to be 97 percent and 89 percent, respectively.
7. Death toll regarding COVID-19
According to the findings from Wordometer, 6,904,567 people have passed away due to COVID-19. According to investigations conducted by Johns Hopkins University, the countries with the highest COVID-19 mortality rates were Peru at 4.9%, Mexico at 4.5%, Ukraine at 2.1%, Iran at 1.9%, Brazil at 1.9%, and Poland at 1.8% of deaths occurring per 100,000 people. The term “excess mortality” refers to the difference between the reported number of deaths in a given week or month (depending on the country) in 2020–2023 and an estimate of the number of deaths that would have been expected for that period if the COVID-19 pandemic had not occurred. This measurement is used to determine the total impact of the pandemic on deaths rather than relying solely on the confirmed death count from COVID-19. The largest rates of excess mortality were recorded in South Africa, Mexico, and Russia.
8. COVID-19 booster dose administration
The longevity of COVID-19 vaccines and previous data supported decreasing host immunity after vaccination is still unclear. There is evidence documenting a drop in COVID-19 antibody titers within the human body over time. Additionally, it has been proven that lower antibody titers may be associated with reduced levels of protection. The requirement for booster doses of the COVID-19 vaccine presented a logistical hurdle for global health policymakers. As of January 1, 2021, no evidence of booster administration was reported. It is revealed from Our word in data that persons in China (92.81%), Brazil (89.10%), the United States (81.81%), Indonesia (74.45%), India (74.44%), and Pakistan (73.43%) received at least 1 COVID-19 vaccination dose. The 2 companies have committed to supply up to 1.8 billion doses to the EU from December through 2023, in addition to the 600 million doses bought this year. Up to April of next year, the US government has ordered 700 million for Americans and 500 million for gifts to the poorest nations. 3 billion vaccinations will be produced this year by Pfizer and BioNTech, and 4 billion the next year. The data presented in Our World in Data demonstrates approximately 72.3% of the world's population, or approximately 5.55 billion individuals, have received at least 1 dose of the Covid-19 vaccine. Globally, approximately 2.72 billion booster doses have been administered.
9. New COVID-19 vaccines
According to the most updated data, 344 COVID-19 vaccines have been constructed, with some still undergoing development. Of the 31 vaccines that have found regulatory approval by the concerned authorities, most of them have employed at least 5 distinct technologies [58]. However, 2 new approaches in vaccine construction have found light recently. These vaccines are the recombinant plant-based adjuvant vaccine [58] and the RBD dimer-based ZF2001 vaccine.
However, 2 new approaches in vaccine construction have recently emerged. These vaccines include the recombinant plant-based adjuvant vaccine (Bacillus subtilis), and the RBD dimer-based ZF2001 vaccine. The majority of COVID-19 vaccines require sophisticated storage capabilities, such as extremely low storage temperatures. In contrast, these 2 vaccines are created to not include those standards, which makes them appropriate for utilization in countries with lower and middle financial status [58]. More importantly, the phase 3 trials of these vaccines were conducted in such countries when available data showed the prevalence of several SARS-CoV-2 variants. However, these data excluded the omicron variant of the virus, indicative of the variant's existence after the trials.
The plant-based recombinant vaccine, which combines adjuvant system 03 (AS03) and prefusion-spike protein from the original strain of the virus, during phase 3 trials, this vaccine was given in 2 doses in various countries including Mexico, Canada, the UK, Argentina, Brazil, and the USA. The vaccine demonstrated effectiveness rates of 78.8% (95% confidence interval [CI], 55.8–90.8) for moderate to severe infection, 74.0% (95% CI, 62.1–82.5) for individuals who were seronegative at the beginning of the trial, and 69.5% against symptomatic infection confirmed by polymerase chain reaction (95% CI, 56.7–78.8) [66].
During the phase 3 trials conducted in Mexico, Canada, the UK, Argentina, Brazil, and the USA, a plant-based recombinant vaccine was given in 2 doses. The vaccine combined an adjuvant system (AS03) and prefusion-spike protein from the original strain of the virus. The vaccine was found to have an efficacy rate of 78.8% (95% CI, 55.8–90.8) against moderate to severe infection, 74.0% (95% CI, 62.1–82.5) in people who had a negative result for antibodies at the start of the trial, and 69.5% against symptomatic infection confirmed by polymerase chain reaction (95% CI, 56.7–78.8). On the other hand, the RBD dimer-based vaccine has shown to be an improvement compared to the plant-based vaccine with a vaccine efficacy of 87% against serious complications, onset just 7 days after the third dose, and 75.7% against COVID-19 in trials conducted by Shen et al. and Jackson et al. [67], [68]. However, both trials excluded the elderly group, breaking the promise to lessen the complications for the elderly who were considered the most prioritized group for vaccination by the WHO.
Given the surging conditions in China, there is a need for better understanding and techniques for generating new vaccine tools. World leaders should consider this, and some initiatives have been taken by the African nations' leaders to combat the disease in case of future complications (mRNA Vaccine Technology Transfer Hub, WHO). Additionally, some plant species have antiviral properties and have been used during the COVID-19 pandemic as home remedies [69].
10. Future foresight for the COVID-19 pandemic
The omicron subvariants that are currently circulating may look different from the prevalent SARS-CoV-2 variants over the upcoming autumn and winter seasons [46], [70]. The immune system individuals already have from their first vaccines, along with an updated booster that more accurately matches today's omicron subvariants, is anticipated to offer improved protection in the future. It could need less frequent boosting as long as omicron sub-lineages are dominant. In the coming weeks, the Food and Drug Administration will convene to choose the fall boosters so that manufacturers can prepare the shots [71]. Vaccine manufacturers like Moderna is currently assessing the immune response to newly developing strains and testing their booster prospects on humans. The results of the tests will likely determine what will be utilized to prepare for an autumn or winter surge [72].
Optionally, the universal coronavirus vaccination strategies that have already shown promise in animal studies might be added to the vaccine booster method [73], [74]. The aim of the research is to develop a universal vaccination that works against all strains. Some researchers are concentrating on chimeric spikes, which mix components of the spikes of various coronaviruses into a single vaccine, to increase protective immunity [75]. Others are experimenting with immunizations using nanoparticles that tell the immune system to concentrate on the areas where the coronavirus increase is most likely to occur [76].
The lab has shown these methods to be successful against resistant SARS-CoV-2 strains. They are also effective against zoonotic coronaviruses from bats that might jump to humans and produce a future SARS-CoV-3 outbreak, as well as the original SARS virus, which caused an outbreak in the early 2000s. The proteins found in SARS-CoV-2 can be classified into 2 types of polyproteins, ORF1a and ORF1ab, which are then divided into 16 nonstructural proteins [77]. There are also 4 structural proteins, such as the S glycoprotein, E glycoprotein, M glycoprotein, and N protein. Additionally, 8 accessory proteins can be found in SARS-CoV-2, namely ORF3a, ORF3b (not present in SARS CoV-2), ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b (also not present in SARS CoV-2). These proteins play a significant role in the replication and pathogenesis of SARS-CoV-2 [58].
11. Conclusion
COVID-19, a viral illness, has altered the planet and continues to do so in some countries. Although COVID-19 cases and death have declined, however, it is still causing widespread illness and death. It circulates new mutations that give rise to new variants with potential implications for vaccine efficacy and public health measures. Viral properties and new mutations of SARS-CoV-2 produce new variants, and they plays a critical role in determining the effectiveness of current vaccines. Immunization protects oneself from infectious diseases, such as SARS-CoV-2, which cannot be stopped with vaccines and booster doses due to new mutations. As a result, more research and updated technologies are required to develop an active COVID-19 vaccine. Different research studies are conducted to evaluate the activeness of vaccines against variants, and few of them revealed variable results. Following booster doses, one must practice proper cleanliness and guidelines and impose natural immunity through nutrition, physical activity, and a healthy lifestyle. In order to mitigate the global transmission of such infectious viruses and safeguard public health, it is imperative to sustain ongoing study and surveillance endeavors, alongside the advancement of prophylactic vaccinations.
Acknowledgments
Funding
Not available.
Acknowledgments
Author acknowledged all of the institutions in this manuscript. Author would also like to acknowledge the 2 anonymous reviewers for their critical comments and thoughtful insights, which has significantly improved the manuscript.
Declaration of competing interest
Author declared that no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data available statement
There is no data for this review.
Ethics statement
No ethical statement is required for this review article.
Informed consent
Not available.
References
- 1.Islam M.A., Shahi S., Al Marzan A., et al. Variant-specific deleterious mutations in the SARS-CoV-2 genome reveal immune responses and potentials for prophylactic vaccine development. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1090717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik JA, Ahmed S, Mir A, Shinde M, Bender O, Alshammari F, Ansari M, Anwar S. The SARS-CoV-2 mutations versus vaccine effectiveness: New opportunities to new challenges. J. Infect. Public Health. 2022;15(2):228–240. doi: 10.1016/j.jiph.2021.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLean G., Kamil J., Lee B., Moore P., Schulz T.F., Muik A., Sahin U., Türeci Ö., Pather S. The Impact of Evolving SARS-CoV-2 Mutations and Variants on COVID-19 Vaccines. MBio. 2022;13(2) doi: 10.1128/mbio.02979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suleiman A.S., Islam M.A., Akter M.S., et al. A meta-meta-analysis of co-infection, secondary infections, and antimicrobial resistance in COVID-19 patients. J. Infect. Public Health. 2023;16(10):1562–1590. doi: 10.1016/j.jiph.2023.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Hu B., Guo H., Zhou P., et al. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Islam M.A., Kaifa F.H., Chandran D., et al. XBB.1.5: a new threatening SARS-CoV-2 Omicron subvariant. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1154296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraiman J., Erviti J., Jones M., Greenland S., Whelan P., Kaplan R.M., Doshi P. Serious adverse events of special interest following mRNA COVID-19 vaccination in randomized trials in adults. Vaccine. 2022;40(40):5798–5805. doi: 10.1016/j.vaccine.2022.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty C., Bhattacharya M., Chopra H., et al. Recently emerged omicron subvariant BF.7 and its R346T mutation in the RBD region reveal increased transmissibility and higher resistance to neutralization antibodies: need to understand more under the current scenario of rising cases in China and fears of driving a new wave of the COVID-19 pandemic. Int. J. Surg. 2023;109(4):1037–1040. doi: 10.1097/JS9.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhama K., Chandran D., Chopra H., et al. SARS-CoV-2 emerging Omicron subvariants with a special focus on BF.7 and XBB.1.5 recently posing fears of rising cases amid ongoing COVID-19 pandemic. J. Exp. Biol. Agric. Sci. 2022;10(6):1215–1221. doi: 10.18006/2022.10(6).1215.1221. [DOI] [Google Scholar]
- 10.Soheili M., Khateri S., Moradpour F., et al. The efficacy and effectiveness of COVID-19 vaccines around the world: a mini-review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2023;22:42. doi: 10.1186/s12941-023-00594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas JW, Bender FL, Ballou S, et al. Frequency of Adverse Events in the Placebo Arms of COVID-19 Vaccine Trials: A Systematic Review and Meta-analysis. JAMA Netw. Open. 2022;5(1) doi: 10.1001/jamanetworkopen.2021.43955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran V, Perrodeau E, Saldanha J. et alEfficacy of first dose of covid-19 vaccine versus no vaccination on symptoms of patients with long covid: target trial emulation based on. ComPaRe e-cohortBMJ Medicine. 2023;2 doi: 10.1136/bmjmed-2022-000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakib M.M.H., Nishat A.A., Islam M.T., et al. Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenforde M.W., Link-Gelles R., Patel M.M. Long-term Protection Associated With COVID-19 Vaccination and Prior Infection. JAMA. 2022;328(14):1402. doi: 10.1001/jama.2022.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakraborty C., Chatterjee S., Bhattacharya M., et al. The D614G mutation helps to increase the transmissibility and reduce the virulence of SARS-CoV-2 variants through natural selection. Int. J. Surg. 2023;109(2):171–174. doi: 10.1097/JS9.0000000000000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suvvari T.K., Kandi V., Mohapatra R.K., et al. The re-emergence of measles is posing an imminent global threat owing to decline in its vaccination rates amid COVID-19 pandemic: a special focus on recent outbreak in India - a call for massive vaccination drive to be enhanced at global level. Int. J. Surg. 2023;109(2):198–200. doi: 10.1097/JS9.0000000000000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasan M.N., Islam M.A., Sangkham S., et al. Insight into vaccination and meteorological factors on daily COVID-19 cases and mortality in Bangladesh. Groundw. Sustain. Dev. 2023;21 doi: 10.1016/j.gsd.2023.100932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song C., Li Z., Li C., Huang M., Liu J., Fang Q., Cao Z., Zhang L., Gao P., Nie W., Luo X., Kang J., Xie S., Lyu J., Zhu X. SARS-CoV-2: The Monster Causes COVID-19. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.835750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotshild V., Hirsh-Raccah B., Miskin I., et al. Comparing the clinical efficacy of COVID-19 vaccines: a systematic review and network meta-analysis. Sci. Rep. 2021;11:22777. doi: 10.1038/s41598-021-02321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C.J.A.…Zuidewind P. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosseini R., Askari N. A review of neurological side effects of COVID-19 vaccination. Eur. J. Med. Res. 2023;28:102. doi: 10.1186/s40001-023-00992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konishi T. Mutations in SARS-CoV-2 are on the increase against the acquired immunity. PLoS One. 2022;17(7) doi: 10.1371/journal.pone.0271305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tai W., He L., Zhang X., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q., Xiang R., Huo S., et al. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Sig Transduct Target Ther. 2021;6(233) doi: 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chilamakuri R., Agarwal S. COVID-19: Characteristics and Therapeutics. Cells. 2021;10(2):206. doi: 10.3390/cells10020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., Park Y.J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calligari P., Bobone S., Ricci G., et al. Molecular investigation of SARS-CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12(4):445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe Y., Allen J.D., Wrapp D., et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thacker P.D. Covid-19: Researcher blows the whistle on data integrity issues in Pfizer’s vaccine trial: Video 1. BMJ. 2021;2635 doi: 10.1136/bmj.n2635. [DOI] [PubMed] [Google Scholar]
- 34.Huang C., Lokugamage K.G., Rozovics J.M., et al. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7(12) doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai Y., Kawachi K., Terada Y., et al. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomar S., Johnston M.L., St John S.E., et al. Ligand-induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3clpro): implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015;290(32):19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.te Velthuis A.J.W., van den Worm S.H.E., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun K., Wang W., Gao L., et al. Transmission heterogeneities, kinetics, and controllability of SARS-CoV-2. Science. 2021;371(6526):eabe2424. doi: 10.1126/science.abe2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao S., Ge X., Wang X., et al. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res. 2015;195:217–224. doi: 10.1016/j.virusres.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y., Wu L., Shaw N., et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. USA. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subissi L., Posthuma C.C., Collet A., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Case J.B., Ashbrook A.W., Dermody T.S., et al. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90(16):7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhardwaj K., Sun J., Holzenburg A., et al. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 2006;361(2):243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirchdoerfer R.N., Wang N., Pallesen J., et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Decroly E., Debarnot C., Ferron F., et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7(5) doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashte S., Gulbake A., El-Amin III S.F., et al. COVID-19 vaccines: rapid development, implications, challenges and future prospects. Human Cell. 2021;34:711–733. doi: 10.1007/s13577-021-00512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan R., Zhang Y., Li Y., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Islam M.R., Hasan M., Nasreen W., Tushar M.I., Bhuiyan M.A. The COVID-19 vaccination experience in Bangladesh: Findings from a cross-sectional study. Int. J. Immunopathol. Pharmacol. 2021;35 doi: 10.1177/20587384211065628. 205873842110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plante J.A., Mitchell B.M., Plante K.S., et al. The variant gambit: COVID-19’s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groves D.C., Rowland-Jones S.L., Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem. Biophys. Res. Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamey G., Schäferhoff M., Hatchett R., et al. Ensuring global access to COVID-19 vaccines. Lancet. 2020;395(10234):1405–1406. doi: 10.1016/S0140-6736(20)30763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bagcchi S. The world's largest COVID-19 vaccination campaign. Lancet Infect. Dis. 2021;21(3):323. doi: 10.1016/S1473-3099(21)00081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hacisuleyman E., Hale C., Saito Y., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384(23):2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hager K.J., Pérez Marc G., Gobeil P., et al. Efficacy and safety of a recombinant plant-based adjuvanted covid-19 vaccine. N. Engl. J. Med. 2022;386(22):2084–2096. doi: 10.1056/nejmoa2201300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blakney A.K., Bekker L.-G. DNA vaccines join the fight against COVID-19. The Lancet. 2022;399(10332):1281–1282. doi: 10.1016/S0140-6736(22)00524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding Y., Fan F., Xu X., Zhao G.., Zhang X., Zhao H., Wang L., Wang B., Gao X.M. A COVID-19 DNA Vaccine Candidate Elicits Broadly Neutralizing Antibodies against Multiple SARS-CoV-2 Variants including the Currently Circulating Omicron BA.5, BF.7, BQ.1 and XBB. Vaccines (Basel) 2023;11(4):778. doi: 10.3390/vaccines11040778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heidary M., Kaviar V.H., Shirani M., Ghanavati R., Motahar M., Sholeh M., Ghahramanpour H., Khoshnood S. A Comprehensive Review of the Protein Subunit Vaccines Against COVID-19. Front Microbiol. 2022;13:927306. doi: 10.3389/fmicb.2022.927306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forman R., Shah S., Jeurissen P., Jit M., Mossialos E. COVID-19 vaccine challenges: What have we learned so far and what remains to be done? Health Policy. 2021;125(5):553–567. doi: 10.1016/j.healthpol.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohkami M., Talaeipour M. Investigation of the Chemical Structure of Carboxylated and Carboxymethylated Fibers From Waste Paper Via Xrd and Ftir Analysis. BioResources. 2011;6(2):1988–2003. doi: 10.15376/biores.6.2.1988-2003. [DOI] [Google Scholar]
- 64.Yang M.-C., Wang C.-C., Tang W.-C., Chen K.-M., Chen C.-Y., Lin H.-H., Hsieh Y.-C., Wang N.-H., Kuo Y.-C., Chu P.-T., Tung H.-Y., Wu Y.-C., Sun J.-L., Liu S.-Y., Li W.-F., Lee W.-H., Lai J.-S., Chang M., Lai M.-T. Immunogenicity of a spike protein subunit-based COVID-19 vaccine with broad protection against various SARS-CoV-2 variants in animal studies. PLOS ONE. 2023;18(3) doi: 10.1371/journal.pone.0283473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.CDC, 2023; SARS-CoV-2 Variant Classifications and Definitions; Accessed from: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html; (accessed on 7 May 2023).
- 66.Kaur S.P., Gupta V. COVID-19 Vaccine: a comprehensive status report. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abu Turab Naqvi A., Fatima K., Mohammad T., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poirier M.C., Divi R.L., Al-Harthi L., et al. Long-term mitochondrial toxicity in HIV-uninfected infants born to HIV-infected mothers. J. Acquir. Immune Defic. Syndr. 2003;33(2):175–183. doi: 10.1097/00126334-200306010-00010. [DOI] [PubMed] [Google Scholar]
- 69.Voysey M., Clemens S.A.C., Madhi S.A., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen C., Wang Z., Zhao F., et al. COVID-19 With Convalescent Plasma. JAMA. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A.…Beigel J.H. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dey S.K., Rahman M.M., Shibly K.H., Siddiqi U.R., Howlader A. Epidemic trend analysis of SARS-CoV-2 in South Asian Association for Regional Cooperation countries using modified susceptible-infected-recovered predictive model. Eng. Rep. 2022 doi: 10.1002/eng2.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22(9):1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L., Borgnia M.J., Haynes B.F., Acharya P. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konishi T. Mutations in SARS-CoV-2 are on the increase against the acquired immunity. PLoS One. 2022;17(7) doi: 10.1371/journal.pone.0271305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu F.C., Guan X.H., Li Y.H., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396(10249):479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thacker P.D. Covid-19: Researcher blows the whistle on data integrity issues in Pfizer’s vaccine trial: Video 1. BMJ. 2021;2635 doi: 10.1136/bmj.n2635. [DOI] [PubMed] [Google Scholar]