Abstract
Yeast flocculation is a phenomenon which is believed to result from an interaction between a lectin-like protein and a mannose chain located on the yeast cell surface. The FLO1 gene, which encodes a cell wall protein, is considered to play an important role in yeast flocculation, which is inhibited by mannose but not by glucose (mannose-specific flocculation). A new homologue of FLO1, named Lg-FLO1, was isolated from a flocculent bottom-fermenting yeast strain in which flocculation is inhibited by both mannose and glucose (mannose/glucose-specific flocculation). In order to confirm that both FLO1 and Lg-FLO1 are involved in the yeast flocculation phenomenon, the FLO1 gene in the mannose-specific flocculation strain was replaced by the Lg-FLO1 gene. The transformant in which the Lg-FLO1 gene was incorporated showed the same flocculation phenotype as the mannose/glucose-specific flocculation strain, suggesting that the FLO1 and Lg-FLO1 genes encode mannose-specific and mannose/glucose-specific lectin-like proteins, respectively. Moreover, the sugar recognition sites for these sugars were identified by expressing chimeric FLO1 and Lg-FLO1 genes. It was found that the region from amino acid 196 to amino acid 240 of both gene products is important for flocculation phenotypes. Further mutational analysis of this region suggested that Thr-202 in the Lg-Flo1 protein and Trp-228 in the Flo1 protein are involved in sugar recognition.
Cell adhesion is generally caused by the recognition of sugar chains located on cell surfaces by lectin-like proteins (38). A similar adhesion phenomenon (flocculation) is also observed in yeast, in which the cells adhere in clumps, resulting in sedimentation from the medium (28). It is thought that yeast flocculation occurs as a result of an interaction between a sugar-binding protein (lectin-like protein) and a mannose chain present on the yeast cell surface for the following reasons (18). (i) The flocculation phenomenon is proteinase sensitive, suggesting the contribution of a protein. (ii) Flocculation is inhibited by the presence of saccharides, such as mannose, suggesting the existence of a protein which recognizes saccharides. (iii) The flocculation phenomenon is Ca2+ dependent, suggesting the existence of a lectin-like protein, such as the C-type animal lectins and lectins of the plant group Leguminosae, which require Ca2+ ion for binding activity (7, 23).
Stratford and Assinder have classified yeast flocculation types into two groups distinguished by sugar inhibition: the NewFlo phenotype, which is inhibited by mannose, glucose, maltose, and sucrose but not by galactose, and the Flo1 type, which is inhibited by mannose but not by glucose, maltose, sucrose, or galactose (29). These two distinct phenotypes are thought to be caused by two different lectin-like proteins. Many attempts have been made to isolate the lectin-like protein involved in flocculation. At least two groups have succeeded in purification of mannose-specific lectins from cell surfaces (22, 31). There is no proof that these lectins are involved in flocculation. On the other hand, it has been reported that yeast flocculation is affected by many genes (27). One dominant flocculation gene, FLO1, has been cloned from a Flo1-type flocculation strain (32, 36). This gene was demonstrated to confer Flo1-type flocculation ability on cells (36) and to encode a cell wall protein (2, 5, 21). Therefore, it is thought that this gene encodes a lectin-like protein involved in flocculation.
In order to clarify the contribution of the FLO1 gene to the yeast flocculation phenomenon, we isolated a new FLO1 homologue, Lg-FLO1, from a NewFlo phenotype strain and replaced the FLO1 gene in a Flo1 phenotype strain with the Lg-FLO1 gene. Here, we show that the FLO1 and Lg-FLO1 genes encode a mannose-specific lectin-like protein and mannose/glucose-specific lectin-like protein, respectively. The region responsible for sugar recognition in these two gene products is also described.
(Part of this work has been presented at the European Brewery Convention in Brussels [Kobayashi et al., Proceedings of the European Brewery Convention Congress, Brussels, Belgium, p. 361–368, 1995].)
MATERIALS AND METHODS
Strains and media.
Saccharomyces pastorianus KBY001 is a bottom-fermenting yeast strain in commercial use. Flocculent and nonflocculent meiotic segregants were obtained from KBY001 according to the method described by Bilinski et al. (4). Saccharomyces cerevisiae KY644, which had a FLO1 gene disrupted by insertion of pRS405 (Stratagene), was constructed from YF191 (a ura3 ade2 leu2 FLO8) (13) and used as a host to introduce the full-length Lg-FLO1 or FLO1 gene. S. cerevisiae KY794 was also constructed from YF191 by replacement of the sequence from position +1 to position +3997 to the open reading frame of the FLO1 gene with pRS405 and used as a host to express the FLO1/Lg-FLO1 chimeric genes.
YPD medium and SD medium containing amino acid supplements were used for yeast cultivation under nonselective and selective conditions, respectively. Both media were prepared according to the method of Sherman et al. (24), except that 2% glucose was replaced by 5% galactose. E. coli was grown in LB medium (19).
Plasmids and transformation.
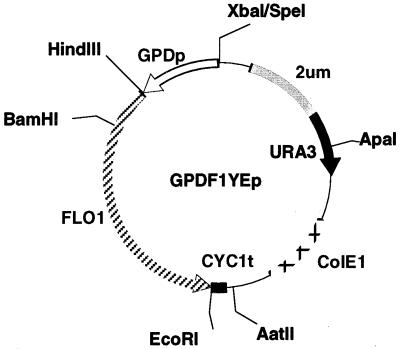
A YCp-type plasmid, pYT71, in which the APH3′II gene of pUC4K (Pharmacia) controlled by the TDH3 promoter (17) was inserted into pYT37 (13), was used as a vector for the cloning of the full-length Lg-FLO1 gene. pKB635, in which the full-length FLO1 gene of ATCC 60715 is inserted into pYT37, was obtained from a genomic library described previously (13) on the basis of homology between this gene and the FLO1 gene of ABXL-1D (32) and used for the expression of the Lg-FL01 gene. For the expression of a series of the FLO1/Lg-FLO1 chimeric gene, a YEp-type plasmid, GPDF1YEp (Fig. 1), which contains the ColE1 origin from pBR322, the 2 μm origin, and the CYC1 terminator from pYES2.0 (Invitrogen), was used. In the FLO1 gene in GPDF1YEp, the KpnI site encoding amino acid residues 240 and 241 of the Flo1 protein was replaced by the BamHI site by a recombinant PCR technique (35) and is controlled by the TDH3 promoter from YEp13K (26). All FLO1/Lg-FLO1 chimeric genes were constructed by amplification of a 240-amino-acid N-terminal region by a recombinant PCR technique, a region that was then substituted for the corresponding region in GPDF1YEp with the HindIII-BamHI site. The N termini of the chimeras were amplified by PCR, and their DNA sequences were determined directly to confirm the constructions. The FLO1/Lg-FLO1 chimeras with amino acid substitutions were constructed in the same way as the FLO1/Lg-FLO1 chimera, with a primer having the following sequence: 5′ GCG GGA TCC ATC TGG CAA TRY CAC ACT AAC AGG AAG TST ASC CMA GRM TMY AGC ATT TGA ATA AAC AAT TTT 3′.
FIG. 1.
Construction of GPDF1YEp.
All plasmids, except for pKB635, containing the functional Lg-FLO1, FLO1, or its modified genes were constructed in yeast cells directly because of the instability of these genes in Escherichia coli. Construction of these plasmids was confirmed by Southern analysis or a combination of PCR and DNA sequencing. For every construction of the FLO1/Lg-FLO1 chimeric genes, except for those with amino acid substitutions, at least two transformants containing the proper construct were selected and tested.
E. coli transformation was carried out as described by Hanahan (10). Transformation of yeast was carried out by the lithium acetate procedure (12).
Flocculation assay.
Cells grown for 48 h at 20°C with shaking were harvested and washed twice with 0.1 M EDTA and twice with sterile water. The flocculation assay was performed according to the method described by Smit et al., with some modifications (25). Cells were resuspended in 3 ml of flocculation buffer (50 mM sodium acetate, 0.1% CaCl2, pH 4.5) in a 1.0-cm cuvette, with or without 0.1% CaCl2, to a final concentration equivalent to an A600 of 2.0. After 5 min of vigorous agitation, the optical density of the yeast suspensions in the presence or absence of Ca2+ was measured. Flocculation ability was determined by the following equation: C = 1 − B/A, where A is the A600 without Ca2+, B is the A600 with Ca2+, and C is the flocculation ability.
Negative values were regarded as being equal to zero. To investigate sugar inhibition, a final concentration of 1 M sugar was added in flocculation buffer unless some other concentration is described.
For rough determinations, the yeast cells were washed twice with 0.1 M EDTA and twice with sterile water and resuspended in flocculation buffer with or without a final concentration of 1 M sugar; flocculation ability was then determined with the naked eye as follows: ++, all cells in buffer aggregate into flocs; +, some cells aggregate into flocs, but cells suspended without aggregation remained; −, no flocs can be observed.
DNA sequencing.
DNA sequencing was performed by the dideoxy chain-termination method (20) with a DNA Sequencing System (Perkin Elmer). Oligonucleotides were synthesized and used as primers. The results of sequencing were analyzed with the DNASIS program (Hitachi Software Engineering, Yokohama, Japan).
Southern and Northern analysis.
Total DNA for use in Southern analysis was prepared according to Hereford et al. (11). HyBond-N+ (Amersham) was used for Southern blotting according to the manufacturer’s instructions. Total RNA for Northern analysis was isolated by the method of Eliton and Warner (8) from cells grown for 48 h at 20°C with shaking. GeneScreen Plus (DuPont) was used for Northern blotting according to the manufacturer’s instructions. A DNA fragment of approximately 1 kb containing the proximal C-terminal region of the FLO1 gene was used to probe the FLO1 gene transcript. As a probe for the ACT1 gene, the coding region of the ACT1 gene was amplified by PCR and used.
Nucleotide sequence accession numbers.
The DNA sequences of the Lg-FLO1 gene (5′ and 3′ regions) are available in the DDBJ, EMBL, and GenBank databases under accession numbers D89860 and AB003521, respectively.
RESULTS
Genetic analysis of the gene homologous to FLO1 in the NewFlo-type flocculation strains.
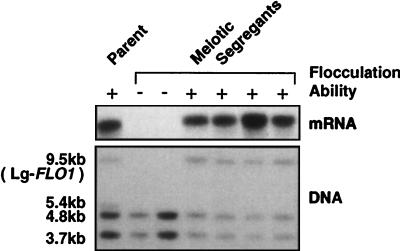
Since little was known about the FLO1 gene or its homologue in NewFlo-type flocculation strains, genetic analysis of the FLO1 gene in a NewFlo-type flocculation strain was carried out. It has been reported that most of bottom-fermenting yeast strains are amphiploid (6) and aneuploid (3, 4), and sporulation efficiencies of these strains are low. As a result of rare meiosis of the NewFlo-type flocculation strain KBY001, both flocculent and nonflocculent segregants were obtained. Southern and Northern analyses of these strains were performed with the FLO1 gene as a probe. The results are shown in Fig. 2. Four HindIII fragments of approximately 9.5, 5.4, 4.8, and 3.7 kb were observed in the parental KBY001 DNA. Two HindIII fragments of 4.8 and 3.7 kb were found to be common to all segregants. A 5.4-kb HindIII fragment could not be found in any segregants. However, a 9.5-kb HindIII fragment was detected only in flocculent segregants. The gene encoded on this fragment that was homologous to FLO1 was named Lg-FLO1 (Lager-type FLO1 gene). Moreover, mRNA homologous to the FLO1 gene was detected only in the flocculent segregants. The size of this mRNA was larger than that of the FLO1 gene in S. cerevisiae. These results suggest that the Lg-FLO1 gene plays an important role in the NewFlo-type flocculation phenotype. The restriction map of the Lg-FLO1 gene was constructed with the results of Southern analysis of the flocculent and nonflocculent segregants (Fig. 3). The restriction maps of the FLO1 and Lg-FLO1 genes differed from each other, suggesting that Lg-FLO1 is a novel gene.
FIG. 2.
Southern and Northern analyses of the FLO1 homologous gene in NewFlo-type flocculation strains. Meiotic segregants were obtained from the parent strain. Plus and minus symbols indicate flocculent and nonflocculent segregants, respectively.
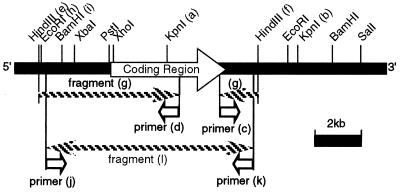
FIG. 3.
Restriction map and cloning strategy for the Lg-FLO1 gene. Details are described in the text.
Cloning and characterization of the Lg-FLO1 gene.
Attempts to clone the full-length Lg-FLO1 gene using probes homologous to the known DNA sequence of the FLO1 gene ended in failure, because this gene could not be maintained in the E. coli strains generally used in genetic manipulation. Therefore, we obtained this gene by a combination of inverse PCR and PCR. The cloning strategy is illustrated in Fig. 3. By using homology with the C terminus of the FLO1 gene, the 5.6-kb KpnI fragment from region a to region b (Fig. 3) was cloned and the DNA sequence of the fragment from region a to region f was determined. With this result, a set of primers labeled c and d (Fig. 3) were synthesized and used for inverse PCR to amplify the 8.2-kb fragment labeled g, in which two HindIII sites of e and f were ligated to each other. The DNA sequence from region h to region i was partially determined, and a set of primers labeled j and k were synthesized and used to amplify the 9-kb fragment labeled l. The restriction map of this fragment coincided well with the theoretical map shown in Fig. 3, suggesting that a fragment containing the full-length Lg-FLO1 gene was obtained.
To confirm whether the Lg-FLO1 gene confers NewFlo-type flocculation ability on yeast cells, the 9-kb fragment labeled l in Fig. 3 was ligated into pYT71 and introduced into the flo1-disruptant strain KY644. The flocculation phenotype of the resulting transformant was compared with that of a transformant with the FLO1 gene-bearing plasmid pKB635. The result is shown in Table 1. The transformant with the Lg-FLO1 gene revealed Ca2+-dependent flocculation ability. Furthermore, flocculation of this transformant was inhibited by mannose, glucose, and maltose but not by galactose, which is typical of NewFlo-type flocculation strains. On the other hand, flocculation of the transformant with the FLO1 gene was inhibited by mannose but not by glucose, maltose, or galactose, which is typical of Flo1-type flocculation. The transformant with the vector revealed no flocculation. These results suggest that the Lg-FLO1 gene in the NewFlo-type flocculation strain is responsible for this phenotype.
TABLE 1.
Flocculation ability and its inhibition by sugar in the transformants
| Clone | Introduced gene | % Flocculation ability with:
|
||||
|---|---|---|---|---|---|---|
| No sugar | Mannose | Glucose | Maltose | Galactose | ||
| KY650 | Lg-FLO1 | 0.702 | 0 | 0 | 0 | 0.458 |
| KY651 | FLO1 | 0.853 | 0.099 | 0.765 | 0.727 | 0.732 |
| KY652 | Vector (pYT71) | 0.001 | 0 | 0 | 0 | 0.003 |
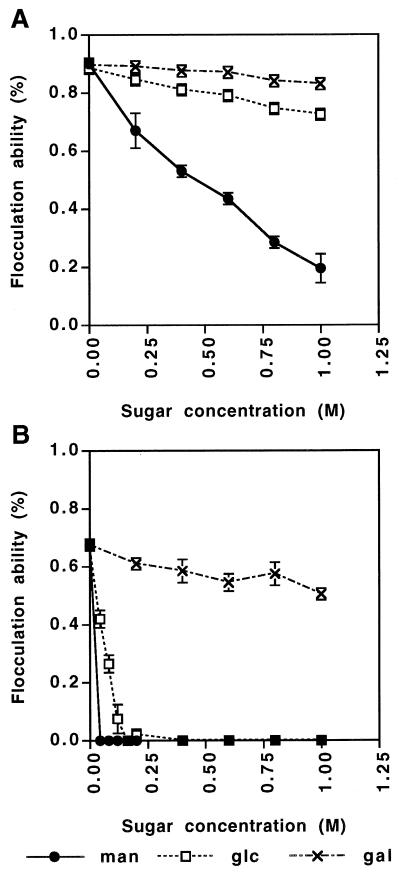
The relationship between sugar concentration and flocculation inhibition of two phenotype transformants was also investigated. The results are shown in Fig. 4. Flocculation of NewFlo-type transformant KY650 was inhibited completely by 0.04 M mannose (Fig. 4B), while 22% flocculation of the Flo1-type transformant KY651 still occurred in the buffer with 1 M mannose compared with flocculation without mannose (Fig. 4A). This result suggested that flocculation of the NewFlo-type transformant is inhibited more easily than that of the Flo1-type transformant by mannose. Furthermore, the concentrations of mannose and glucose required for flocculation inhibition of KY650 differed; the concentrations of mannose and glucose required for complete inhibition of flocculation were 0.04 M and 0.16 M, respectively (Fig. 4B). This result suggested that affinity of the lectin-like protein involved in NewFlo-type flocculation for mannose is stronger than that for glucose.
FIG. 4.
Sugar inhibition of flocculation of transformants carrying the FLO1 gene (KY651) (A) or the Lg-FLO1 gene (HY650) (B). Each datum point represents the mean of three experiments. man, mannose; glc, glucose; gal, galactose.
DNA sequences of the N and C termini of the Lg-FLO1 gene.
Since the full-length Lg-FLO1 gene could not be maintained in E. coli, only partial DNA sequences of the N and C termini were determined. The sequence data are shown in Fig. 5 and Fig. 6. The 694-bp N terminus contains a 636-bp open reading frame which has 61.7% identity with the corresponding region of the reported FLO1 gene, while the 2,903-bp C terminus contains a 2,550-bp open reading frame which has 55.1% identity with the corresponding region of the reported FLO1 gene. By use of these DNA sequences and the restriction map of the Lg-FLO1 gene shown in Fig. 3, the length of the open reading frame of this gene was calculated to be 5.8 kb, which is longer than the coding region of the reported FLO1 gene (4,611 bp) (37). This result is consistent with the Northern analysis, in which the Lg-FLO1 gene was larger than the FLO1 gene (Fig. 2). Both 5′ and 3′ flanking regions of the Lg-FLO1 open reading frame share no significant homology with the corresponding regions of the FLO1 gene. The sequences of S. cerevisiae S288C with the highest homology in these regions were found to be in the 5′ noncoding region of open reading frame YHR211 (chromosome VIII) and in the 3′ noncoding region of open reading frame YAL065 (chromosome I), respectively.
FIG. 5.
Partial DNA sequence of the 5′ region of the Lg-FLO1 gene and predicted amino acid sequence.
FIG. 6.
Partial DNA sequence of the 3′ region of the Lg-FLO1 gene and predicted amino acid sequence.
Since the sugar-binding domain of the Flo1 and Lg-Flo1 proteins has been supposed to be in the N termini (32, 37), the putative amino acid sequences of this region of these proteins were compared. The 240-amino-acid sequences of this region of Flo1 and the corresponding region of Lg-Flo1 are shown in Fig. 7. Amino acids 26 to 40 of the Lg-Flo1 protein had complete identity with the reported partial amino acid sequence of the cell wall protein flocculin, which was purified from the flocculent bottom-fermenting strain MPY1 (30). Some differences were found between the N-terminal amino acid sequences of the Flo1 and Lg-Flo1 proteins, including a 27-amino-acid deletion in the Lg-Flo1 protein. However, the two proteins had a high degree of homology.
FIG. 7.
Comparison of putative amino acid sequences of the N termini of the Flo1 and Lg-Flo1 proteins. Upper and lower sequences represent Flo1p and Lg-Flo1p, respectively. Amino acid residues on black and gray backgrounds indicate identical residues and similar residues between two sequences, respectively.
Region responsible for NewFlo-type flocculation in the Lg-Flo1 protein.
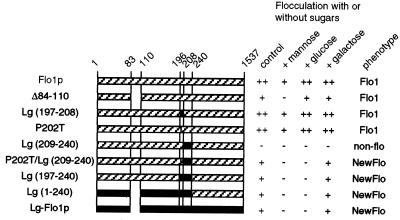
To investigate the region which determines the pattern of sugar recognition in the Flo1 and Lg-Flo1 proteins, FLO1/Lg-FLO1 chimeric genes were constructed and expressed in the FLO1 disruptant strain KY794. The flocculation phenotypes of the resulting transformants are shown in Fig. 8. All substitutions were performed by replacing Flo1 amino acid sequences with the corresponding region of the Lg-Flo1 protein. All amino acid positions are numbered as for the Flo1 protein.
FIG. 8.
Relationship between flocculation with sugars and the FLO1/Lg-FLO1 chimera. Control indicates flocculation without sugar. ▨ and ▪, Flo1 and Lg-Flo1 proteins, respectively.
Substitution of the N-terminal 240 amino acids conferred a NewFlo-type flocculation similar to that conferred by the full-length Lg-Flo1 protein [see clone Lg(1-240)]. Deletion of the region from amino acid 84 to amino acid 110 of the Flo1 protein, which was found in the Lg-Flo1 protein, did not change the flocculation phenotype (see clone Δ84-110). The region which contributes most to sugar recognition in the Lg-Flo1 protein lies between amino acids 197 and 240. Substitution with this region caused NewFlo-type flocculation similar to that observed with the full-length Lg-Flo1 protein [see clone Lg(197-240)]. Substitution of amino acids 197 to 208 did not cause NewFlo-type flocculation, while substitution of amino acids 209 to 240 caused loss of flocculation ability [see clones Lg(197-208) and Lg(209-240)]. Since at least two transformants with each construct were tested and replacement of proline 202 of Lg(209-240) with threonine caused recovery of flocculation ability, it is unlikely that loss of flocculation ability in Lg(209-240) was caused by failure of proper expression.
To investigate the amino acid residue of the region from amino acid 197 to amino acid 208 of the Lg-Flo1 protein responsible for binding to glucose, serine 199, leucine 200, or proline 202 was replaced with alanine, alanine, or threonine, respectively. Replacement of serine 199 or leucine 200 with alanine had little effect on sugar recognition (data not shown). On the other hand, clone P202T/Lg(209-240), in which proline 202 of clone Lg(209-240) was replaced with threonine, showed NewFlo-type flocculation. However, threonine 202 of the Lg-Flo1 protein has little effect without the presence of amino acids 209 to 240 of this protein (see clone P202T). These results suggest that both threonine 202 and the region from amino acids 209 to 240 of the Lg-Flo1 protein are required for NewFlo-type flocculation.
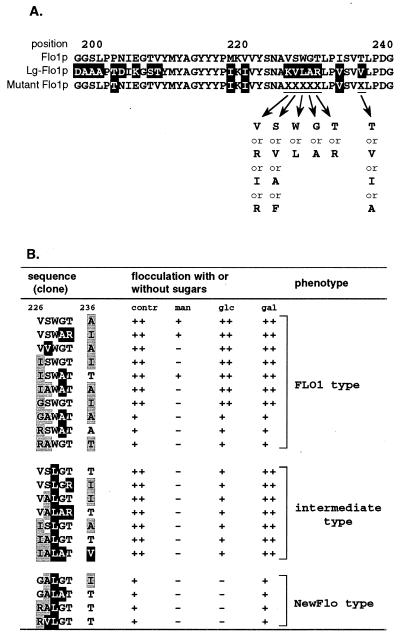
There is little difference in the sequences from amino acids 209 to 240 of the Flo1 and Lg-Flo1 proteins (Fig. 7). However, residues from amino acid 226 to amino acid 230 of these sequences are not homologous. Furthermore, threonine 236 in the Flo1 protein, which has the potential to make a hydrogen bond, is replaced with valine in the Lg-Flo1 protein, which does not make a hydrogen bond. To determine which residues are involved in NewFlo-type flocculation, mutations were introduced into this region by PCR in clone P202T/Lg(209-240). Chimeric FLO1/Lg-FLO1 genes were then introduced into KY794, and the flocculation phenotypes of the transformants were investigated. Furthermore, the 5′ region of the chimeric FLO1/Lg-FLO1 genes in the transformants were amplified by PCR, and their DNA sequences were determined. The results are summarized in Fig. 9. Flocculation of transformants could be classified into three phenotypes: Flo1 type, NewFlo type, and an intermediate phenotype in which flocculation was partially inhibited by glucose. All transformants which showed NewFlo-type or an intermediate phenotype flocculation possessed chimeric genes with leucine 228. On the other hand, all transformants which showed Flo1-type flocculation possessed chimeric genes with tryptophan 228. Additionally, NewFlo-type flocculation transformants possessed chimeric genes in which the amino acid residue 226 was G or R, while the intermediate phenotype flocculation transformants possessed chimeric genes in which the amino acid residue 226 was V or I. On the basis of these results, it is suggested that leucine 228 is required for NewFlo-type flocculation and that the amino acid residue 226 also affects the flocculation phenotype.
FIG. 9.
Relationship between flocculation in the presence of sugars and mutations in the FLO1/Lg-FLO1 chimera. (A) Comparison of the amino acid sequence from amino acid 197 to amino acid 240 of the Flo1 protein, the Lg-Flo1 protein, and mutant Flo1 proteins. Amino acid residues which could be found at positions 226, 227, 228, 229, 230, and 236 (underlined) in mutant Flo1 proteins are indicated. (B) Relationship between amino acid residues actually found at positions 226, 227, 228, 229, 230, and 236 in mutant Flo1 proteins in transformants and flocculation inhibition by sugars. Control indicated flocculation without sugar. In both panels, amino acids without any background indicate amino acids identical to those in the Flo1 protein or in common between Flo1 and Lg-Flo1 proteins, those on a black background indicate amino acids identical to those in the Lg-Flo1 protein, and those on a gray background indicate amino acid residues not present in either the Flo1 or Lg-Flo1 protein.
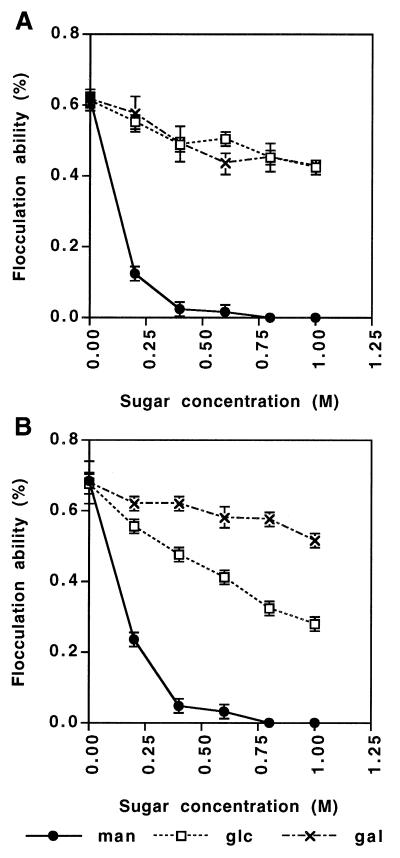
To investigate sugar inhibition of intermediate phenotypeflocculation in detail, concentrations of sugars required for flocculation in VSWGT-A (Flo1 type) and VSLGT-T (intermediate phenotype) were compared. The results are shown in Fig. 10. No significant differences were found between inhibition of flocculation of these strains by mannose and inhibition by galactose. On the other hand, inhibition of flocculation of these strains by glucose differed; flocculation inhibition of VSWGT-A by glucose was as little as inhibition by galactose, while flocculation of VSLGT-T was inhibited to 41% by 1 M glucose compared with flocculation without glucose (compare Fig. 10A and Fig. 10B).
FIG. 10.
Sugar inhibition of flocculation of transformants carrying the FLO1 gene with mutation VSWGT-A (A) and VSLGT-T (B).
DISCUSSION
Yeast flocculation is considered to result from an interaction between a lectin-like protein and a mannose chain present on the cell surface. Though the FLO1 gene is supposed to encode the lectin-like protein involved in flocculation, direct proof has not yet been reported. Much effort has gone into the study of the FLO1 gene of Flo1 phenotype strains, while little is known of the FLO1 homologue of NewFlo phenotype strains. In this study, attention was focused on the FLO1 homologue in strains with distinct flocculation phenotypes distinguished by sugar inhibition.
A new homologue, Lg-FLO1, is strongly believed to govern flocculation ability in NewFlo-type strains for the following reasons. (i) The result of Southern analysis showed a close correlation between the existence of Lg-FLO1 and flocculation ability in meiotic segregants. (ii) mRNA homologous to FLO1 was detected only in the flocculent segregants.
Attempts to clone either the full-length Lg-FLO1 gene from a genomic phage library of KBY001 or the 9.5-kb HindIII fragment shown in Fig. 2 ended in failure. Although a plasmid bearing the full-length Lg-FLO1 gene could be constructed in S. cerevisiae, transformation of E. coli DH5 by this plasmid was not successful (unpublished data). Therefore, it was concluded that the full-length Lg-FLO1 gene could not be maintained in E. coli. It has been reported that the copy number of the highly repetitive sequence in the FLO1 gene is reduced easily in E. coli (2, 32, 37). Since either N- or C-terminal region of the Lg-FLO1 gene could be maintained stably in E. coli, the highly repetitive sequence in the middle of this gene is supposed to be unstable in E. coli.
The DNA sequence highly homologous to the 5′ and 3′ flanking regions of the Lg-FLO1 gene was found in chromosomes VIII and I, respectively, of S. cerevisiae S288C. In the genome of S. cerevisiae S288C, there are seven open reading frame sequences sharing homology with the FLO1 gene (33); YAR050, which is the FLO1 gene, and YAR061/062 on the right arm of chromosome I; YAL063 and its pseudogene YAL065, on the left arm of chromosome I; YHR211, which has been suggested to be the FLO5 gene (1, 34), and its pseudogene YHR213, on the right arm of chromosome VIII; and YKR102, on the right arm of chromosome XI. Furthermore, Southern analysis of the chromosomes separated by pulsed-field gel electrophoresis showed that the Lg-FLO1 gene is located on the chromosome that is the same size as chromosome VIII (unpublished data). Therefore, it is supposed that the Lg-FLO1 gene originated from a recombination between YHR211 and YAL065 after these homologues had been generated by duplication and translocation.
Recent studies have suggested that the Flo1 protein is a cell wall protein located on the cell surface (2, 5, 30). In this study, the FLO1 and Lg-FLO1 genes conferred the Flo1-type and NewFlo-type flocculation phenotypes, respectively (Table 1). Since Flo1-type flocculation is inhibited by mannose but not by glucose, while NewFlo-type flocculation is inhibited by both of these sugars (29), this result strongly suggests that the FLO1 and Lg-FLO1 genes encode a mannose-specific protein and a mannose/glucose-specific lectin-like protein, respectively.
Amino acids 26 to 40 of the Lg-Flo1 protein had complete identity with the reported partial amino acid sequence of the cell wall protein flocculin (30), suggesting that the Lg-Flo1 protein and flocculin are identical. The putative amino acid sequence of the Lg-Flo1 protein contains an additional 24 amino acid residues in the N-terminal region, suggesting that this region is a signal sequence for secretion. Although Straver et al. could not detect mannose-binding activity in flocculin, there is a possibility that more sensitive methods may detect this activity with the Flo1 or Lg-Flo1 protein.
Replacement of the N terminus of the Flo1 protein by the corresponding region of the Lg-Flo1 protein caused conversion from Flo1-type to NewFlo-type flocculation, suggesting that the N-terminal regions are responsible for sugar recognition in the Flo1 and Lg-Flo1 proteins (Fig. 8). This finding coincided with the previous suggestion that the Flo1 protein exposes its N-terminal region to the exterior which is involved in flocculation (32, 37). Therefore, we investigated the region responsible for sugar recognition in the N termini of Flo1/Lg-Flo1 proteins by comparing the differences between the sequences and the characteristics of these proteins. The region spanning amino acids 197 to 240 of the Lg-Flo1 protein appears essential for recognition of glucose. Moreover, the regions from amino acids 197 to 208 and from amino acids 209 to 240 are considered to be required for the recognition of glucose. The loss of flocculation ability in clone Lg(209-240) suggests that the region from amino acid 209 to amino acid 240 of the Flo1 protein is essential for mannose recognition and that this region of the Lg-Flo1 protein is unable to recognize either mannose or glucose. The phenotypes of clones P202T and P202T/Lg(209-240) suggest that threonine 202 interacts with mannose and glucose when the region from amino acid 209 to amino acid 240 is unable to bind mannose. The flocculation phenotypes of the FLO1/Lg-FLO1 chimeras with amino acid replacements suggest that tryptophan 228 must be replaced by leucine to produce a flocculation phenotype inhibited by glucose (Fig. 9). However, it is also suggested that the amino acid residues neighboring leucine 228, especially amino acid residue 226, affect the flocculation phenotype.
During detailed investigation of sugar inhibition of flocculation, it was observed that flocculation of either the NewFlo type or the intermediate phenotype can be inhibited more by mannose than by glucose (Fig. 4 and 10), suggesting that the difference between these phenotypes was caused only by strength of flocculation ability. The amino acids of the Flo1 protein in VSLGT-T that are different from those in VSWGT-A are only leucine 228 and threonine 236, which caused partial inhibition by glucose. Since threonine 236 was found in the original Flo1 protein, which confers Flo1-type flocculation ability, it is suggested that this amino acid residue is not responsible for flocculation of VSLGT-T inhibited by glucose. Therefore, this result also suggests that tryptophan 228 must be replaced by leucine to produce a flocculation phenotype inhibited by glucose. Numerical data could not be obtained for the flocculation ability of the NewFlo-type strains with amino acid substitutions, because the flocculation ability of the NewFlo-type strains, in which the FLO1/Lg-FLO1 chimera was controlled by the TDH3 promoter, was weak. Therefore, it is possible that the amino acid replacement in the FLO1/Lg-FLO1 chimeras has not produced a NewFlo-type flocculation which is inhibited by mannose more easily than by glucose.
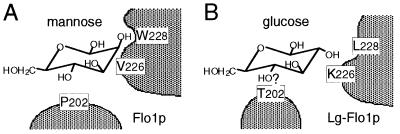
From the results of this study, we propose the following model for sugar recognition by the Flo1 and Lg-Flo1 proteins (Fig. 11). The only difference between the structure of mannose and that of glucose is the orientation of the C-2 hydroxyl group. The domain formed by tryptophan 228 and its neighboring amino acid residues in the Flo1 protein recognizes the C-2 hydroxyl group of mannose but does not recognize the C-2 hydroxyl group of glucose. In the Lg-Flo1 protein, the domain formed by leucine 228 and its neighboring amino acid residues does not recognize the C-2 hydroxyl group of either mannose or glucose, while threonine 202 probably interacts with another hydroxyl group, allowing recognition of both mannose and glucose. The C-4 hydroxyl group is supposed to be one of the candidates with which threonine 202 interacts, because galactose cannot inhibit NewFlo-type flocculation. The difference between the structure of glucose and that of galactose is the orientation of the C-4 hydroxyl group.
FIG. 11.
Model for sugar recognition by the Flo1 and Lg-Flo1 proteins.
In this study, it was found that modification of two regions was required to change the mannose-specific sugar recognition pattern of the Flo1 protein to the mannose/glucose-specific pattern. In other words, function is well conserved between the Flo1 and Lg-Flo1 proteins. It is known that lectins have diverse molecular architectures (38), and it has been reported that the topological similarity between two lectins, which share no homology, is conserved (16). Therefore, it is suggested that the Flo1 and Lg-Flo1 proteins have important biological functions in nature. Smit et al. reported that flocculation of S. cerevisiae is induced by nutrient limitation (25). Furthermore, it should be noted that some of flocculation genes are also involved in filamentous growth (14, 15), which is induced by nitrogen starvation (9). Accordingly, yeast flocculation is supposed to be an important characteristic in an environment with scarce nutrients. However, further work is needed to determine the role of flocculation in nature.
ACKNOWLEDGMENT
We thank Yukio Tamai for providing plasmids. We also thank Takashi Nakamura for his suggestions.
REFERENCES
- 1.Bindard F, Blondin B, Dequin S, Vezinhet F, Barre P. Cloning and analysis of the FLO5 flocculation gene from S. cerevisiae. Curr Genet. 1994;25:196–201. doi: 10.1007/BF00357162. [DOI] [PubMed] [Google Scholar]
- 2.Bidard F, Bony M, Blondin B, Dequin S, Barre P. The Saccharomyces cerevisiae FLO1 flocculation gene encodes for a cell wall surface protein. Yeast. 1995;11:809–822. doi: 10.1002/yea.320110903. [DOI] [PubMed] [Google Scholar]
- 3.Bilinski C A, Russell I, Stewart G G. Analysis of sporulation in brewer’s yeast: induction of tetrad formation. J Inst Brew. 1986;92:594–598. [Google Scholar]
- 4.Bilinski C A, Russell I, Stewart G G. Physiological requirements for induction of sporulation in lager yeast. J Inst Brew. 1987;93:216–219. [Google Scholar]
- 5.Bony M, Thines-Sempoux D, Barre P, Blondin B. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein. Flo1p. J Bacteriol. 1997;179:4929–4936. doi: 10.1128/jb.179.15.4929-4936.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Børsting C, Hummel R, Schultz E R, Rose T M, Pedersen M B, Knudsen J, Kristiansen K. Saccharomyces carlsbergensis contains two functional genes encoding the acyl-coA binding protein, one similar to the ACB1 gene from S. cerevisiae and one identical to the ACB1 gene from S. monacensis. Yeast. 1997;13:1409–1421. doi: 10.1002/(SICI)1097-0061(199712)13:15<1409::AID-YEA188>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Drickamer K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J Biol Chem. 1988;263:9557–9560. [PubMed] [Google Scholar]
- 8.Eliton E A, Warner J R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- 9.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Hereford L, Fahrner K, Woolford J J, Rosbash M. Isolation of yeast histone genes H2A and H2B. Cell. 1979;18:1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- 12.Ito H, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations or thiol compounds. Agric Biol Chem. 1984;48:341–347. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi O, Suda H, Ohtani T, Sone H. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Mol Gen Genet. 1996;251:707–715. doi: 10.1007/BF02174120. [DOI] [PubMed] [Google Scholar]
- 14.Lambrechts M G, Bauer F F, Marmur J, Pretorius I S. MucI, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Styles C A, Fink G R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobsanov Y D, Gitt M A, Leffler H, Barondes S H, Rini J M. X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-Å resolution. J Biol Chem. 1993;268:27034–27038. doi: 10.2210/pdb1hlc/pdb. [DOI] [PubMed] [Google Scholar]
- 17.McAlister L, Holland M J. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985;260:15019–15027. [PubMed] [Google Scholar]
- 18.Miki B L A, Poon N H, James A P, Seligy V L. Possible mechanism for flocculation interactions in Saccharomyces cerevisiae. J Bacteriol. 1982;150:890–899. doi: 10.1128/jb.150.2.890-899.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreuder M P, Mooren A T A, Toschka H Y, Verrips C T, Klis F M. Immobilizing proteins on the surface of yeast cells. Trends Biotechnol. 1996;14:115–120. doi: 10.1016/0167-7799(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 22.Shankar C S, Umesh-Kumar S. A surface lectin associated with flocculation in brewing strains of Saccharomyces cerevisiae. Microbiology. 1994;140:1097–1101. doi: 10.1099/13500872-140-5-1097. [DOI] [PubMed] [Google Scholar]
- 23.Sharon N, Lis H. Legume lectins: a large family of homologous proteins. FASEB J. 1990;4:3198–3208. doi: 10.1096/fasebj.4.14.2227211. [DOI] [PubMed] [Google Scholar]
- 24.Sheman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 25.Smit G, Straver M H, Lugtenberg J J, Kijne J W. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl Eviron Microbiol. 1992;58:3709–3714. doi: 10.1128/aem.58.11.3709-3714.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone H, Fujii T, Kondo K, Shimizu F, Tanaka J, Inoue T. Nucleotide sequence and expression of the Enterobacter aerogenes alpha-acetate decarboxylase gene in brewer’s yeast. Appl Environ Microbiol. 1988;54:38–42. doi: 10.1128/aem.54.1.38-42.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stratford M. Yeast flocculation: recognition of physiological and genetic viewpoints. Yeast. 1992;8:25–38. doi: 10.1002/yea.320080103. [DOI] [PubMed] [Google Scholar]
- 28.Stratford M. Yeast flocculation: flocculation onset and receptor availability. Yeast. 1993;9:85–93. doi: 10.1002/yea.320090111. [DOI] [PubMed] [Google Scholar]
- 29.Stratford M, Assinder S. Yeast flocculation: Flo1 and NewFlo phenotypes and receptor structure. Yeast. 1991;7:559–574. doi: 10.1002/yea.320070604. [DOI] [PubMed] [Google Scholar]
- 30.Straver M H, Smit G, Kijne J W. Purification and partial characterization of a flocculin from brewer’s yeast. Appl Eviron Microbiol. 1994;60:2754–2758. doi: 10.1128/aem.60.8.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straver M H, Traas V M, Smit G, Kijne J W. Isolation and partial purification of mannose-specific agglutinin from brewer’s yeast involved in flocculation. Yeast. 1994;10:1183–1193. doi: 10.1002/yea.320100906. [DOI] [PubMed] [Google Scholar]
- 32.Teunissen A W R H, Holub E, van den Hucht J, van der Berg J A, Steensma H Y. Sequence of the FLO1 gene from Saccharomyces cerevisiae. Yeast. 1993;9:423–427. doi: 10.1002/yea.320090413. [DOI] [PubMed] [Google Scholar]
- 33.Teunissen A W R H, Steensma H Y. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast. 1995;11:1001–1013. doi: 10.1002/yea.320111102. [DOI] [PubMed] [Google Scholar]
- 34.Teunissen A W R H, van den Berg J A, Steensma H Y. Localization of the dominant flocculation genes FLO5 and FLO8 of Saccharomyces cerevisiae. Yeast. 1995;11:735–745. doi: 10.1002/yea.320110805. [DOI] [PubMed] [Google Scholar]
- 35.Vallette F M, Mege E, Reiss A, Adesnik M. Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res. 1989;17:723–733. doi: 10.1093/nar/17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watari J, Takata Y, Ogawa M, Nishikawa N, Kamimura M. Molecular cloning of a flocculation gene in Saccharomyces cerevisiae. Argic Biol Chem. 1989;53:901–903. [Google Scholar]
- 37.Watari J, Takata Y, Ogawa M, Sahara H, Koshino S, Onnela M-L, Airaksinen U, Jaatinen R, Penttila M, Keränen S. Molecular cloning and analysis of the yeast flocculation of gene FLO1. Yeast. 1994;10:211–225. doi: 10.1002/yea.320100208. [DOI] [PubMed] [Google Scholar]
- 38.Weis W I, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]