Abstract
The Escherichia coli gapB gene codes for a protein that is very similar to bacterial glyceraldehyde-3-phosphate dehydrogenases (GAPDH). In most bacteria, the gene for GAPDH is located upstream of the pgk gene encoding 3-phosphoglycerate kinase (PGK). This is the case for gapB. However, this gene is poorly expressed and encodes a protein with an erythrose 4-phosphate dehydrogenase activity (E4PDH). The active GAPDH is encoded by the gapA gene. Since we found that the nucleotide region upstream of the gapB open reading frame is responsible for part of the PGK production, we analyzed gapB promoter activity in vivo by direct measurement of the mRNA levels by reverse transcription. We showed the presence of a unique transcription promoter, gapB P0, with a cyclic AMP (cAMP) receptor protein (CRP)-cAMP binding site centered 70.5 bp upstream of the start site. Interestingly, the gapB P0 promoter activity was strongly enhanced when glucose was used as the carbon source. In these conditions, deletion of the CRP-cAMP binding site had little effect on promoter gapB P0 activity. In contrast, abolition of CRP production or of cAMP biosynthesis (crp or cya mutant strains) strongly reduced promoter gapB P0 activity. This suggests that in the presence of glucose, the CRP-cAMP complex has an indirect effect on promoter gapB P0 activity. We also showed that glucose stimulation of gapB P0 promoter activity depends on the expression of enzyme IIGlc (EIIGlc), encoded by the ptsG gene, and that the gapA P1 promoter is also activated by glucose via the EIIGlc protein. A similar glucose-mediated activation, dependent on the EIIGlc protein, was described by others for the pts operon. Altogether, this shows that when glucose is present in the growth medium expression of the E. coli genes required for its uptake (pts) and its metabolism (gapA and gapB-pgk) are coordinately activated by a mechanism dependent upon the EIIGlc protein.
Most bacteria are able to sense the availability of nutrients in their growth environment. In the presence of different carbon sources, and in order to preserve energy, unnecessary genes are not expressed, either by inactivation of some specific transport system and/or by regulation of the intracellular cyclic AMP (cAMP) level (for a review, see reference 30). Regulation of Escherichia coli lac operon expression during growth on glucose and lactose is often cited as the paradigm of such an adaptation (references 26 and 30 and references therein).
Regulation of the E. coli genes required for glucose permeation and metabolism is less documented. Glucose uptake has been shown to activate transcription of the pts operon (12–14). The ptsH, ptsI, and crr genes, encoding the histidine-containing phosphocarrier protein Hpr, enzyme I (EI), and the cytoplasmic protein EIII (EIIAGlc), respectively, constitute the pts operon. The promoter region upstream of the ptsH gene contains at least two transcription start sites (P0 and P1) separated by 100 bp. Initiation at P0 is enhanced by the presence of exogenous glucose (12), whereas in the absence of glucose, initiation depends strictly on the presence of the complex formed between the cAMP receptor protein (CRP) and cAMP (CRP-cAMP complex) (14, 20). The glucose-mediated and CRP-cAMP-mediated activations arise independently of each other (13, 14, 33). It was proposed that the glucose-mediated activation occurs through a signal transduction mechanism dependent upon the phosphorylation state of the EIIGlc protein (EIIBCGlc) produced by the ptsG gene (13). Nevertheless, the molecular mechanism of the positive effect of glucose on pts transcription has not been determined. Domain C of the ptsG product is membrane embedded, whereas domain B, carrying the phosphorylation sites, is cytoplasmic (for a review, see reference 30). No DNA binding property was reported for either of these two domains. This suggests an indirect involvement of the EIIGlc protein in transcription activation.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (EC 1.2.1.12) is a key enzyme of glucose metabolism. It plays a crucial role in catabolic and anabolic carbohydrate metabolism, catalyzing the reversible oxidation of d-glyceraldehyde-3-phosphate into 1,3-diphosphoglycerate (22). In most bacteria studied so far, the GAPDH-encoding gene (gap) is found upstream of the pgk gene encoding phosphoglycerate kinase in a cluster of genes encoding other glycolytic enzymes (e.g., see references 6, 10, 17, 19, and 36). E. coli is unusual in this respect. It has two GAPDH-encoding genes, designated gapA and gapB (2). The gapB gene is localized upstream of the pgk gene at 61.9 min on the chromosome (38). Despite the fact that the gapB open reading frame (ORF) codes for a protein that is very similar to bacterial GAPDHs (2), no GAPDH activity was measured for this gene product (3, 39). Instead, the protein was shown to be expressed at a very low level and displayed a nonphosphorylating erythrose 4-phosphate dehydrogenase (E4PDH) activity (3, 39). The gapA gene was located in a completely different region of the E. coli chromosome, at 39.3 min (24), and codes for a protein that is more similar to eukaryotic than to eubacterial GAPDHs (4). Since mutations in the gapA gene abolish the production of GAPDH activity, the gapA gene is considered to be the only active GAPDH-encoding gene in E. coli (5, 11, 24). The gapA gene is transcribed from at least four promoters (8). Three are well identified: P1 and P3 are transcribed by the Eς70 holoenzyme and P2 is transcribed by the heat shock RNA polymerase Eς32. In addition, P3 is regulated by catabolic repression. We have shown previously that these three transcription start sites are activated differentially in cultures in rich medium (8).
No extensive study of chromosomal gapB gene expression has been reported. As Alefounder and Perham (2) localized by sequence analysis a putative CRP-cAMP binding sequence, centered 202 nucleotides (nt) upstream of the gapB ATG initiation codon, catabolic repression might be implicated in gapB regulation (see Fig. 2B). Gel shift experiments showed the binding of a CRP-cAMP complex to this sequence (31). A putative FruR binding site was also localized in the gapB promoter region, and FruR was shown to repress expression of a gapB-lacZ fusion (31). Based on sequence analysis and transposon insertion mutagenesis, the gapB and pgk genes seem to be transcribed into a bicistronic mRNA (2, 28). However, the two genes are expressed at very different levels: the GAPB protein is present at very low levels (3, 39), whereas PGK, like GAPDH and other glycolytic enzymes, is expressed at high levels in cells. Using E. coli strains transformed with two distinct plasmids, one (pPBK500) containing the entire gapB-pgk gene cluster and including a 812-nt sequence upstream of gapB and one (pBK200) missing this upstream sequence, we showed that 59% of the PGK production depends upon the nucleotide sequence upstream of gapB. This prompted us to start a detailed analysis of the transcriptional properties of the DNA sequence upstream of gapB, and the results obtained are described in this paper.
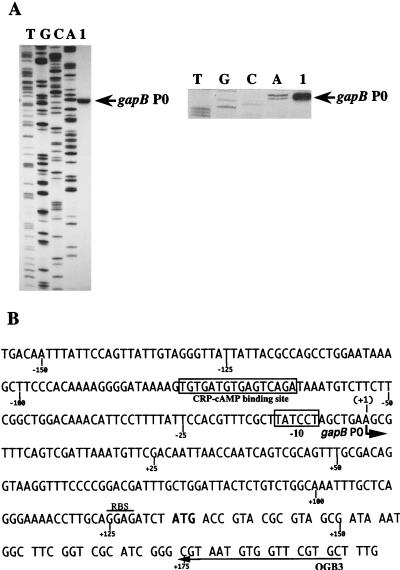
FIG. 2.
Localization of the in vivo gapB transcription start site. (A) Primer extension was performed with oligonucleotide OGB3 and with total RNA extracted from TG1 cells grown on M63 medium supplemented with glucose as the sole carbon source (lane 1). Total RNA extraction and primer extension were performed as described in Materials and Methods. Markers (lanes A, C, G, and T) were made by sequencing plasmid pPBK100 with oligonucleotide OGB3 and T7 DNA polymerase. The position of the initiation site is indicated (gapB P0) on the gel autoradiogram (left) and on the enlargement of the area corresponding to the initiation site (right). (B) DNA sequence of the transcriptional regulatory region of the E. coli gapB gene (2). The in vivo transcription initiation site gapB P0 identified in panel A is marked with a bent arrow. The first nucleotide of the gapB mRNA is numbered +1 (the preceding one is numbered −1). The −10 region and the CRP-cAMP binding sequence are boxed. The arrow labeled OGB3 shows the sequence which is complementary to the primer oligonucleotide used for primer extension analysis. The ribosome binding sequence (RBS) is overlined.
The data presented in this paper were obtained by primer extension analysis of in vivo-produced mRNAs and by measurement of the specific activities of GAPDH and PGK enzymes in soluble fraction of E. coli cells. Using these approaches, we showed that the DNA region upstream of gapB contains a single promoter, P0, with interesting features. Both glucose and the CRP-cAMP complex are required for its optimal activity. Based on previous results for the pts operon, which are mentioned above, we tested whether the observed glucose-mediated activation effect depended on the EIIGlc protein (the product of the ptsG gene). We found that that was indeed the case. Since expression of the pgk and gapA genes has to be coordinated, we also checked whether gapA gene expression was also subject to ptsG-dependent glucose induction. Based on the results obtained, we propose that the EIIGlc protein is a key factor in the glucose activation of genes involved in glucose uptake and metabolism.
MATERIALS AND METHODS
Bacterial strains.
E. coli TG1 (supE hsdΔ5, thi, Δ[lac-proAB] F′ [traD36 proAB+ lacIq lacZΔM15]) was used as a source of chromosomal DNA. The E. coli strains TG1 and TP2503 (F− xyl argH1 ilvA) (12) were used to study the in vivo transcriptional activity of the gapB, gapA, and ptsH genes. The influences of CRP and cAMP were studied in E. coli POP4129ΔcrpT8zhd::Tn10 (a generous gift from A. Kolb) and TP2006 (F− xyl lacΔX74 Δcya glp-8306) (32), respectively. The role of the ptsG gene product was analyzed in strains TP2504 (F−, xyl argH1 ilvA zcf-229::Tn10 ptsG22) and TP2512 (F− xyl argH1 ilvA zcf-229::Tn10 ptsG22 Δcya glp8306) (12).
Medium and growth conditions.
The growth medium used was either Luria broth (LB) or the minimal M63 medium (27) supplemented with thiamine (10 μg/ml) and 0.1% (wt/vol) Casamino Acids and with either glucose or pyruvate (0.4% [wt/vol]) or a succinate and glycerol mixture (0.4% [wt/vol] and 0.08% [vol/vol], respectively). When required, arginine was added (100 μg/ml). The effect of cAMP in the medium was tested at a 1 mM final concentration. Ampicillin (100 μg/ml) was added for the growth of transformants. Cells were grown aerobically at 37°C, and growth was monitored by measuring the optical density at 600 nm (OD600).
Plasmids.
Standard methods of molecular biology were used for plasmid constructions (35). The genomic DNA of E. coli TG1 was extracted, and DNA fragments containing the gapB-pgk cluster were amplified by PCR with the oligonucleotide primers OGB1 and OGB5. OGB1 (5′ TGGAATAAAGCTTCCCACAA 3′) is complementary to nucleotide positions −243 to −224 of the gapB gene (see Fig. 1A and 2B) (the A residue of the initiation codon is designated position +1). A HindIII nuclease recognition site is present between positions −231 and −236. OGB5 (5′ TCACCAGTGATTACGCCAG 3′) is complementary to positions 47 to 29 of the fda gene (see Fig. 1A). After amplification, the OGB1-OGB5 fragment was cloned blunt end in plasmid pBluescript(SK)+ by using the HincII site. The recombinant plasmid was designated pPBK100. The nucleotide sequences of the cloned gapB-pgk genes were checked by the dideoxynucleotide chain termination method for double-stranded DNA sequencing. A plasmid carrying a longer nucleotide sequence upstream of the gapB gene was also constructed. A DNA fragment was amplified by PCR with oligonucleotides OGB6 and OGB3 (Fig. 1A). OGB6 (5′ TGGGAATATCTCGAGCAACAAGAC 3′) is complementary to positions −812 to −789 upstream of the gapB gene and carries a XhoI restriction site (underlined in the sequence). OGB3 (5′ GCACGAACCACATTACG 3′) is complementary to nucleotide positions 59 to 43 of the gapB coding sequence. The PCR fragment was digested by endonucleases XhoI and HindIII and was cloned in plasmid pBK100 between the XhoI and HindIII sites to create plasmid pPBK500. The 2.5-kb BglII-XbaI fragment from plasmid pPBK100, carrying the gapB and pgk genes without the gapB promoter region, was cloned between the BamHI and XbaI sites of plasmid pBluescript(SK)+ to create plasmid pBK200. An M13mp9 phage with the same insert as plasmid pPBK100 was used for directed mutagenesis of the CRP-cAMP binding site by the Kramer method (35). The wild type (WT) (5′ to 3′) sequence TGTGATGTGAGTCAGA was changed into the sequence AACGTGGATCCTACGT. The HindIII-XbaI fragment carrying the mutated sequence was cloned between the HindIII and XbaI sites of plasmid pPBK500 to create plasmid pPBK500mutCRP. Plasmids pBR322 and PTSG10 (18) were used to transform strain TP2512.
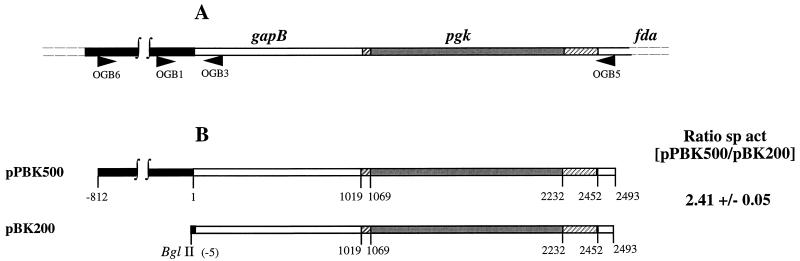
FIG. 1.
Role of the gapB promoter region in PGK protein production. (A) The gapB-pgk-fda gene cluster. Positions of the oligonucleotides OGB1, OGB3, OGB5, and OGB6 used for PCR amplification of the gapB and pgk genes are represented by horizontal arrows. The gapB upstream region is represented in black and the gapB and pgk coding regions are shown in white and grey, respectively. The intergenic regions between gapB and pgk and between pgk and fda are striped. (B) Fragments cloned in the plasmids used to analyze pgk gene expression. Construction of plasmids pPBK500 and pBK200 is described in Materials and Methods. Cells transformed with the plasmids pPBK500 and pBK200 were grown in M63 medium supplemented with glucose in the presence of ampicillin at 37°C until stationary growth phase. After sonication, PGK activity was measured in the soluble fraction as described in Materials and Methods. Specific activities were expressed in nanomoles of NADH per minute per OD280 unit per OD600 unit at which cells were harvested. The ratio of the specific activities (sp act [pPBK500/pBK200]) is the average value of the ratios determined for three different cultures.
RNA isolation and analysis.
Total RNA was isolated by the hot phenol procedure (1) from cells harvested at an OD600 of 0.5 to 0.55. RNA extractions were always performed with the same amount of cells (about 5 × 108 cells). Synthetic oligonucleotides OGB3, complementary to the gapB gene (see Fig. 2B), OG1, complementary to the gapA gene (8), and H12, complementary to the ptsH gene (14), were 5′ end labeled with [γ-32P]ATP (3,000 Ci/mmol) (Amersham) and T4 polynucleotide kinase (Boehringer-Mannheim). They were used as primers for cDNA synthesis with reverse transcriptase. For annealing, 5 ng of 5′ end-labeled oligonucleotide and 5 μg of total RNA extracted from transformed cells or 20 μg of total RNA from untransformed cells were heated to 70°C for 10 min in 10 μl of 50 mM Tris hydrochloride (pH 8.3)–40 mM KCl–6 mM MgCl2 and then cooled slowly to room temperature (20 min). The cDNA synthesis was performed as previously described (8), and cDNAs were fractionated on a 7% polyacrylamide–8 M urea sequencing gel and compared with dideoxy sequencing reaction products obtained with the same primer and plasmid pPBK100 as the template. Direct sequencing of mRNA was carried out in the same conditions as for primer extension, except for the presence of 2′, 3′-dideoxyribonucleoside triphosphates (ddNTPs) at a dNTP/ddNTP ratio of 1:5 in the reaction mixture (8).
Protein extract preparation and analysis.
Protein extractions were made on cells harvested at an OD600 of 0.5 or 3. After centrifugation, the cell pellet was washed and sonicated. The GAPDH activity of the soluble fraction was measured as previously described (5). PGK activity was measured by monitoring at 340 nm the decrease of NADH in a coupled enzymatic system with the Bacillus stearothermophilus GAPDH. To determine specific activities of both enzymes, the measured activities were divided by the OD280 value of the soluble fraction. For both enzymes, the specific activities were divided by the OD600 value of the cell culture used for the enzymatic assay.
RESULTS
Part of PGK expression depends upon the gapB promoter region.
No characteristic transcription termination signal is present between the gapB and pgk coding sequences (2). Hence, it was tempting to postulate that the transcriptional promoter localized upstream of gapB contributed to PGK production. To test this hypothesis, two constructs were produced (Fig. 1B): pPBK500, containing the entire gapB-pgk cluster including the nucleotide sequence upstream of the gapB coding sequence, and pBK200, containing the gapB and pgk coding sequences without the gapB upstream region. E. coli TG1 cells were transformed by these plasmids and grown at 37°C in M63 medium supplemented with glucose as the carbon source. When cultures reached an OD600 of 3, PGK specific activities were measured as described in Materials and Methods. They were found to be diminished by a factor of 2.41 when the nucleotide sequence upstream of gapB was absent (Fig. 1B). These results indicated that PGK production depended on the promoter region upstream of gapB. We next focused our attention on the transcriptional properties of the gapB upstream region.
The gapB gene is transcribed from a single start site.
We used the primer extension method to identify the 5′ extremities of gapB transcripts (Fig. 2). Total RNA was extracted from E. coli TG1 cells cultured at 37°C in M63 medium supplemented with glucose as the carbon source. The 32P-labeled OGB3 oligonucleotide, complementary to a sequence at the beginning of the gapB coding region (Fig. 2B), was used as the primer for reverse transcription. Direct RNA sequencing with primer OGB3 confirmed the specific hybridization of this oligonucleotide to gapB transcripts (data not shown). After fractionation of the reaction products, a single cDNA band was observed. By comparison of the 3′ end of this cDNA product with DNA dideoxy-sequencing reaction products, obtained with primer OGB3 and plasmid pPBK100 as the template (Fig. 2A), the 5′ end of the gapB transcripts was found to correspond to a position 132 bp upstream of the translation initiation site of gapB (Fig. 2B). Consistent with the presence of a transcription start site, a TATCCT sequence, located 6 nt upstream of this position, fits the consensus sequence of the −10 element of promoters recognized by the E. coli Eς70 RNA polymerase (23). No sequence that might correspond to a −35 element is present. The center of symmetry of the putative CRP-cAMP binding site, which was already noticed by Alefounder and Perham (2), is localized 70.5 nt upstream of the transcription start site. This is a similar location to that found for malT and certain other CRP-controlled genes (7, 9). Thus, the nucleotide sequence between positions −79 and +1 explained a transcription initiation at position +1. The corresponding promoter was named gapB P0.
Influence of the carbon source on the amount of gapB transcripts.
To get more information on the control of promoter gapB P0 by the carbon source, we measured the amount of gapB transcripts in E. coli TG1 cells grown in the presence of either glucose or a succinate-glycerol mixture as the sole carbon source (Fig. 3A). Similar experiments were also made with either pyruvate or glycerol as the sole carbon source (Fig. 4 and data not shown). The amount of the gapB P0 transcript varied considerably with the nature of the carbon source (compare lanes 1 and 3 in Fig. 3A and lanes 1 and 2 in Fig. 4A). The highest levels were obtained for growth on glucose. This amount was decreased by factors of 20 and 10 for growth on the succinate-glycerol mixture and on pyruvate, respectively. Although we could not exclude the possibility that mRNA stability varies depending upon the carbon source, our results strongly suggested that gapB P0 transcription is reduced during growth on poor carbon sources compared to growth on glucose. Hence, due to the presence of a CRP consensus binding site at a functional distance from the gapB P0 start site, we had to test for the involvement of the CRP-cAMP complex on gapB P0 activity.
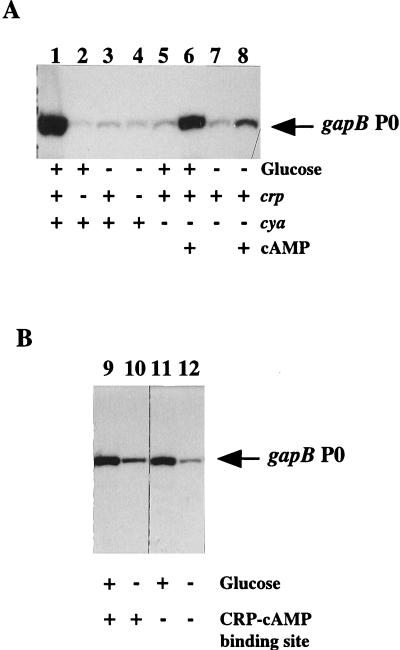
FIG. 3.
Effects of glucose, CRP, and cAMP on gapB mRNA level. (A) Primer extension reactions using total RNA extracts were conducted as described in Materials and Methods. E. coli TG1 (lanes 1 and 3), POP4129Δcrp (lanes 2 and 4), and TP2006 (cya) (lanes 5 to 8) were grown at 37°C to mid-log phase in the presence of glucose (lanes 1, 2, 5, and 6) or succinate-glycerol (lanes 3, 4, 7, and 8) as the sole carbon source. cAMP was added to TP2006 cultures at 1 mM (lanes 6 and 8). (B) E. coli TG1 cells transformed with plasmid pPBK500 (lanes 9 and 10) or pPBK500mutCRP (lanes 11 and 12) were grown in M63 medium supplemented with ampicillin and with glucose (lanes 9 and 11) or a succinate-glycerol mixture (lanes 10 and 12) as the sole carbon source. Primer extension reactions were done with 5 μg of total RNA extracts.
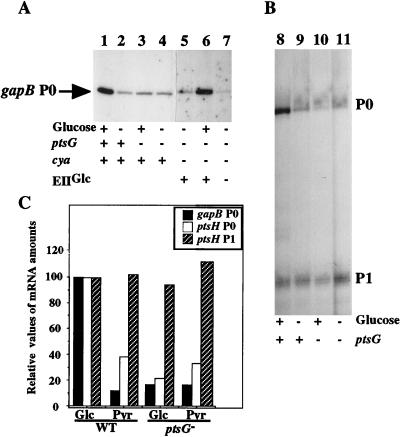
FIG. 4.
Effect of pstG gene expression on gapB P0 promoter activity. Cells from TP2503 (lanes 1, 2, 8, and 9) and TP2504 (lanes 3, 4, 10, and 11) strains were grown in M63 medium supplemented with glucose (lanes 1, 3, 8, and 10) or pyruvate (lanes 2, 4, 9, and 11) as the sole carbon source. TP2512 cells (lanes 5 to 7) were transformed with plasmid PTSG10 (lanes 5 and 6) or pBR322 (lane 7) and grown in M63 medium supplemented with ampicillin and either glucose (lane 6) or pyruvate (lane 5 and 7). Overexpression of protein EIIGlc by plasmid PSTG10 is indicated by the plus signs below lanes 5 and 6, and cells transformed with pBR322 are indicated by the minus sign below lane 7. The RNA extracts were prepared as described in the legend for Fig. 3 and analyzed using either primer OGB3 for gapB P0 transcripts (A) or primer H12 for ptsH transcripts initiated at promoters P0 and P1 (B). (C) Quantification of the RNA amounts were achieved by phosphorimaging (ImageQuant software; Molecular Dynamics). The values for each promoter are expressed relative to a value of 100 for the RNA amount in WT cells grown in glucose.
Influence of CRP and cAMP on the level of gapB transcripts in total RNA extracts.
To test for the roles of CRP and cAMP on gapB P0 promoter activity, we looked for the amount of gapB transcripts in E. coli POP4129Δcrp, which is deficient in CRP production, and in the E. coli TP2006cya mutant strain, defective in cAMP production (32). The bacteria were grown in a Casamino Acids-containing medium in the presence of glucose or a succinate-glycerol mixture (see Materials and Methods). For cells grown in a glucose-containing medium, the amount of the gapB P0 transcript was dramatically reduced in the absence of CRP (compare lanes 1 and 2 in Fig. 3A), whereas the absence of CRP had no marked effect on the low level of gapB P0 mRNA in cells grown in a succinate-glycerol-containing medium (compare lanes 3 and 4 in Fig. 3A). Similarly, there was no stimulation of the gapB P0 transcript by glucose in the cya mutant strain in the absence of cAMP (Fig. 3a, compare lanes 5 and 7 with lane 1). The addition of cAMP to a glucose-containing medium produced a strong increase in the level of gapB P0 transcript in the cya mutant strain (Fig. 3A, compare lanes 5 and 6). In contrast, in the absence of glucose, supplementation of the culture with cAMP produced only a slight increase in the level of gapB P0 transcript (Fig. 3A, lanes 7 and 8). We concluded from these results that the maximum utilization of promoter gapB P0 strongly depends on the presence of glucose in the medium and requires the presence of the CRP-cAMP complex in the cells. Since the cAMP concentration is expected to be low in cells grown on glucose as the carbon source (for a review, see reference 30), it was difficult to understand how transcription could have been activated directly by the CRP-cAMP complex and also be glucose activated. Altogether, this suggested an indirect role of the CRP-cAMP complex.
Role of the CRP-cAMP binding sequence on promoter gapB P0 activity.
To test whether the CRP-cAMP dependence observed in the presence of glucose was due to direct binding of this complex to its binding site on the gapB P0 promoter region, we analyzed the effect of a mutation in the CRP-cAMP binding sequence on promoter gapB P0 activity. For this purpose, the WT sequence (TGTGATGTGAGTCAGA) was replaced by the AACGTGGATCCTACGT sequence by site-directed mutagenesis (see Materials and Methods). E. coli TG1 cells were transformed with plasmid pPBK500, containing a WT gapB-pgk operon, and plasmid pBK500mutCRP, containing an operon with a mutation in the CRP binding site. Primer extension analyses were performed on 5 μg of total RNA extracted from the transformed cells. With these RNA amounts, no detectable reverse transcription products were obtained with RNA from the untransformed cells (data not shown). The results obtained (Fig. 3B) show that for growth on glucose, mutation of the CRP-cAMP binding sequence had no marked effect on gapB P0 promoter activity (compare lanes 9 and 11 in Fig. 3B). A possible explanation was that the CRP-cAMP complex activates the expression of a gene whose product is involved in gapB P0 activation in the presence of glucose.
Role of the ptsG gene product in gapB transcription.
Based on previous data for the pts operon (13), we checked whether the ptsG gene, which is activated by the CRP-cAMP complex (25, 29), was involved in the glucose-mediated activation of promoter gapB. gapB and ptsH mRNAs were analyzed in the E. coli ptsG mutant strain TP2504, isogenic to strain TP2503 (12). The bacteria were grown in a glucose- or a pyruvate-containing medium. In the ptsG mutant strain grown on glucose, the amount of the gapB P0 transcript was eight times lower than that of the isogenic ptsG+ strain (Fig. 4A and C). In agreement with previous experiments based on the measurement of β-galactosidase activity produced by gene fusions (13), we found a 2.4-fold increase in the amount of ptsH mRNA initiated at the P0 promoter with glucose as the carbon source compared to that with pyruvate as the carbon source (Fig. 4B and C). The stimulatory effect of glucose decreased fourfold in a ptsG mutant context. Also in agreement with a previous report (12), no similar regulation was found for the ptsH P1 promoter (Fig. 4B and C) or for the crr promoter (data not shown). These results showed, first, that the primer extension method analysis we have used gives results that are in agreement with those obtained by other approaches (12, 13). Second, they strongly suggest that ptsG is involved in the glucose-mediated regulation of gapB P0 promoter activity. To confirm this hypothesis, we transformed strain TP2512 (ptsG, cya) with plasmid PTSG10, a high-copy-number plasmid derived from pBR322 and carrying the ptsG gene (18). We compared the amounts of gapB P0 mRNA in the transformed cells grown on glucose versus pyruvate (Fig. 4A, lanes 6 and 5). The amount of gapB P0 transcripts was increased 20-fold in the presence of glucose. Thus, the glucose stimulation of the gapB P0 promoter activity was only observed in cells producing the EIIGlc protein. Importantly, when ptsG was overexpressed, gapB transcription was stimulated by glucose even in the absence of cAMP (Fig. 4A, lane 6). In conclusion, these results show that expression of the EIIGlc protein is required for glucose stimulation of both the gapB P0 and ptsH P0 promoters.
Regulation of PGK production by glucose and EIIGlc protein.
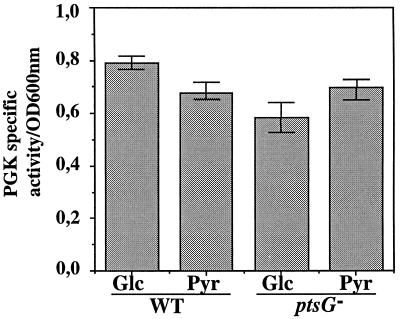
Since part of PGK production appeared to depend on the gapB P0 promoter activity, we tested whether the glucose-mediated activation of promoter gapB P0 also regulated pgk expression. We monitored the rate of PGK production by measuring the cellular PGK specific activity (see Materials and Methods). Experiments were performed with E. coli TP2503 (WT) and TP2504 (ptsG) strains grown until mid-log phase (Fig. 5). The PGK specific activity was slightly lower for growth on pyruvate compared to that for growth on glucose. Also, the PGK specific activity for growth on glucose was decreased by a factor of 0.75 in a ptsG mutant strain compared to that for a ptsG+ strain. Thus, expression of PGK is regulated by the presence of glucose and by a mechanism that involves the EIIGlc protein.
FIG. 5.
Effects of carbon source and expression of the ptsG gene on PGK production. TP2503 and TP2504 strains were cultured as described in Materials and Methods and harvested at mid-log phase. PGK activity was measured on the soluble fraction obtained after sonication by using a coupled enzymatic reaction (see Materials and Methods). Specific activities are the average values for three different cultures (error bars represent the range of the three specific activities that were measured) and are expressed as described in the legend for Fig. 1C. The carbon source used for the culture and the genetic background (WT or mutant ptsG) are indicated at the bottom.
gapA expression is also stimulated by glucose in an EIIGlc-dependent manner.
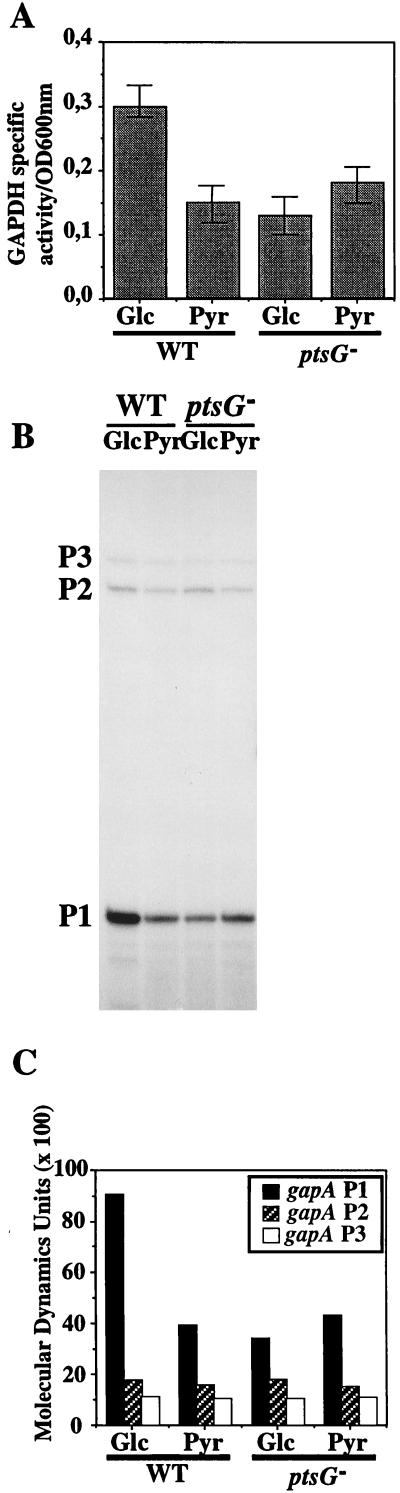
As mentioned in the introduction, GAPDH and PGK have the peculiarity of being produced from two distinct loci in E. coli. However, the syntheses of these two enzymes, which act at successive steps during glycolysis, have to be coordinated. Hence, we addressed the question of whether production of GAPDH by the gapA gene might also be stimulated by glucose and by a mechanism depending upon the EIIGlc protein. We analyzed gapA gene expression in isogenic ptsG and ptsG+ strains. The rate of gapA gene expression was monitored by measuring the GAPDH specific activity of cells harvested during exponential growth phase (see Materials and Methods). As can be seen in Fig. 6A, the GAPDH specific activity during growth on pyruvate is half of that found during growth on glucose. Then, we verified that this effect was due to differences in the gapA transcription level. As can be seen in Fig. 6B, this is indeed the case: the amount of RNA initiated at promoter P1 varied depending upon the nature of the carbon source, whereas the amounts of mRNAs initiated at promoters P2 and P3 did not vary. Differences observed in the P1 mRNA level parallel the differences observed for GAPDH specific activity. The amount of RNA initiated at the P1 promoter increased by a factor of 3.6 during growth on glucose compared to that observed during growth on pyruvate. Both GAPDH specific activity and gapA mRNA levels were then analyzed in the ptsG mutant strain grown either in a glucose- or a pyruvate-containing medium. As found for gapB, neither GAPDH expression nor the gapA P1 mRNA level was stimulated by growth on glucose in the ptsG mutant strain (Fig. 6). We also confirmed that the absence of glucose stimulation of the gapA P1 promoter was overcome when the EIIGlc protein was overexpressed in a ptsG mutant background (data not shown). We conclude that transcription of the gapA gene is stimulated by glucose at the P1 promoter and that this activation is dependent upon EIIGlc protein expression.
FIG. 6.
Effects of carbon source and expression of the ptsG gene on gapA gene expression. Soluble crude extracts prepared and used as described in the legend for Fig. 5 were analyzed for GAPDH activity. (A) GAPDH activity was measured by using the test of Ferdinand on the soluble fraction obtained after cell sonication (5). Specific activities are the average values for three different cultures and are expressed in nanomoles of NADH per minute per OD280 unit per OD600 unit at which the cells were harvested. The total RNA extracts analyzed for Fig. 4 were also used for the gapA transcription analysis. (B) In vivo levels of the gapA P1, P2, and P3 mRNAs were analyzed by primer extension with oligonucleotide OB1 as the primer, as described in Materials and Methods. (C) Quantification of mRNA levels shown in panel B achieved by phosphorimaging (ImageQuant software; Molecular Dynamics). The carbon source used for each culture and the genetic background (WT or mutant ptsG) are indicated either at the bottom (panels A and C) or the top (panel B) of each panel.
DISCUSSION
The results of the experiments presented in this report provide new information concerning the expression of the gapB-pgk gene tandem and the gapA gene. More importantly, our results show a coregulation of several genes encoding enzymes for glucose uptake and metabolism, i.e., gapB-pgk, gapA, and pts.
The gapB gene is transcribed from a single start site located 132 nt upstream of the gapB coding sequence. More than half of the PGK expression also depends on this transcriptional activity. This is evidenced by the PGK specific activity in the presence and absence of promoter gapB P0 (Fig. 1) and confirmed by the fact that variations of cellular PGK activity reflect variations in gapB P0 promoter activity (Fig. 4A and 5). The absence of any obvious transcription terminator, either rho dependent or rho independent, in the segment preceding the pgk open reading frame (ORF) (2) is in agreement with this observation, implying a cotranscription of the two genes. However, this does not exclude the possibility that another transcriptional promoter located upstream of the PGK ORF participates in transcription of the pgk gene.
One interesting observation is the very efficient initiation at the gapB P0 promoter. In spite of this high transcriptional activity, the GAPB protein is present in very low amounts (3, 39). This discrepancy indicates that a posttranscriptional event is responsible for the low level of expression of the GAPB protein. A similar discrepancy in the expression of two adjacent gap and pgk genes was also described for Zymomonas mobilis (16). In this organism, a bicistronic gap-pgk transcript is produced, whereas different amounts of GAPDH and PGK proteins are observed. This differential gene expression is explained by endonucleolytic cleavages in the gap-pgk transcript producing individual gap and pgk mRNAs with different stabilities (15). Whether a similar mechanism is responsible for the poor gapB expression remains to be investigated.
Like the ptsH P0 promoter, the gapB P0 promoter region possesses a CRP-cAMP binding sequence. The role of the CRP-cAMP complex has been demonstrated for ptsH P0 promoter activity (13, 14, 20, 33). Though binding of the CRP-cAMP complex to the gapB CRP-cAMP binding sequence occurs in vitro (31), an activation role was only detected for the low activity shown by promoter gapB P0 in the absence of glucose. No marked difference of promoter gapB P0 activity was detected upon substitution of the CRP-cAMP binding sequence by a very different sequence in the presence of glucose (Fig. 3B). The low cis activation by the CRP-cAMP complex observed in the absence of glucose probably participates in the PGK production when glucose is absent (Fig. 3B, lanes 10 and 12). This basal level of pgk expression may be required to rapidly resume glycolysis when glucose becomes available and/or for neoglucogenesis. In accord with this hypothesis, it should be noted that a promoter regulated by catabolic repression (gapA P3) is also present in the gapA promoter region (8). In contrast, by using strains impaired in CRP or cAMP production, we showed that the CRP-cAMP complex has a strong indirect effect on glucose-mediated activation of the gapB P0 promoter (Fig. 3A). With these strains, a residual low gapB P0 activity is observed, which is the same as that in a crp+ or cya+ background during growth in the presence of pyruvate or succinate-glycerol.
On the one hand, we showed that the glucose stimulation of the gapB P0 promoter activity depends upon the expression of the ptsG gene (Fig. 4). On the other hand, ptsG expression depends on the CRP-cAMP complex (25, 29). Hence, we can reasonably conclude that the absence of glucose stimulation of the gapB P0 promoter in mutant crp and cya genetic contexts is likely due to the reduction of ptsG expression in these two genetic backgrounds. Consistently, overexpression of the ptsG gene in a cya mutant strain restores the glucose effect (Fig. 4).
What is the advantage for the cell to induce gapA, gapB-pgk, and pts gene expression during growth on glucose? In addition to that of the pts operon, the level of ptsG gene expression is also increased in the presence of glucose (18, 29, 37). Consequently, the production of all the specific components of the glucose uptake machinery is increased in the presence of glucose, which should lead to a greater uptake of glucose and production of high levels of glucose-6-phosphate. To prevent its accumulation, the enzymes acting downstream in the glycolytic pathway need to be highly expressed. The fact that at least the expression of GAPDH and PGK can be stimulated by glucose is in agreement with such a model. As pointed out earlier (21), glycolysis and protein synthesis may be connected. It was proposed that transcription of the rrn operons monitors the translation rate by responding to the ATP pool (21). GAPDH is one of the key enzymes responsible for ATP production through its conversion of glyceraldehyde-3-phosphate into 1,3-diphosphoglycerate, a high-energy compound. Hence, altogether, the glucose-mediated activation of pts, gapA, and gapB-pgk transcription probably plays an important role for the high growth rate that E. coli shows on glucose.
The next question is whether the three promoters are activated by the same mechanism (and, if so, what is the molecular basis of this mechanism). In the case of ptsH P0, De Reuse and Danchin proposed that the glucose activation acts like a two-component system, with EIIGlc acting as the sensor protein and with an as yet unknown regulator protein activating the ptsH P0 transcription (13). In this system, glucose is considered as the external signal changing the phosphorylation state of the sensor protein, which transmits a transduction signal to the regulator. In this view, the regulator can be either a transcriptional activator, whose DNA-binding activity is enhanced upon glucose activation, or a repressor, whose DNA-binding activity is decreased during growth on glucose. The role of a glucose-inducible repressor was proposed to explain the glucose effect on the ptsH P0 promoter (33). FruR has a binding site within the gapB and ptsH P0 promoter regions (31, 34). However, the glucose stimulatory effect is conserved in a fruR mutant strain (34). Recently, glucose induction has been correlated with the activity of the Mlc repressor (29). We are planning to test the direct or indirect role of Mlc on gapB P0 and gapA P1 promoter activities. Similar mechanisms for activation of the ptsH P0, gapA P1, and gapB P0 promoters would imply a specific binding of the regulator in the promoter region. Nevertheless, no evident common sequence has been found by nucleotide sequence comparison. An alternative mechanism is the implication of a cellular factor that modifies the DNA structure, whose activity or abundance would be modified upon growth on glucose. Work is in progress to test each of these possibilities.
ACKNOWLEDGMENTS
This work was supported by the Ministère de l’Enseignement Supérieur et de la Recherche, the Centre National de la Recherche Scientifique, the Pôle Technologique Régional Lorrain: Protéines et Biotechnologie, and the Programme d’Intérêt Régional pour la Lorraine: Génie des Protéines. B.C. and V.B. were fellows of the Ministère de l’Enseignement Supérieur et de la Recherche.
We warmly thank J. Plumbridge for her critical reading of the manuscript. H. De Reuse is thanked for her generous gift of strains TP2503, TP2504, and TP2512 and plasmid PTSG10. A. Kolb is thanked for her generous gift of Δcrp and cya mutant strains. The Service Commun de Biophysicochimie des Interactions Moléculaires de l’Université H. Poincaré-Nancy I is acknowledged for computer facilities.
REFERENCES
- 1.Aïba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Alefounder P R, Perham R N. Identification, molecular cloning and sequence analysis of a gene cluster encoding the class II fructose 1,6-bisphosphate aldolase, 3-phosphoglycerate kinase and a putative second glyceraldehyde-3-phosphate dehydrogenase of Escherichia coli. Mol Microbiol. 1989;3:723–732. doi: 10.1111/j.1365-2958.1989.tb00221.x. [DOI] [PubMed] [Google Scholar]
- 3.Boshi-Muller S, Azza S, Pollastro D, Corbier C, Branlant G. Comparative enzymatic properties of gapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and phosphorylating glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1997;272:15106–15112. doi: 10.1074/jbc.272.24.15106. [DOI] [PubMed] [Google Scholar]
- 4.Branlant G, Branlant C. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of d-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1985;150:61–66. doi: 10.1111/j.1432-1033.1985.tb08988.x. [DOI] [PubMed] [Google Scholar]
- 5.Branlant G, Flesch G, Branlant C. Molecular cloning of the glyceraldehyde-3-phosphate dehydrogenase genes of Bacillus stearothermophilus and Escherichia coli, and their expression in Escherichia coli. Gene. 1983;25:1–7. doi: 10.1016/0378-1119(83)90161-0. [DOI] [PubMed] [Google Scholar]
- 6.Branlant C, Oster T, Branlant G. Nucleotide sequence determination of the DNA region coding for Bacillus stearothermophilus glyceraldehyde-3-phosphate dehydrogenase and of the flanking DNA regions required for its expression in Escherichia coli. Gene. 1989;75:145–155. doi: 10.1016/0378-1119(89)90391-0. [DOI] [PubMed] [Google Scholar]
- 7.Busby S, Kolb A. The CAP regulon. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 255–279. [Google Scholar]
- 8.Charpentier B, Branlant C. The Escherichia coli gapA gene is transcribed by the vegetative RNA polymerase holoenzyme Eς70 and by the heat shock RNA polymerase Eς32. J Bacteriol. 1994;176:830–839. doi: 10.1128/jb.176.3.830-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway T, Ingram L O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988;170:1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Della Seta F, Boshi-Muller S, Vignais M L, Branlant G. Characterization of Escherichia coli strains with gapA and gapB genes deleted. J Bacteriol. 1997;179:5218–5221. doi: 10.1128/jb.179.16.5218-5221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Reuse H, Danchin A. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferse system: a complex operon with several modes of transcription. J Bacteriol. 1988;170:3827–3837. doi: 10.1128/jb.170.9.3827-3837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Reuse H, Danchin A. Positive regulation of the pts operon of Escherichia coli: genetic evidence for a signal transduction mechanism. J Bacteriol. 1991;173:727–733. doi: 10.1128/jb.173.2.727-733.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Reuse H, Kolb A, Danchin A. Positive regulation of the expression of the Escherichia coli pts operon. J Mol Biol. 1992;226:623–635. doi: 10.1016/0022-2836(92)90620-y. [DOI] [PubMed] [Google Scholar]
- 15.Eddy C K, Keshav K F, Haejung An E A, Mejia J P, Ingram L O. Segmental message stabilization as a mechanism for differential expression from the Zymomonas mobilis gap operon. J Bacteriol. 1991;173:245–254. doi: 10.1128/jb.173.1.245-254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy C K, Mejia J P, Conway T, Ingram L O. Differential expression of gap and pgk genes within the gap operon of Zymomonas mobilis. J Bacteriol. 1989;171:6549–6554. doi: 10.1128/jb.171.12.6549-6554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eikmanns B J. Identification of a Corynebacterium glutamicum gene cluster encoding the three enzymes glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and triosephosphate isomerase. J Bacteriol. 1992;174:6076–6086. doi: 10.1128/jb.174.19.6076-6086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erni B, Zanolari B. Glucose-permease of the bacterial phosphotransferase system. Gene cloning, overproduction, and amino acid sequence of enzyme IIGlc. J Biol Chem. 1986;261:16398–16403. [PubMed] [Google Scholar]
- 19.Fabry S, Heppner P, Dietmaier W, Hansel R. Cloning and sequencing the gene encoding 3-phosphoglycerate kinase from mesophilic Methanobacterium bryantii and thermophilic Methanothermus fervidus. Gene. 1990;91:19–25. doi: 10.1016/0378-1119(90)90157-m. [DOI] [PubMed] [Google Scholar]
- 20.Fox D K, Presper K A, Adhya S, Roseman S, Garges S. Evidence for two promoters upstream of the pts operon: regulation by the cAMP receptor protein regulatory complex. Proc Natl Acad Sci USA. 1992;89:7056–7059. doi: 10.1073/pnas.89.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 22.Harris J I, Waters M. Glyceraldehyde-3-phosphate dehydrogenase. In: Boyer P D, editor. The enzymes. Vol. 13. New York, N.Y: Academic Press; 1976. pp. 1–49. [Google Scholar]
- 23.Hawley D K, Mc Clure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillman J D, Fraenkel D G. Glyceraldehyde-3-phosphate dehydrogenase mutants of Escherichia coli. J Bacteriol. 1975;122:1175–1179. doi: 10.1128/jb.122.3.1175-1179.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcription regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Nellemann L J, Holm F, Atlung T, Hansen F G. Cloning and characterization of the Escherichia coli phosphoglycerate kinase (pgk) gene. Gene. 1989;77:185–191. doi: 10.1016/0378-1119(89)90373-9. [DOI] [PubMed] [Google Scholar]
- 29.Plumbridge, J. Expression of ptsG, the gene for the major PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol. Microbiol., in press. [DOI] [PubMed]
- 30.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramseier T M, Bledig S, Michotey V, Faghali R, Saier M H. The global regulatory protein, FruR, modulates the direction of carbon flow in Escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 32.Roy A, Danchin A. The cya locus of Escherichia coli. Mol Gen Genet. 1982;188:465–471. doi: 10.1007/BF00330050. [DOI] [PubMed] [Google Scholar]
- 33.Ryu S, Garges S. Promoter switch in the Escherichia coli pts operon. J Biol Chem. 1994;269:4767–4772. [PubMed] [Google Scholar]
- 34.Ryu S, Ramseier T M, Michotey V, Saier M H, Jr, Garges S. Effect of the FruR regulator on transcription of the pts operon in Escherichia coli. J Biol Chem. 1995;270:2489–2496. doi: 10.1074/jbc.270.6.2489. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schläpfer B S, Zuber H. Cloning and sequencing of the genes encoding glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase and triosephosphate isomerase (gap operon) from mesophilic Bacillus megaterium: comparison with corresponding sequences from thermophilic Bacillus stearothermophilus. Gene. 1992;122:53–62. doi: 10.1016/0378-1119(92)90031-j. [DOI] [PubMed] [Google Scholar]
- 37.Stock J B, Waygood E B, Meadow N D, Postma P, Roseman S. Sugar transport by the bacterial phosphotransferase system. The glucose receptors of the Salmonella typhimurium phosphotransferase system. J Biol Chem. 1982;257:14543–14552. [PubMed] [Google Scholar]
- 38.Thomson J, Gerstenberg P D, Goldberg D E, Gociar E, de Silva A O, Fraenkel D G. ColE1 hybrid plasmids for Escherichia coli genes of glycolysis and the hexose monophosphate shunt. J Bacteriol. 1979;137:502–506. doi: 10.1128/jb.137.1.502-506.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao G, Pease A J, Bharani N, Winkler M E. Biochemical characterization of gapB-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli K-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J Bacteriol. 1995;177:2804–2812. doi: 10.1128/jb.177.10.2804-2812.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]