Abstract
The arginine deiminase pathway enables Bacillus licheniformis to grow anaerobically on arginine. Both the presence of arginine and anaerobiosis are needed to trigger induction of the pathway. In this study we have cloned and sequenced the arc genes encoding the pathway. They appear clustered in an operon-like structure in the order arcA (arginine deiminase), arcB (ornithine carbamoyltransferase), arcD (putative arginine-ornithine antiporter), arcC (carbamate kinase). It was found that B. licheniformis has an arginine repressor, ArgR, homologous to the B. subtilis arginine repressor AhrC. Mutants affected in argR were isolated. These mutants have lost both repression by arginine of the anabolic ornithine carbamoyltransferase and induction of the arginine deiminase pathway. Electrophoretic band shift experiments and DNase I footprinting revealed that in the presence of arginine, ArgR binds to a site upstream from the arc promoter. The binding site is centered 108 nucleotides upstream from the transcription start point and contains a single Arg box.
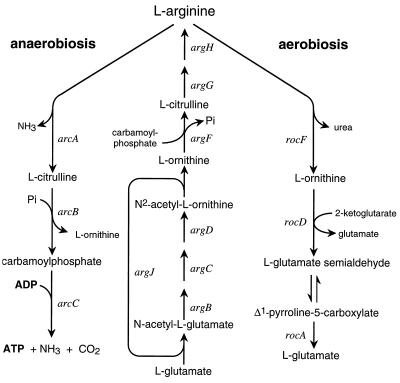
Bacillus licheniformis is a facultative anaerobic bacterium. In the absence of oxygen, it can derive the energy needed for growth from fermentation of glucose, respiration of nitrate, or degradation of l-arginine (7). The arginine deiminase pathway enabling arginine-dependent anaerobic growth consists of three enzymatic steps. In the last step, catalyzed by carbamate kinase, the phosphate group of carbamoylphosphate is used to phosphorylate one ADP to ATP (see Fig. 1). A second pathway of l-arginine catabolism, the arginase pathway (Fig. 1), enables the cell to use arginine as a carbon and nitrogen source (46). Although arginine is required for induction of either pathway, additional regulations prevent the simultaneous presence of both pathways at high levels (7): good aeration favors the arginase pathway, while anaerobic conditions repress it and induce the arginine deiminase pathway (7). Regulation of the latter has been extensively studied in the gram-negative bacterium Pseudomonas aeruginosa, where the pathway is encoded by four genes organized in an operon: arcDABC (5, 38, 47). Expression of the arc genes in P. aeruginosa is up-regulated by the Anr protein, an activator belonging to the Fnr family of transcriptional regulators (20, 21). Anr mediates induction by anaerobiosis. In contrast to the situation in B. licheniformis, induction of the arginine deiminase pathway does not require arginine in P. aeruginosa (41).
FIG. 1.
Pathways of l-arginine biosynthesis and catabolism in B. licheniformis. The genes encode the following enzymes: arginine deiminase (arcA), catabolic ornithine carbamoyltransferase (arcB), carbamate kinase (arcC), N-acetylglutamate synthetase (argJ), N-acetylglutamate 5-phosphotransferase (argB), N-acetylglutamate 5-semialdehyde reductase (argC), N-acetylornithine aminotransferase (argD), anabolic ornithine carbamoyltransferase (argF), argininosuccinate synthetase (argG), argininosuccinase (argH), arginase (rocF), ornithine aminotransferase (rocD), pyrroline-5-carboxylate dehydrogenase (rocA).
Regulation of the aerobic arginase pathway in Bacillus subtilis has been studied. Arginine is a source of nitrogen but not of carbon for B. subtilis, and arginase is its only pathway of arginine catabolism. The operons rocABC (25) and rocDEF (23), which encode the enzymes and permeases of the pathway, have promoter elements at −12 and −24, and their transcription is driven by a specialized sigma factor, a product of the sigL gene, and the B. subtilis homologue of sigma54 (16). Transcription from the promoters of these operons is dependent on the product of the rocR gene (9, 22, 23) and on AhrC (22, 34, 43), initially characterized as the repressor of the biosynthetic genes (43). AhrC is homologous to the Escherichia coli arginine repressor ArgR (44). Several other gram-positive bacteria are reported to possess an ArgR- or AhrC-like protein (18, 45, 48), and it seemed likely that there is one in B. licheniformis, since arginine availability controls the levels of the arginine biosynthetic enzymes (6). We have recently resumed the study of the B. licheniformis arginine deiminase to understand the mechanism of dual regulation by arginine and oxygen. Because AhrC has an activator function in B. subtilis, we tested the hypothesis that an ArgR- or AhrC-like protein plays a similar role in induction of the arginine deiminase pathway in B. licheniformis. This led us to characterize the ArgR protein of B. licheniformis and investigate its role in induction of the arginine deiminase pathway. The arc genes encoding the pathway were cloned and sequenced. The transcription initiation point was identified, and the sequences upstream from it were analyzed to see if some might be involved in regulation.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| Bacillus licheniformis | ||
| ATCC 14580 | Wild type | ATCCa |
| Ahr200-6 | argR | This work |
| Ahr400-6 | argR | This work |
| Ahr400-7 | argR | This work |
| AhrO-3 | argR | This work |
| AhrO-7 | argR | This work |
| Escherichia coli | ||
| C600 OTC− | r− m− Δ(pro argF lac) argI | This laboratory |
| HMS174(DE3)plys S | F−recA rK− mK+ Rifr (DE3) | Novagen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac F′[proAB+ lacIqlacZΔM15 Tn10] | Stratagene |
| Plasmids | ||
| pCR2.1 | T overhang vector for PCR product cloning, Apr Kmr | Invitrogen |
| pET-3a | T7 expression plasmid, Apr | Promega |
| pET-3a.argR | Expression vector for the production of ArgR, Apr | This work |
| YEp52 | Yeast expression vector, LEU2 GAL10 Apr | F. Lacroute |
| pGEM-7Zf(+) | Cloning vector, Apr | Promega |
| pHin4a | pGEM-7Zf(+)::10 kb HindIII fragment containing B. licheniformis arc genes | This work |
| pHin4b | pGEM-7Zf(+)::5 kb HindIII fragment from pHin4a | This work |
ATCC, American Type Culture Collection.
Cultures and growth conditions.
All cultures were grown at 37°C on a rotary shaker. Two culture media were used: Luria-Bertani (LB) broth and minimal medium 154 (57) supplemented with thiamine (1 μg/ml); carbon and nitrogen sources were added to minimal medium to the final concentration of 20 mM, except for arginine, which, in oxygen-limited or anaerobic cultures, was added to the concentration of 40 mM. Two types of culture were used for the enzymatic assays. Well-aerated cultures were obtained by growing the cells in a volume of medium equal to 1/10 of the total volume of the culture flask. Growth was checked by monitoring the optical density at 660 nm, and the cells were harvested in mid-exponential phase. Limited aeration (promoting good induction of the arginine deiminase pathway in the wild type) was achieved as follows. Each strain to be tested was grown as a well-aerated culture on medium 154 plus glutamine to the early exponential phase (3 × 107 cells/ml), and then arginine was added and the culture was poured into a smaller culture flask with a capacity not exceeding 125% of the culture volume. The culture was incubated further for 18 h with shaking. The aerobic start ensured a good cell yield even for mutants impaired in anaerobic growth.
Selection of transformants.
Selection of ampicillin-resistant E. coli transformants was done on LB agar plates supplemented with 50 μg of ampicillin per ml. Selection for argF complementation in transformants of the E. coli C600 derivative was done on solid minimal glucose-ammonium medium supplemented with ampicillin (50 μg/ml) and proline (1 mM).
Selection of hydroxamate-resistant mutants.
Arginine-hydroxamate-resistant colonies appeared spontaneously when the wild-type strain was streaked on solid 154 medium supplemented with glucose, ammonium, and arginine hydroxamate (200 or 400 mM). The resistance mutants were purified on the same medium. Their capacity to grow anaerobically on arginine was tested on solid 154 medium with 40 mM arginine and 20 mM pyruvate. The plates were incubated in a GasPak jar with Anaerocult A (Merck) reagents to produce the anaerobic environment.
Ornithine carbamoyltransferase assay.
Although they function in opposite directions in the cell, both the anabolic and the catabolic ornithine carbamoyltransferases were assayed in vitro for the production of citrulline in the presence of carbamoylphosphate and ornithine. Since they have the same optimum pH, they are distinguishable only by their kinetic behavior (36). The anabolic enzyme has a characteristic saturation curve for ornithine. Its activity peaks at 3 mM ornithine and decreases as the ornithine concentration is further raised, the activity at 50 mM representing only 30% of the activity at 3 mM. The catabolic enzyme displays a typical Michaelis-Menten curve. It is saturated at 8 mM ornithine, and its activity is maintained beyond 50 mM. Ornithine carbamoyltransferase activity was therefore always assayed at two ornithine concentrations (3 and 50 mM) in order to assess whether it was due to the catabolic or the anabolic enzyme. Assays were performed at 37°C according to the method of Broman et al. (6) in a reaction mixture containing 50 mM EDTA-NaOH buffer (pH 8.5), 3 or 50 mM ornithine, 5 mM carbamoylphosphate, and sonicated cell extract. Archibald’s colorimetric assay (2) was used to measure the amount of citrulline formed. Protein concentrations were determined by Lowry’s procedure.
In what follows, specific activities are expressed in micromoles of product formed per hour and per milligram of protein.
DNA manipulations.
Restriction reactions, ligation, and cell transformation were done according to standard procedures (50). Plasmid DNA was purified on Nucleobond AX PC100 columns (Macherey-Nagel).
Determination of the transcription start site.
Primer extension was performed according to the method of Débarbouillé and Raibaud (15) with an RNeasy kit from Qiagen, Moloney murine leukemia virus reverse transcriptase, and 50 to 100 μg of total RNA extracted from oxygen-limited cultures grown on glutamine-arginine medium. Two oligoprimers were used: Adi−P3 (5′-CGTGTATCGGTGTTGTCATGA-3′), which hybridizes with nucleotides 25 to 5 of the arcA gene, and ArcD-C−13 (5′-CCGATGACAAGCGCGATGAGCGCA-3′), which corresponds to nucleotides 53 to 30 of the arcD gene.
PCR conditions.
PCRs were performed with Taq polymerase in a Techne thermocycler (Progene). A typical procedure consisted of 30 cycles (30 s at 94°C, 30 s at 50°C, and 2 min at 72°C), preceded by 3 min at 94°C and followed by 10 min at 72°C. During inverse PCR, the elongation time was adapted to the length of the fragment to be amplified. The reaction mixture (100 μl) composition was as recommended by the manufacturer of the polymerase (Boehringer Mannheim). Genomic DNA to be used as a template in the PCR was purified on a Qiagen RNA/DNA maxi kit column according to the manufacturer’s instructions.
Cloning of a 5′ arcA′ fragment.
The sequence located upstream from the HindIII site and believed to contain the ATG codon and the beginning of the arcA gene was obtained by PCR amplification applied to a genomic library of Sau3A fragments cloned at the BamHI site of plasmid YEp52b. An oligonucleotide (ADI-p, 5′-CGGATCTCTTGTGAAATAAAGGTTCGGC-3′) that hybridizes with nucleotides 464 to 492 of the B. licheniformis arcA gene, combined with an oligonucleotide (Leu-2, 5′-ATAATGGTGAAAGTTCCCTCAAGA-3′) that matches part of the Saccharomyces cerevisiae LEU2 gene carried by this plasmid and located 570 bp from the BamHI cloning site, was used to amplify a fragment of about 1,300 bp, which was cloned in plasmid pCR2.1.
Cloning of the argR gene.
B. subtilis and B. licheniformis being closely related, it was expected that if an AhrC homologue exists in B. licheniformis, it would be very similar to the B. subtilis protein. Therefore, two degenerate oligonucleotides were designed on the basis of the B. subtilis ahrC (44) and Bacillus stearothermophilus argR sequences (18): Dahrc+1 (5′-TGAACAAAGGSCARAGGCATATT-3′) and Dahrc−2 (5′-CCGGCARATRATTAAAMWCGTAT-3′). With this pair of primers, we amplified a 398-bp fragment, which was then cloned in plasmid pCR2.1 and sequenced. Sequence similarities between the fragment and the B. subtilis ahrC gene confirmed that we had indeed cloned part of a homologous gene, which we called argR. Inverse PCR was used to complete the sequence. Genomic DNA was restricted with HindIII. The circular fragments obtained after ligation served as templates for PCR amplification with a pair of divergent primers, ArgR+4 (5′-CTCAGCCAGCCATCTGATTGTGTT-3′) and ArgR−3 (5′-TTCAATTTCATTCGCGGTGATAAT-3′), which match parts of the B. licheniformis sequence. The 1,300-bp fragment amplified in this experiment contained the remainder of the argR sequence. Two primers matching sequences upstream and downstream from the gene (Nd-argR+5, 5′-GCCATATGAACAAAGGTCAAAGG-3′, and Nd-argR−6, 5′-GCCATATGTCAGGCCTTCGTTTA-3′) were then used to amplify the complete gene, which was subsequently cloned in plasmid pCR2.1.
Production and purification of the ArgR protein.
The NdeI sites present on the Nd-argR+5 and Nd-argR−6 oligoprobes used to amplify the argR gene before cloning allowed excision of the argR insert and its subcloning into the NdeI cloning site of the expression vector pET-3a. The correct orientation of the insert was confirmed by the SmaI restriction pattern. The recombinant plasmid was stable in E. coli HMS174(DE3)pLysS. For production of ArgR, the strain was grown in LB broth to mid-exponential phase. Overexpression was then triggered by addition of IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) and allowed to proceed for another three hours before cells were harvested. After sonication of the cells in 10 mM Tris-HCl, pH 8, ArgR was recovered in the centrifugation pellet and solubilized in the extraction buffer by addition of NaCl (final concentration, 1.5 M) according to the procedure used to purify the B. subtilis AhrC protein (14). The protein appeared highly purified after this single step, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the preparations revealed a single band at the expected Mr of 16,800 after the gel was stained with Coomassie brilliant blue. The yield was typically 2 mg of protein for a 100-ml culture. Protein preparations were stored at 4°C.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoresis under native conditions were performed on a Phast System (Pharmacia) with gels in a gradient of 8 to 25% polyacrylamide.
In vitro binding of ArgR.
A 272-bp fragment corresponding to nucleotides −221 to +51 with respect to the transcription initiation point was produced by PCR from genomic DNA; the oligonucleotides used were Arc+P1 (5′-CAGCTTTTTTTGCCCTTCAA-3′) and Adi−P3 (5′-CGTGTATCGGTGTTGTCATGA-3′). One of the two oligoprimers (according to the DNA strand to be visualized in the DNase I footprinting experiments) was labeled at its 5′ end with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase before PCR. The labeled fragment produced by PCR was purified from a nondenaturing 6% (wt/vol) polyacrylamide gel. Band shift experiments and DNase I footprinting (19) were performed on this fragment with purified ArgR protein, as described by Charlier et al. (10).
DNA sequencing.
All DNA sequences were read on both strands of plasmid DNA by the enzymatic chain termination method (51) with a T7 polymerase sequencing kit (Pharmacia) and [α-35S]dATP (Amersham). When clones resulted from ligation of a PCR amplification product (as with the wild-type and mutant argR genes), sequencing was repeated on two different clones obtained from two independent PCRs. For sequencing of the HindIII-BamHI arc fragment, overlapping deletions were created with exonuclease III and S1 nuclease (50). The antiparallel strand was sequenced with synthetic oligonucleotide primers designed to hybridize with the first sequence.
Sequence analysis.
Database searches were done with the BLAST program (1), and multiple alignments of protein sequences were done with CLUSTAL W (58).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this work have been deposited with the DDBJ/EMBLGenBank databases under the accession no. Y17554 (arc sequences) and Y17553 (argR).
RESULTS
Cloning of the arcB gene.
B. licheniformis possesses two ornithine carbamoyltransferases, one specifically associated with arginine biosynthesis and the other associated with arginine catabolism (6) (Fig. 1). In vitro, both of these enzymes catalyze citrulline synthesis with equal efficiencies (6), since the equilibrium constant of the reaction favors citrulline synthesis (3, 56). Therefore, complementation of an argF mutation in E. coli seemed an appropriate strategy for selecting a clone bearing arcB, the gene encoding the catabolic ornithine carbamoyltransferase. Hence, plasmid pHin4a was selected from a genomic library of HindIII restriction fragments in plasmid pGEM-7Zf for its ability to restore growth of an argF argI derivative of E. coli C600 on arginine-free minimal medium. The enzyme produced from the plasmid clearly displayed the kinetic features of the B. licheniformis catabolic carbamoyltransferase. Plasmid pHin4a was found to contain a 10-kb insert. A restriction map of this HindIII restriction fragment was made, and a 5-kb HindIII-BamHI fragment having retained the ability to complement the argF mutation was subcloned (Fig. 2). The HindIII-BamHI fragment of the resulting plasmid pHin4b was entirely sequenced. Sequence analysis revealed three possible open reading frames, preceded by a truncated one, all transcribed in the same direction. The first complete open reading frame (1,008 bp) nearest the HindIII site was easily identified as the arcB gene, which encodes the catabolic ornithine carbamoyltransferase, as the 5′ end of its translated sequence exactly matched the first 23 residues of the N-terminal portion of the purified protein (60a). Furthermore, extensive similarities were found between the B. licheniformis arcB gene product and all other available ornithine carbamoyltransferases, upon alignment of their sequences. The arcB product (335 amino acids, 37.6 kDa) was notably compared with the sequence of the much-studied catabolic ornithine carbamoyltransferase of P. aeruginosa (4) (Fig. 3). The motifs S-T-R-T-R (residues 57 to 61 in P. aeruginosa) and H-P-T-Q (residues 135 to 138 in P. aeruginosa), conserved among carbamoyltransferases (4, 33), are present in the B. licheniformis sequence. Also present are several residues located within and between the S-T-R-T and H-P-T-Q motifs, whose involvement in catalysis or carbamoylphosphate binding has been demonstrated in studies of other carbamoyltransferases: Ser-57, Arg-59, Thr-60, Lys-88, Arg-108, Gly-129, His-135, and Gln-138 (26, 32, 35). The ornithine binding site is present as well, i.e., the Cys-274 residue (27, 40) surrounded by the conserved motif F-M/L-H-C-L-P (residues 271 to 276 in P. aeruginosa).
FIG. 2.
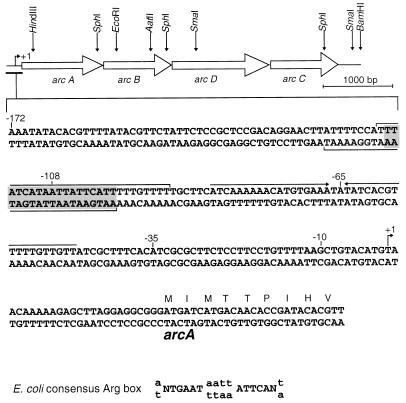
Organization of the arc genes and DNA sequence of the 5′ end of arcA. The transcription initiation site is indicated by a bent arrow at position +1. Nucleotides protected by the ArgR protein are indicated by horizontal brackets. The shaded area contains the Arg box sequence. Convergent arrows mark the inverted repeat containing a Crp- or Fnr-like binding site.
FIG. 3.
Sequence alignment of the catabolic ornithine carbamoyltransferases of B. licheniformis (B.lich.) and P. aeruginosa (P.aeru.) (4). Residues that are identical in the two sequences appear in boldface. Dots mark the conservative substitutions.
Identification of the arcA, arcC, and arcD genes.
Homology searches enabled us to locate the arcC and arcD genes on the same HindIII-BamHI restriction fragment (Fig. 2). The 1,407-bp open reading frame immediately downstream from arcB encodes a 50-kDa protein. The high similarity (46% identity) observed between this protein and the arginine-ornithine antiporter of P. aeruginosa (38) suggests that this open reading frame corresponds to the arcD gene. The third complete open reading frame (951 bp) was identified as the arcC gene. It encodes a 316-residue, 33.7-kDa protein 39% identical to the P. aeruginosa carbamate kinase (5). Analysis of the sequences upstream from the arcB gene revealed the existence of a fourth possible open reading frame, truncated at the HindIII site. Translation of the 1,009 bp of this incomplete open reading frame indicated similarity between its product and P. aeruginosa arginine deiminase, the product of the arcA gene (5). We thus used inverse PCR as detailed in Materials and Methods to obtain the sequence upstream from the HindIII site expected to contain the ATG codon and the beginning of the arcA gene. The initial sequence was extended by 478 bp upstream from the HindIII site, and the ATG codon of the arcA gene was found 230 bp upstream from the site. Sequence analysis showed that the arcA gene (1,242 bp) encodes a 413-residue, 47.4-kDa protein showing 34% identity with the arginine deiminase of P. aeruginosa (5).
Homology searches were carried out with the remaining sequences: the 248 bp upstream from arcA and the 321 bp separating the TAA stop codon of arcC from the downstream BamHI site. No additional open reading frames were identified.
Thus, similarity to P. aeruginosa genes alone enabled us to identify the four open reading frames of the HindIII-BamHI restriction fragment. Even higher similarities were observed when the B. licheniformis sequences were compared with those of the homologous proteins of gram-positive bacteria: Lactobacillus sake (62), with 58% identity in arcA, 67% in arcB, 51% in arcC, and 43% in arcD, and Clostridium perfringens (45), with 53% identity in arcA, 56% in arcB, 51% in arcD, and 42% in arcC.
Analysis of the arc cluster.
Sequencing of the HindIII-BamHI fragment showed that the four arc genes encoding the entire arginine deiminase pathway are clustered on the B. licheniformis chromosome. They are transcribed in the same direction and separated by small intergenic spaces: 28 bp between arcA and arcB, 53 bp between arcB and arcD, and 21 bp between arcD and arcC. A typical ribosome binding site was recognized upstream from each gene: GAGG upstream from the ATG codons of arcA and arcD, GGAGAG (arcB), and AGAGG (arcC). Primer extension experiments were carried out to locate transcriptional start points in the cluster. Two different oligoprimers were used: Adi−P3 (which hybridizes in arcA) and ArcD-C−13 (which hybridizes in arcD). The choice of an oligonucleotide complementary to arcD was justified by the somewhat larger intergenic space and the presence of an inverted repeat at the end of arcB that might belong to a terminator. No transcription start point was found in the 200 bp immediately upstream from the arcD gene. Mapping of the extension products indicated that the adenine located 25 nucleotides upstream from the arcA ATG codon is the transcriptional starting point (data not shown).
Two interesting features were discovered upstream from the transcription initiation point, with possible implications for transcriptional regulation of the arc genes (Fig. 2). In the sequence 5′-TTATCATAATTATTCATT-3′ spanning positions −116 to −99 with respect to the transcription start point, 15 of the 18 nucleotides match the E. coli consensus Arg box (10, 13, 39) (Fig. 2). Similar Arg boxes are found on the biosynthetic argCJBD-cpa-argF operons of B. subtilis and B. stearothermophilus (52, 53), where they are the targets of the arginine repressor.
Downstream from the putative Arg box, in the middle of a zone of dyad symmetry centered at −65.5, the imperfect palindromic sequence 5′-ACATGTGAAATATATCACGTTT-3′ is very close to the Crp consensus sequence of E. coli (5′-AA-TGTGA--T---TCACA-T-T-3′) (24) and to the putative Fnr binding site of B. subtilis (5′-A-A-TTGAT--A-ATCAAT----3′) (12). The Fnr and Crp proteins belong to the same family of regulatory proteins and bind to similar, well-conserved sequences (55). The existence of two possible binding sites very near the arc promoter suggests the involvement of at least two regulatory proteins, one of the ArgR or AhrC type and one Fnr family member.
Sequence analysis of the argR gene.
The strategy used to clone the B. licheniformis argR gene was based on the assumed high homology between the arginine repressors of B. licheniformis and B. subtilis. This homology was confirmed by alignment of the complete sequences: ArgR is 89% identical to its B. subtilis homologue (44) and 71% identical to its B. stearothermophilus homologue (18). Other gram-positive ArgR proteins are much less similar, the percentages of identical residues being in the same range as for the proteins of gram-negative bacteria; the B. licheniformis protein is 34% identical to Mycobacterium tuberculosis ArgR (accession no. Z85982), 31% identical to the C. perfringens protein (45), and 32% identical to the E. coli protein (37).
The B. licheniformis arginine regulator contains both highly conserved domains already revealed by aligning the B. subtilis (44) and B. stearothermophilus (18) sequences with the E. coli sequence (37) (Fig. 4). One domain, located in the N-terminal part of the protein, is the DNA binding site (28), which contains, notably, the SR motif (residues 47 and 48 in E. coli) involved in DNA binding (60). The second domain, in the C-terminal region, is the arginine-binding domain, defined by successive studies of E. coli (8, 60, 61). In the E. coli protein, this domain spans residues 80 to 156 and contains residues A105, D128, and D129, the substitution of which affects arginine binding (8, 60), and G123, involved in oligomerization (11, 60). All the corresponding amino acids are conserved in both B. licheniformis and B. subtilis.
FIG. 4.
Sequence alignment of the B. licheniformis ArgR protein (B.l) with the arginine repressors AhrC of B. subtilis (B.s) (44) and ArgR of E. coli (E.c) (37). Asterisks mark the identical residues in the three sequences, and dots mark the conservative substitutions. Residues known to be involved in DNA binding in E. coli are indicated by circles, those involved in arginine binding are indicated by filled squares, and gly-124, required for oligomerization, is indicated by an open square. The amino acids modified in the argR mutants selected during this study appear in boldface.
Characterization of the ArgR protein.
The 447 bp of the argR gene encodes a 149-residue polypeptide chain with a calculated molecular mass of 16.82 kDa. Purified ArgR protein electrophoresed under native conditions revealed a major band at 100 kDa. This result is consistent with a hexameric structure, as for the B. subtilis (14) and E. coli (37) arginine repressors. A very faint band at 50 kDa may be due to the presence of the trimeric form in small proportion.
ArgR binds in vitro to a region upstream from the arc promoter.
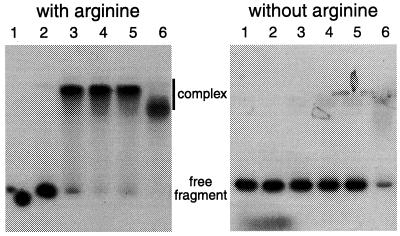
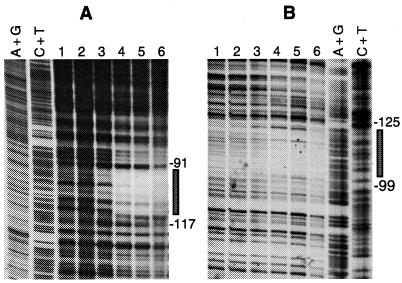
By producing and purifying the B. licheniformis ArgR protein we were able to test whether its target is indeed the putative Arg box upstream from the arc promoter. Band shift assays clearly showed that purified ArgR binds to a 272-bp DNA fragment bearing the arc promoter (nucleotides −221 to +51 with respect to the transcription start site), which reduces its electrophoretic mobility, and that this binding is arginine dependent (Fig. 5). In footprinting experiments with the same 272-bp fragment, binding of ArgR protected a single stretch of 27 nucleotides on each strand against DNase I digestion (Fig. 2 and 6). There was only a 19-bp overlap, however, between the regions protected on the two strands. This stretch, centered at −108, contains the putative Arg box revealed by sequence analysis. While the limits of the Arg box and of the protected region differ by no more than 1 nucleotide at the 5′ end, protection is extended by 8 or 9 nucleotides beyond the limit of the Arg box towards the 3′ end. The difference is probably too large to be solely attributed to the recognition and cutting mode of DNase I in the minor groove of the DNA helix. Similar differences have been observed at other ArgR binding sites (18, 34). The length of the protected region (27 bp) is markedly smaller than the region protected by ArgR in the different E. coli operators (on average, 40 bp), where two Arg boxes are present (10, 59). In the operators of the B. subtilis genes involved in arginine catabolism, only one Arg box has been identified and AhrC again protects smaller regions: 17 nucleotides in OrocD and 19 to 21 nucleotides in OrocA (42).
FIG. 5.
Gel mobility shift assays with ArgR in the absence and presence of arginine (10 mM). The 272-bp fragment encompasses nucleotides −221 to +51 with respect to the transcription start site. The ArgR concentrations were as follows: none (lane 1), 0.05 μM (lane 2), 0.2 μM (lane 3), 0.4 μM (lane 4), 0.9 μM (lane 5), and 4.3 μM (lane 6).
FIG. 6.
DNase I footprinting of B. licheniformis ArgR protein binding to the same 272-bp operator fragment as that shown in Fig. 5. (A) Upper strand; (B) lower strand. Footprinting was done in the presence of arginine (10 mM) with the following protein concentrations: none (lane 1), 4.5 nM (lane 2), 45 nM (lane 3), 90 nM (lane 4), 450 nM (lane 5), and 900 nM (lane 6). Reference ladders (A+G and C+T) were generated by the enzymatic chain termination method (A) and by Maxam and Gilbert sequencing (B). The protected region on each strand is indicated by a rectangle.
Induction of the arginine deiminase pathway does not occur in argR mutants.
In B. subtilis, mutations in the gene coding for the AhrC arginine repressor confer resistance to arginine hydroxamate, an arginine analogue. In ahrC mutants, the arginine-biosynthetic enzymes are present at high, constitutive levels and their synthesis is not repressed in the presence of exogenous arginine (29, 43). Arginine hydroxamate is also a growth inhibitor of B. licheniformis. Spontaneously arising mutants displaying resistance to arginine hydroxamate were selected on analogue-supplemented minimal glucose-ammonium medium. Five such mutants (Ahr200-6, Ahr400-6, Ahr400-7, AhrO-3, and AhrO-7) had a phenotype consistent with an argR or ahrC mutation, showing high constitutive ornithine carbamoyltransferase activity on well-aerated glucose-ammonium medium (Table 2). The ratio of the activity measured at 50 mM ornithine to that measured at 3 mM ornithine confirmed that the activity was due to the anabolic ornithine carbamoyltransferase. Although some mutants (Ahr200-6, Ahr400-6, and Ahr400-7) displayed a somewhat different activity when arginine was added to the glucose-ammonium medium, this was probably not significant, as the standard deviation of such high activities can represent up to 23% of the specific activity measured (Table 2). All constitutive mutants were impaired in their ability to grow anaerobically on arginine-pyruvate medium, apparently as a consequence of their inability to induce the arginine deiminase pathway (Table 2). During oxygen-limited growth, they displayed low specific ornithine carbamoyltransferase activity, resulting from the presence of residual catabolic and anabolic enzyme.
TABLE 2.
Phenotypes of the argR mutants
| Strain | Mutation | Anabolic OCT sp acta in:
|
Catabolic OCT sp acta under oxygen-limited conditionsc | Arginine-dependent anaerobic growth | |
|---|---|---|---|---|---|
| Minimal mediumb | Minimal medium + arginine | ||||
| ATCC 14580 | Wild type | 16 (3.4) | 3 (0.5) | 6,155 (435) | + |
| Ahr200-6 | G124R | 144 (20) | 152 (19) | 40 (9.3) | − |
| Ahr400-6 | Q142stop | 122 (24) | 160 (25) | 44 (6.5) | − |
| Ahr400-7 | A103V | 108 (7.5) | 150 (35) | 68 (19) | − |
| AhrO-3 | D111G | 94 (13) | 79 (9.5) | 14 (5.3) | − |
| AhrO-7 | C132Y | 159 (20) | 159 (5) | 11 (5.7) | − |
OCT sp act, ornithine carbamoyltransferase specific activities. Specific activities are defined as micromoles of citrulline formed per hour per milligram of protein. Standard deviations are given within parentheses.
Minimal medium is medium 154 supplemented with 20 mM glucose and 10 mM (NH4)2SO4.
Oxygen-limited conditions were achieved in medium 154 with glutamine as described in Materials and Methods.
The argR genes of the mutants were cloned and sequenced in the same manner as the wild-type gene, after amplification by PCR. Sequencing confirmed that mutants Ahr200-6, Ahr400-6, Ahr400-7, AhrO-3, and AhrO-7 had each acquired a mutation by substitution of 1 nucleotide in the argR gene (Table 2 and Fig. 4).
The absence of arginine deiminase pathway induction in the argR mutants is a direct consequence of a mutated ArgR. Resistance to arginine hydroxamate cannot in itself explain it, since two of the arginine hydroxamate-resistant mutants studied displayed high but repressible specific anabolic ornithine carbamoyltransferase activity in aerobic cultures and normal induction of the arginine deiminase pathway after oxygen depletion (data not shown). These mutants turned out, upon sequencing, to have an intact argR gene.
All five argR mutations were located in the C-terminal part of the protein, near the putative arginine-binding site. In the following description, residue numbers refer to the B. licheniformis sequence. Three mutations, G124R in Ahr200-6, A103V in Ahr400-7, and D111G in AhrO-3, affect conserved residues of the arginine binding site (Fig. 4). Previous studies of the E. coli arginine repressor have proved the involvement of the A103 residue (A105 in E. coli) in arginine binding (60). The existence of mutant Ahr400-6, whose ArgR lacks its last eight residues, confirms the importance of the C-terminal part. The C132Y substitution in mutant AhrO-7 may affect a residue related to specific properties of the Bacillus arginine repressors, as this cysteine residue is present in other Bacillus species (Fig. 5) but not in E. coli or other gram-negative bacteria.
DISCUSSION
By cloning and sequencing the arc genes encoding the arginine deiminase pathway of Bacillus licheniformis, we have shown that these genes are clustered on the genome of this species. The arc genes appear in the same order as in another gram-positive bacterium, C. perfringens (45): arcA-arcB-arcD-arcC. A different order, (arcD)-arcA-arcB-arcC, is found in the gram-negative bacteria P. aeruginosa (4, 5, 38) and Rhizobium etli (17). Mycoplasma pneumoniae (31) and Halobacterium salinarium (49) also display a different organization of the arc genes.
In B. licheniformis, a protein homologous to the arginine regulator AhrC of B. subtilis acts as a repressor of the arginine-biosynthetic pathway. The DNase I footprinting data show that the purified protein, called ArgR, binds to a single Arg box, revealed by analysis of the long noncoding sequence preceding arcA. We have isolated five argR mutants by selection for resistance to the arginine analogue arginine hydroxamate. Their phenotype confirms that in the presence of arginine, B. licheniformis ArgR is both a repressor of the anabolic ornithine carbamoyltransferase and an activator of the arginine deiminase pathway. The inability of the argR mutants to induce the arginine deiminase pathway after oxygen depletion strongly suggests that induction of the arginine deiminase pathway by arginine requires binding of the ArgR protein to the Arg box located upstream from the arc promoter.
Several differences between activation of the arc genes by ArgR in B. licheniformis and activation of the rocABC and rocDEF operons by AhrC in B. subtilis are apparent. In the roc operons, the region recognized by AhrC is immediately adjacent to the transcription start point (34, 42), while in B. licheniformis, ArgR binds 108 bp upstream from the arc transcription start point. Moreover, the promoters of the roc operons are of the −12/−24 type; their transcription requires a specific sigma factor, a product of the sigL gene (16), and binding of the RocR activator to two UAS sequences (9, 22, 23). It has been suggested that activation results from DNA bending induced by AhrC binding (34, 42), but on the other hand the recent evidence presented by Gardan et al. (22) suggests that interaction between the AhrC and RocR proteins directly causes activation. The arc promoter of B. licheniformis bears no resemblance to the −12/−24 roc promoters, and there are no stretches resembling the Bacillus ςA-dependent consensus promoter sequence at −10 and −35 (30). The Bacillus licheniformis promoter does contain, between the ArgR binding site and the site covered by RNA polymerase, an imperfect palindromic sequence similar to the E. coli Crp and B. subtilis Fnr consensus sequences. This finding suggests the involvement of a second regulatory protein, possibly belonging to the Crp or Fnr family. Since many Crp or Fnr family members are involved in anaerobic regulations (54), it is tempting to hypothesize that the second regulatory protein is the anaerobic activator of the system.
ACKNOWLEDGMENTS
This work was supported by the Fonds pour l’Encouragement de la Recherche (Université Libre de Bruxelles) and the Belgian Fund for Joint Basic Research.
We thank J.-P. Ten Have for his skillful assistance with the figures.
REFERENCES
- 1.Altschul S, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Archibald R M. Determination of citrulline and allantoin and demonstration of citrulline in blood plasma. J Biol Chem. 1944;156:121–142. [Google Scholar]
- 3.Baur H, Tricot C, Stalon V, Haas D. Converting catabolic ornithine carbamoyltransferase to an anabolic enzyme. J Biol Chem. 1990;265:14728–14731. [PubMed] [Google Scholar]
- 4.Baur H, Stalon V, Falmagne P, Lüthi E, Haas D. Primary and quaternary structure of the catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa. Eur J Biochem. 1987;166:111–117. doi: 10.1111/j.1432-1033.1987.tb13489.x. [DOI] [PubMed] [Google Scholar]
- 5.Baur H, Lüthi E, Stalon V, Mercenier A, Haas D. Sequence analysis and expression of the arginine deiminase and carbamate kinase genes of Pseudomonas aeruginosa. Eur J Biochem. 1989;179:53–60. doi: 10.1111/j.1432-1033.1989.tb14520.x. [DOI] [PubMed] [Google Scholar]
- 6.Broman K, Stalon V, Wiame J-M. The duplication of arginine catabolism and the meaning of the two ornithine carbamoyltransferases in Bacillus licheniformis. Biochem Biophys Res Commun. 1975;66:821–827. doi: 10.1016/0006-291x(75)90583-5. [DOI] [PubMed] [Google Scholar]
- 7.Broman K, Lauwers N, Stalon V, Wiame J-M. Oxygen and nitrate in utilization by Bacillus licheniformis of the arginase and arginine deiminase routes of arginine catabolism and other factors affecting their synthesis. J Bacteriol. 1978;135:920–927. doi: 10.1128/jb.135.3.920-927.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke M, Merican A, Sherratt D. Mutant Escherichia coli arginine repressor proteins that fail to bind l-arginine, yet retain the ability to bind their normal DNA-binding sites. Mol Microbiol. 1994;13:609–618. doi: 10.1111/j.1365-2958.1994.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 9.Calogero S, Gardan R, Glaser P, Schweitzer J, Rapoport G, Débarbouillé M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlier D, Roovers M, Van Vliet F, Boyen A, Cunin R, Nakamura Y, Glansdorff N, Piérard A. Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J Mol Biol. 1992;226:367–386. doi: 10.1016/0022-2836(92)90953-h. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Merican A, Sherratt D. DNA binding of Escherichia coli arginine repressor mutants altered in oligomeric state. Mol Microbiol. 1997;24:1143–1156. doi: 10.1046/j.1365-2958.1997.4301791.x. [DOI] [PubMed] [Google Scholar]
- 12.Cruz-Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunin R, Eckhardt T, Piette J, Boyen A, Piérard A, Glansdorff N. Molecular basis for modulated regulation of gene expression in the arginine regulon of Escherichia coli. Nucleic Acids Res. 1983;11:5007–5019. doi: 10.1093/nar/11.15.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czaplewski L, North A, Smith M, Baumberg S, Stockley P. Purification and initial characterization of AhrC; the regulator of arginine metabolism genes in Bacillus subtilis. Mol Microbiol. 1992;6:267–275. doi: 10.1111/j.1365-2958.1992.tb02008.x. [DOI] [PubMed] [Google Scholar]
- 15.Débarbouillé M, Raibaud O. Expression of the Escherichia coli malPQ operon remains unaffected after drastic alteration of its promoter. J Bacteriol. 1983;153:1221–1227. doi: 10.1128/jb.153.3.1221-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of ς54 from Gram-negative bacteria. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Hooge I, Vander Wauven C, Michiels J, Tricot C, de Wilde P, Vanderleyden J, Stalon V. The arginine deiminase pathway in Rhizobium etli: DNA sequence and functional study of the arcABC genes. J Bacteriol. 1997;179:7403–7409. doi: 10.1128/jb.179.23.7403-7409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dion M, Charlier D, Wang H, Savchenko A, Hallet J-N, Glansdorff N, Sakanyan V. The extremely thermostable arginine repressor of Bacillus stearothermophilus: gene cloning and repressor-operator interaction. Mol Microbiol. 1997;25:385–398. doi: 10.1046/j.1365-2958.1997.4781845.x. [DOI] [PubMed] [Google Scholar]
- 19.Galas D J, Schmitz A. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galimand M, Gamper M, Zimmermann A, Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamper M, Zimmermann A, Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J Bacteriol. 1991;173:4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardan R, Rapoport G, Débarbouillé M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol Microbiol. 1997;24:825–838. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 23.Gardan R, Rapoport G, Débarbouillé M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 24.Gaston K, Kolb A, Busby S. Binding of the Escherichia coli cyclic-AMP receptor protein to DNA fragments containing consensus nucleotide-sequences. Biochem J. 1989;261:649–653. doi: 10.1042/bj2610649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser P, Kunst F, Arnaud M, Coudart M-P, Gonzales W, Hullo M-F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweitzer J, Vertès A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97 kb region from 325° to 333°. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 26.Goldsmith J O, Kuo L C. Protonation of arginine 57 of Escherichia coli ornithine carbamoyltransferase regulates substrate binding and turnover. J Biol Chem. 1993;268:18485–18490. [PubMed] [Google Scholar]
- 27.Goldsmith J O, Lee S, Zambiditis I, Kuo L C. Control of l-ornithine specificity in Escherichia coli ornithine transcarbamoylase. J Biol Chem. 1991;266:18626–18634. [PubMed] [Google Scholar]
- 28.Grandori R, Lavoie T A, Pflumm M, Tian G, Nierbach H, Maas W K, Fairman R, Carey J. The DNA-binding domain of the hexameric arginine repressor. J Mol Biol. 1995;254:150–162. doi: 10.1006/jmbi.1995.0607. [DOI] [PubMed] [Google Scholar]
- 29.Harwood C R, Baumberg S. Arginine hydroxamate-resistant mutants of Bacillus subtilis with altered control of arginine metabolism. J Gen Microbiol. 1977;100:177–188. doi: 10.1099/00221287-100-1-177. [DOI] [PubMed] [Google Scholar]
- 30.Helmann J. Compilation and analysis of Bacillus subtilis ςA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honzatko R B, Lipscomb W N. Interaction of phosphate ligands with Escherichia coli aspartate carbamoyltransferase in the crystalline state. J Mol Biol. 1982;160:265–286. doi: 10.1016/0022-2836(82)90176-0. [DOI] [PubMed] [Google Scholar]
- 33.Houghton J E, Bencini D A, O’Donovan G A, Wild J R. Protein differentiation: a comparison of aspartate transcarbamoylase and ornithine carbamoyltransferase from Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4864–4868. doi: 10.1073/pnas.81.15.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klingel U, Miller C, North A, Stockley P, Baumberg S. A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/activator of arginine metabolism. Mol Gen Genet. 1995;248:329–340. doi: 10.1007/BF02191600. [DOI] [PubMed] [Google Scholar]
- 35.Kuo L C, Miller A W, Lee S, Kozuma C. Site-directed mutagenesis of Escherichia coli ornithine carbamoyltransferase: the role of arginine-57 in substrate binding and catalysis. Biochemistry. 1988;27:8823–8832. doi: 10.1021/bi00424a021. [DOI] [PubMed] [Google Scholar]
- 36.Laishley E J, Bernlohr R W. The regulation and kinetics of the two ornithine transcarbamylase enzymes of Bacillus licheniformis. Biochim Biophys Acta. 1968;167:547–554. doi: 10.1016/0005-2744(68)90044-2. [DOI] [PubMed] [Google Scholar]
- 37.Lim D, Oppenheim J D, Eckhardt T, Maas W K. Nucleotide sequence of the argR gene of Escherichia coli K12 and isolation of its product, the arginine repressor. Proc Natl Acad Sci USA. 1987;84:6697–6701. doi: 10.1073/pnas.84.19.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lüthi E, Baur H, Gamper M, Brunner F, Villeval D, Mercenier A, Haas D. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene. 1990;87:37–43. doi: 10.1016/0378-1119(90)90493-b. [DOI] [PubMed] [Google Scholar]
- 39.Maas W. The arginine repressor of Escherichia coli. Microbiol Rev. 1994;58:631–640. doi: 10.1128/mr.58.4.631-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall M, Cohen P P. Ornithine transcarbamylases. J Biol Chem. 1980;255:7287–7290. [PubMed] [Google Scholar]
- 41.Mercenier A, Simon J P, Vander Wauven C, Haas D, Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980;144:159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller C, Baumberg S, Stockley P. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at the catabolic sites. Mol Microbiol. 1997;26:37–48. doi: 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 43.Mountain A, Baumberg S. Map locations of some mutations conferring resistance to arginine hydroxamate in Bacillus subtilis 168. Mol Gen Genet. 1980;178:691–701. doi: 10.1007/BF00337880. [DOI] [PubMed] [Google Scholar]
- 44.North A K, Smith M C, Baumberg S. Nucleotide sequence of Bacillus subtilis arginine regulatory gene and homology of its product to the Escherichia coli arginine repressor. Gene. 1989;80:29–38. doi: 10.1016/0378-1119(89)90247-3. [DOI] [PubMed] [Google Scholar]
- 45.Ohtani K, Bando M, Swe T, Banu S, Oe M, Hayashi H, Shimizu T. Collagenase gene (colA) is located in the 3′-flanking region of the perfringolysin O (pfoA) locus in Clostridium perfringens. FEMS Microbiol Lett. 1997;146:155–159. doi: 10.1111/j.1574-6968.1997.tb10186.x. [DOI] [PubMed] [Google Scholar]
- 46.Ramaley R F, Bernlohr R W. Apparent induction of ornithine transcarbamylase and arginase by arginine in Bacillus licheniformis. J Mol Biol. 1965;11:842–844. doi: 10.1016/s0022-2836(65)80041-9. [DOI] [PubMed] [Google Scholar]
- 47.Rella M, Mercenier A, Haas D. Transposon insertion mutagenesis of Pseudomonas aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene. 1985;33:293–303. doi: 10.1016/0378-1119(85)90237-9. [DOI] [PubMed] [Google Scholar]
- 48.Rodríguez-García A, Ludovice M, Martín J, Liras P. Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol Microbiol. 1997;25:219–228. doi: 10.1046/j.1365-2958.1997.4511815.x. [DOI] [PubMed] [Google Scholar]
- 49.Ruepp A, Soppa J. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J Bacteriol. 1996;178:4942–4947. doi: 10.1128/jb.178.16.4942-4947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. In Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savchenko A, Charlier D, Dion M, Weigel P, Hallet J-N, Holtam C, Baumberg S, Glansdorff N, Sakanyan V. The arginine operon of Bacillus stearothermophilus: characterization of the control region and its interaction with the heterologous B. subtilis arginine repressor. Mol Gen Genet. 1996;252:69–78. doi: 10.1007/BF02173206. [DOI] [PubMed] [Google Scholar]
- 53.Smith M C M, Czaplewski L, North A K, Baumberg S, Stockley P G. Sequences required for regulation of arginine biosynthesis promoters are conserved between Bacillus subtilis and Escherichia coli. Mol Microbiol. 1989;3:23–28. doi: 10.1111/j.1365-2958.1989.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 54.Spiro S. The Fnr family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 55.Spiro S, Gaston K, Bell A, Roberts R, Busby S, Guest J. Interconversion of the DNA-binding specificities of the two related transcription regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 56.Stalon V, Legrain C, Wiame J-M. Anabolic ornithine carbamoyltransferase of Pseudomonas aeruginosa: the bases of its functional specialization. Eur J Biochem. 1977;74:319–327. doi: 10.1111/j.1432-1033.1977.tb11396.x. [DOI] [PubMed] [Google Scholar]
- 57.Stalon V, Ramos F, Piérard A, Wiame J M. The occurrence of a catabolic and an anabolic ornithine carbamoyltransferase in Pseudomonas fluorescens. Biochim Biophys Acta. 1967;139:91–97. doi: 10.1016/0005-2744(67)90115-5. [DOI] [PubMed] [Google Scholar]
- 58.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian G, Lim D, Carey J, Maas W K. Binding of the arginine repressor of Escherichia coli K12 to its operator sites. J Mol Biol. 1992;226:387–397. doi: 10.1016/0022-2836(92)90954-i. [DOI] [PubMed] [Google Scholar]
- 60.Tian G, Maas W K. Mutational analysis of the arginine repressor of Escherichia coli. Mol Microbiol. 1994;13:599–608. doi: 10.1111/j.1365-2958.1994.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 60a.Vander Wauven, C., and V. Stalon. Unpublished results.
- 61.Van Duyne G, Ghosh G, Maas W, Sigler P. Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J Mol Biol. 1996;256:377–391. doi: 10.1006/jmbi.1996.0093. [DOI] [PubMed] [Google Scholar]
- 62.Zúñiga M, Champomier-Verges M, Zagorec M, Pérez-Martínez G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol. 1998;180:4154–4159. doi: 10.1128/jb.180.16.4154-4159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]