Abstract
The oxidative pentose phosphate pathway is required for function of the alternative pyrimidine biosynthetic pathway, a pathway that allows thiamine synthesis in the absence of the PurF enzyme in Salmonella typhimurium. Mutants that no longer required function of the oxidative pentose phosphate pathway for thiamine synthesis were isolated. Further phenotypic analyses of these mutants demonstrated that they were also sensitive to the presence of serine in the medium, suggesting a partial defect in isoleucine biosynthesis. Genetic characterization showed that these pleiotropic phenotypes were caused by null mutations in yjgF, a previously uncharacterized open reading frame encoding a hypothetical 13.5-kDa protein. The YjgF protein belongs to a class of proteins of unknown function that exhibit striking conservation across a wide range of organisms, from bacteria to humans. This work represents the first detailed phenotypic characterization of yjgF mutants in any organism and provides important clues as to the function of this highly conserved class of proteins. Results also suggest a connection between function of the isoleucine biosynthetic pathway and the requirement for the pentose phosphate pathway in thiamine synthesis.
The increasing number of completed genome sequences has resulted in the identification of new families of hypothetical proteins whose function has yet to be established. The lack of existing mutants defective in these conserved proteins suggests novel, complex, or subtle phenotypes. Through our work on thiamine synthesis in Salmonella typhimurium, we have isolated mutants defective in the recently identified YER057c/YjgF protein family. Our data suggest that defects in this protein result in complex phenotypes involving thiamine and isoleucine biosynthesis.
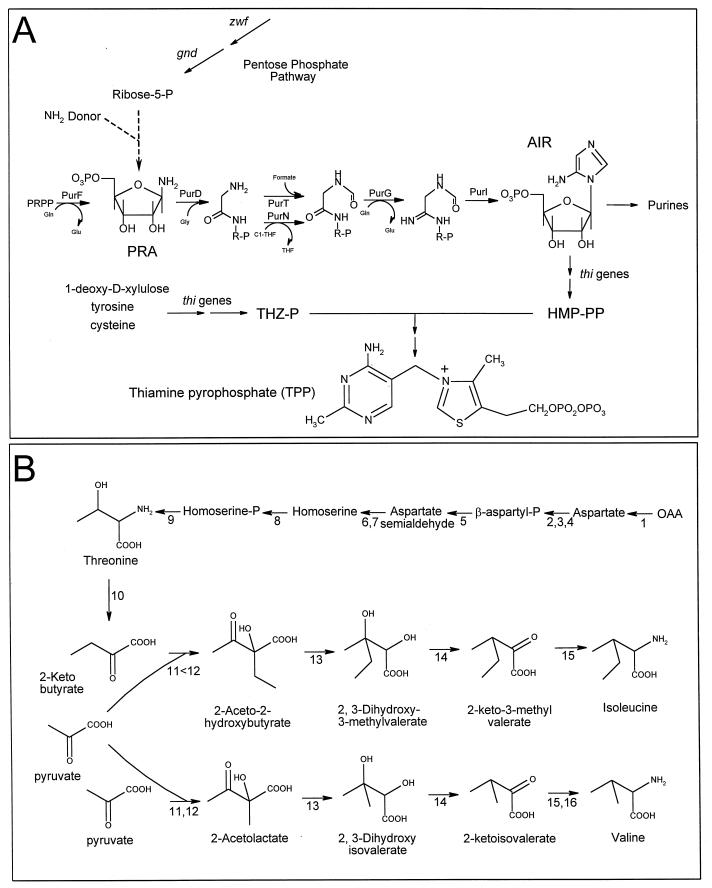
Thiamine pyrophosphate (TPP) serves as an essential cofactor for a number of metabolic reactions involving the removal or transfer of C2 units. Despite the important role of TPP in cellular metabolism, its synthesis and regulation are not well understood in any organism. TPP is formed from two precursors, 4-methyl-5-(β-hydroxyethyl)thiazole phosphate (THZ-P) and 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate (HMP-PP). These compounds are joined and subsequently phosphorylated as shown in Fig. 1A. Although many of the enzymatic steps in both the THZ-P and HMP-PP pathways have not been clearly defined, the major precursor molecules for both of these compounds have been determined by labeling studies (17, 20, 28, 29). In particular, the purine pathway intermediate, aminoimidazole ribotide (AIR), has been shown to provide all of the atoms in HMP (28, 50, 51).
FIG. 1.
Pathway schematics. (A) Biosynthetic pathway for TPP. The involvement of the purine pathway in HMP-PP synthesis is shown with structural intermediates prior to the AIR branch point. Arrows denoted with dotted lines represent proposed steps. Reactions involved in the conversion of AIR to HMP-PP and in the synthesis of THZ-P have not been clearly defined. Genes whose products are required for selected reactions are indicated next to the relevant arrows. Abbreviations: R-P, ribose-5-phosphate, PRPP, phosphoribosylpyrophosphate. (B) Biosynthetic pathways for the branched-chain amino acids isoleucine and valine. Enzymes that catalyze specific steps are as follows: 1, aspartate transaminase; 2, 3, and 4, aspartate kinases I, II, and III, respectively; 5, aspartate semialdehyde dehydrogenase; 6 and 7, homoserine dehydrogenases I and II, respectively; 8, homoserine kinase; 9, threonine synthase; 10, threonine deaminase; 11 and 12, acetohydroxy acid synthases I and II, respectively; 13, acetohydroxy acid isomeroreductase; 14, dihydroxy acid dehydratase; 15, transaminase B; 16, transaminase C. OAA, oxaloacetic acid.
Although the involvement of the purine pathway in the synthesis of HMP is clear, there is substantial genetic and biochemical evidence indicating that the first enzyme of the purine pathway, phosphoribosylpyrophosphate amidotransferase (PurF) (EC 2.4.2.14), is not required for HMP synthesis in S. typhimurium under all conditions. Mutants defective in purF are able to grow in the absence of thiamine when glucose is used as a carbon source if pantothenate is also supplied in the medium (23). Similarly, purF mutants do not require thiamine when grown on a number of nonglucose carbon sources, such as gluconate or ribose (54). The pathway responsible for synthesis of HMP independent of the PurF enzyme has been defined as the alternative pyrimidine biosynthetic (APB) pathway (21, 54); recent biochemical data suggest that phosphoribosylamine (PRA), or a derivative, is an intermediate in this pathway (24).
Significant progress in our understanding of the APB pathway has been made by the isolation and characterization of mutants unable to synthesize thiamine in a purF background. One class of mutants, designated apbA, was defective in a pantothenate biosynthetic enzyme (ketopantoate reductase [PanE]) (32, 33), consistent with previous results implicating a role for pantothenate in PurF-independent thiamine synthesis (23). A second class of these mutants was defective in the oxidative pentose phosphate pathway, affecting either glucose-6-phosphate dehydrogenase (Zwf) or gluconate-6-phosphate dehydrogenase (Gnd) (25, 54). Addition of ribose-5-phosphate (ribose-5-P) restored function of the APB pathway in these mutants, suggesting that the role of these enzymes in HMP synthesis was to supply ribose-5-P. These results led to the model shown in Fig. 1A which implicates ribose-5-P and an amine donor as precursors to PRA. Repeated attempts have failed to identify either the predicted PRA-forming activity or mutants defective in this step (27). There are several possible explanations for this. It is possible that the correct substrates have not been identified and/or that the PRA-forming activity is required for another cellular function.
In this report, we describe the isolation and characterization of mutations that allow function of the APB pathway in the absence of the pentose phosphate pathway. These mutations were found to disrupt a previously uncharacterized open reading frame (ORF) encoding a hypothetical 13.5-kDa protein. We have designated this gene yjgF based on homology to the respective ORF in Escherichia coli. The YjgF protein belongs to the YER057c/YjgF protein family, a class of proteins of unknown function that exhibit striking conservation across a wide range of organisms. Characterization of these mutants revealed that they also were sensitive to the presence of serine in the medium, exhibiting a requirement for isoleucine under this condition. The phenotypes caused by yjgF mutations suggest that the YjgF protein may be involved in regulation or function of the isoleucine biosynthetic pathway. Further, results suggest a connection between isoleucine biosynthesis and function of the APB pathway in thiamine synthesis.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
Unless otherwise noted, all strains used in this study are derivatives of S. typhimurium LT2 and are listed with their respective genotypes in Table 1. MudJ and MudA refer to the Mudl1734 and Mud1-8 transposons, respectively, which have been described elsewhere (11, 39). Tn10d(Tc) refers to the transposition-defective mini-Tn10(Tn10Δ16Δ17) (67).
TABLE 1.
Bacterial strains
| Strain | Genotype |
|---|---|
| LT2 | Wild type |
| DM574 | purF2085 gnd175::Tn10d(Tc) |
| DM730 | purF2085 gnd181 apbA1::MudJ |
| DM1936 | purF2085 |
| DM1231 | purF2085 gnd174::MudJ |
| DM1247 | purF2085 gnd181 apbA1::MudJ yjgF1::Tn10d(Tc) |
| DM1248 | purF2085 gnd181 apbA1::MudJ yjgF2::Tn10d(Tc) |
| DM2944 | purF2085 pyrB2694::MudA yjgF1::Tn10d(Tc) |
| DM2946 | purF2085 pyrB2694::MudA yjgF2::Tn10d(Tc) |
| DM3215 | purF2085 gnd181 apbA7::Tn10d(Tc) |
| DM3477 | purF2085 gnd181 apbA7::Tn10d(Tc) yjgF3::MudJ |
| DM3478 | purF2085 gnd181 apbA7::Tn10d(Tc) yjgF4::MudJ |
| DM3479 | purF2085 gnd181 apbA7::Tn10d(Tc) yjgF5::MudJ |
| DM3480 | yjgF3::MudJ |
| DM3488 | purF2085 gnd181 pgi::Tn5 |
| DM3489 | purF2085 gnd181 pgi::Tn5 yjgF1::Tn10d(Tc) |
| DM3502 | purF2085 zwf23::MudJ |
| DM3503 | purF2085 zwf23::MudJ yjgF1::Tn10d(Tc) |
| DM3660 | yjgF3::MudJ/pT7-5YjgF1 |
| DM4053 | yjgF1::Tn10d(Tc) |
| DM4440 | relA21::Tn10d(Tc) |
| DM4442 | yjgF1::Tn10d(Tc) hisT290::Tn5 |
| DM4762 | purF2085 gnd174::MudJ ilvA595::Tn10 |
| DM4763 | purF2085 gnd174::MudJ |
| DM4765 | purF2085 gnd174::MudJ ilvA219 |
| DM4767 | purF2085 gnd174::MudJ yjgF2::Tn10d(Tc) |
| DM4769 | purF2085 gnd174::MudJ yjgF2::Tn10d(Tc) ilvA219 |
| DM4772 | purF2085 gnd175::Tn10d(Tc) yjgF3::MudJ |
| DM5027 | yjgF3::MudJ/pSU19YjgFa |
| DM5031 | BL21/λDE3/pT7-5YjgFa |
| DM5032 | BL21/λDE3/pT7-6YjgFa |
| BL21/λDE3 | F−ompT hsdSB(rB− mB−) gal dcm (E. coli) |
| TT9534 | pyrB2694::MudA |
| TT10288 | hisD9953::MudJ his-9944::MudA |
| TV086 | ilvA595::Tn10 |
| TV088 | ilvA219 |
Unless otherwise indicated, no-carbon E medium supplemented with 1 mM MgSO4 (18, 66) was used as minimal medium. Carbon sources were used at a concentration of 16 mM, with the exception of pyruvate (24 mM), glycerol (55 mM), and acetate (40 mM). Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) was used as rich medium. Luria broth was used for experiments involving plasmid isolation and protein overexpression. Difco BiTek agar was added (15 g/liter) for solid medium. When present in the culture media, and unless otherwise stated, the compounds were used at the following final concentrations: adenine, 0.4 mM; thiamine, 0.5 μM; pantothenate, 100 μM; serine, 4 mM; isoleucine, 0.3 mM. The final concentrations of the antibiotics in rich and minimal medium were as follows: tetracycline, 20 and 10 μg/ml, respectively; kanamycin, 50 and 125 μg/ml, respectively, and ampicillin, 30 and 15 μg/ml, respectively. Unless otherwise stated, all chemicals were purchased from Sigma Chemical Co., St. Louis, Mo.
Genetic techniques. (i) Transduction methods.
Transductional crosses were performed by using the high-frequency general transducing mutant of bacteriophage P22 (HT105/1, int-201) (58). Methods for transductional crosses, purification of transductants from phage, and identification of phage-free transductants have been described elsewhere (22, 54).
(ii) Mutant isolation.
A P22 lysate grown on a pool of >70,000 cells containing random Tn10d(Tc) insertions was generated as described elsewhere (40, 43) and mutagenized with hydroxylamine (18, 38). This lysate was used to transduce DM730 (purF2085 gnd181 apbA1::MudJ) to tetracycline resistance (Tcr) on nutrient agar plates. The Tcr transductants were screened for growth on glucose medium supplemented with adenine and tetracycline. Upon reconstruction, the mutants isolated in this screen were found to grow weakly on glucose-adenine medium but were indistinguishable from a purF mutant (DM1936) on glucose medium supplemented with adenine and pantothenate. MudJ insertion mutants were isolated as follows. A P22 phage lysate grown on TT10288, which contains a MudJ construct that allows independent transposition events by cis complementation (41), was used to transduce DM3215 [purF2085 gnd181 apbA7::Tn10d(Tc)] to kanamycin resistance (Knr). Knr transductants were screened for those that could grow on glucose medium supplemented with adenine, pantothenate, and kanamycin. All putative mutations [Tn10d(Tc) and MudJ insertions] were reconstructed into the parent strain to confirm the observed phenotype. Although an apbA mutation was present in the strains used for mutant isolation, the phenotypes described are independent of this mutation.
(iii) Construction of strains containing the ilvA feedback-resistant mutation.
A P22 phage lysate grown on TV086 [ilvA595::Tn10d(Tc)] was used to transduce DM1231 (purF2085 gnd174::MudJ) to Tcr, creating DM4762 [purF2085 gnd174::MudJ ilvA595::Tn10d(Tc)]. DM4762 was subsequently transduced with a P22 phage lysate grown on either TV088 (ilvA219) or LT2 (ilvA+). Transductants that grew on glucose medium supplemented only with adenine and thiamine, i.e., Ile+, were saved and tested to confirm they were Tcs, creating strains DM4765 (purF2085 gnd174::MudJ ilvA219) and DM4763 (purF2085 gnd174::MudJ ilvA+). The yjgF2::Tn10d(Tc) insertion was then introduced via P22 transduction into DM4765 and DM4763 by using DM1248 [purF2085 gnd181 apbA1::MudJ yjgF2::Tn10d(Tc)] as a donor and selecting for Tcr.
Phenotypic characterization.
Nutritional requirements of mutants were tested in solid medium by using agar overlays and in liquid medium by using growth curves as previously described (54). Data presented are representative of at least three independent experiments. Compounds that corrected the serine sensitivity of yjgF mutants were identified by spotting pools of overlapping compounds on solid medium (18).
Molecular biology techniques. (i) Plasmid manipulation and recombinant DNA techniques.
Standard methods were used for DNA restriction endonuclease digestion and ligation. All restriction enzymes and ligase were purchased from Promega (Madison, Wis.). Plasmid DNA was isolated with the QIAprep spin plasmid kit, and DNA fragments requiring purification were purified with the Qiaquick gel extraction kit (Qiagen, Chatsworth, Calif.). Plasmids were transferred between strains by electroporation with a Bio-Rad (Richmond, Calif.) E. coli pulser as suggested by the manufacturer. Primers for PCRs and sequencing were generated by the University of Wisconsin Biotechnology Center-Nucleic Acid and Protein Facility (Madison, Wis.) and Genosys Biotechnologies, Inc. (The Woodlands, Tex.).
(ii) Localization of yjgF mutations by PCR amplification.
The Tn10-I (5′ GACAAGATGTGTATCCACCTTAAC 3′) and MuR (5′ GCTTTCGCGTTTTTCGTG 3′) primers, specific to the ends of the Tn10d(Tc) and MudJ insertions, respectively, were used to PCR amplify the DNA between these insertions in DM2944 [purF2085 yjgF1::Tn10d(Tc) pyrB2694::MudA] and DM2946 [purF2085 yjgF2::Tn10d(Tc) pyrB2694::MudA] as described previously (68). Amplification was performed by using Vent (Exonuclease-minus) polymerase (New England Biolabs) in a Thermolyne Temp-Tronic thermocycler. For DM2944, PCR conditions were as follows: denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1.5 min. For DM2946, PCR conditions were as follows: denaturation at 95°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1.75 min. The Tn10d(Tc) proximal end of the 1.1-kb fragment resulting from each PCR amplification was sequenced by using the pyrBI-1 primer (5′ ATTTGGGTTGGTTATGAGAT 3′).
(iii) Cloning of yjgF.
The yjgF gene and adjacent sequences were amplified from LT2 by using primers specific to the ends of the mgtA and pyrI genes, XbamgtA-1 (5′ GCTCTAGAGCGCATATGATCCGTACCCGC 3′) and XbapyrI-1 (5′ GCTCTAGAGCTCGCCCTCAAATGCAAATAC 3′), respectively. Primers were designed with XbaI restriction sites at the ends to facilitate cloning. Amplification was performed as described above. PCR conditions were as follows: denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The resulting 1.4-kb product was digested with XbaI and ligated into the pT7-5 overexpression vector (61) also digested with XbaI. The ligation mix was electroporated into DM3480, and Apr electroporants that grew on glucose medium containing serine were obtained. One plasmid that contained the 1.4-kb insert was designated pT7-5YjgF1.
Following sequence analysis (see below), it was found that an additional ORF (besides yjgF) was present between mgtA and pyrBI in S. typhimurium. To obtain a single-gene clone of yjgF, the yjgF gene was amplified from the pT7-5YjgF1 plasmid by using the XbapyrI-1 primer (above) and a primer specific to sequence immediately following the yjgF gene, yjgF-end (5′ CTGTTGGTGCCCCGAACC 3′). PCR conditions were denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1.5 min. The resulting 600-bp fragment was blunt end ligated into the pSU19 vector (48) previously digested with HincII, creating pSU19YjgFa.
To construct a single-gene clone for overexpression of YjgF, pSU19YjgFa was digested with EcoRI and PstI. The resulting 600-bp fragment containing the yjgF gene was gel purified and ligated into pT7-5 and pT7-6 digested with the same restriction enzymes, creating plasmids pT7-5YjgFa and pT7-6YjgFa, respectively. Restriction fragment digestion patterns with either PvuII/XmnI or HindIII/XmnI confirmed that these two constructs contained the yjgF gene in opposite orientations with respect to the T7 promoter. pT7-6YjgFa contained the yjgF gene in the correct orientation for T7-dependent expression.
(iv) DNA sequencing.
The 1.4-kb insert of pT7-5YjgF1 was sequenced by the University of Wisconsin Biotechnology Center-Nucleic Acid and Protein Facility. Primers in addition to the XbamgtA-1 and XbapyrI-1 primers above were designed as sequence was determined. The additional primers used for sequencing were mgtA-2 (5′ GCCAGCGTGCTTAATCC 3′), pyrBI-2 (5′ GGTCGATCCGAAAACCG 3′), and yjgF promoter (5′ GCCTTCACTAATGGTACG 3′). The entire 1.4-kb insert was sequenced at least three times.
Computer analysis.
DNA sequence was analyzed with BLAST (Basic Local Alignment Search Tool) (1). The DNASTAR alignment program (Madison, Wis.) was used to line up homologous proteins.
Overexpression of YjgF.
The overexpression plasmids pT7-5YjgFa and pT7-6YjgFa were constructed as described above. These plasmids were electroporated into E. coli BL21/λDE3, generating strains DM5031 and DM5032, respectively. These strains contain the T7 RNA polymerase on a lambda lysogen under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. For induction, strains were grown with shaking at 37°C to 60 Klett units in Luria broth containing ampicillin. IPTG (Fischer Biotech, Chicago, Ill.) was added at a final concentration of 400 μM, and incubation was continued at 37°C for 3 h. After this time, 1 ml of cells was removed, pelleted, and resuspended in 100 μl of cracking buffer (3). Proteins were subsequently separated by sodium dodecyl sulfate (SDS)–15% polyacrylamide gel electrophoresis (PAGE) by the Bio-Rad MiniProtean electrophoresis system and visualized by Coomassie blue staining.
Stringent response assays.
Cultures of LT2, DM3480 (yjgF3::MudJ), and DM4440 (relA21::Tn10) were grown overnight in nutrient broth (NB) medium and then subcultured (1/25 dilution) into the same medium. After growth at 37°C to an optical density at 600 nm (OD600) of 0.15 to 0.2, two 0.6-ml aliquots were removed to prewarmed test tubes. Lysine hydroxamate (49) (Sigma) was then added at a final concentration of 400 μg/ml to induce amino acid starvation (time = −5 min). At t = 0, 5 μl of 32Pi (H3PO4, 1 mCi/ml; DuPont) was added. One hundred-microliter aliquots were subsequently removed into 3 ml of ice-cold 10% (wt/vol) trichloroacetic acid (TCA) at t = 5, 15, 30, 45, and 60 min. The TCA-precipitatable material was filtered, washed, and counted as described elsewhere (34). Control cultures were monitored by OD600, and counts were corrected for optical density at each time point. Lysine hydroxamate inhibited cell growth of all of the strains to the same extent, as determined by growth rates.
Nucleotide sequence accession number.
The nucleotide sequence of yjgF has been submitted to GenBank under accession no. AF095578.
RESULTS
Isolation of mutations that suppress the requirement for Gnd in thiamine synthesis.
Both purF and purF apbA mutants can grow on glucose minimal medium containing adenine and pantothenate due to PurF-independent thiamine synthesis, defined as the APB pathway (Fig. 1A) (25). When a gnd mutation is introduced into these strains, it causes a thiamine requirement, presumably because thiamine synthesis is disrupted. To further investigate the role of the pentose phosphate pathway in thiamine synthesis, mutations that allowed PurF-independent thiamine synthesis in the absence of Gnd were isolated. A P22 phage lysate grown on a pool of Tn10d(Tc) insertions was treated with hydroxylamine and used to transduce DM730 (purF2085 apbA1::MudJ gnd181) to tetracycline resistance. Derivatives from this cross that grew on glucose minimal medium supplemented with adenine and pantothenate were saved. Seven mutants with the desired phenotype were isolated from a screen of 16,000 Tcr transductants. Four of these mutants were assumed to be purF+ since they grew on minimal medium, and one was assumed to be gnd+ because it was highly linked to a known gnd insertion. The remaining two mutants, DM1247 and DM1248, were further characterized. The Tn10d(Tc) insertions in both of these mutants were found to be 100% linked to the respective phenotype by P22 transduction, and thus the insertions were presumed to be causative of the growth phenotype.
A second mutant hunt was performed to identify MudJ insertions that caused the same phenotype as that described above. In this hunt, a P22 lysate grown on a strain that allows random insertion of MudJ transposons was used to transduce DM3215 [purF2085 apbA7::Tn10d(Tc) gnd181] to kanamycin resistance. Twelve mutants that could grow on glucose-adenine-pantothenate medium were identified by screening 16,000 transductants. By the criteria described above, 4 of the 12 were assumed to be purF+. The MudJ insertions in the remaining mutants were tested for linkage to the Tn10d(Tc) insertions in strains DM1247 and DM1248. Three of the isolates, DM3477, DM3478, and DM3479, contained MudJ insertions that were >95% linked by P22 transduction to the Tn10d(Tc) insertions in both DM1247 and DM1248. This result suggested that these five insertions were disrupting the same locus, and further analysis of this class of mutants was performed.
Insertion mutations disrupt a gene designated yjgF.
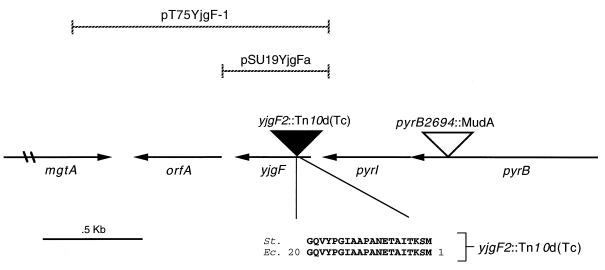
The map location of the Tn10d(Tc) insertions in DM1247 and DM1248 was determined in several steps. Derivatives of DM1247 and DM1248 that were sensitive to tetracycline were isolated by the method of Bochner (6, 47). In each strain, it was possible to identify Tcs derivatives that were auxotrophic for pyrimidines. This result suggested that the Tn10d(Tc) insertions were near a gene required for pyrimidine synthesis. After testing linkage to a number of known pyrimidine loci, we found that the Tn10d(Tc) insertions in both DM1247 and DM1248 were 99% linked by P22 transduction to an insertion in pyrB (pyrB2694::MudA; TT9534). This result placed both Tn10d(Tc) insertions at 96.8 min on the S. typhimurium chromosome. To determine the precise location of the insertions, the DNA between the Tn10d(Tc) insertion and the pyrB::MudA insertion in strains DM2944 [purF2085 yjgF1::Tn10d(Tc) pyrB2694::MudA] and DM2946 [purF2085 yjgF2::Tn10d(Tc) pyrB2694::MudA] was amplified by PCR. Sequence analysis of the resulting 1.1-kb fragment by using a primer to the C terminus of pyrI determined that the Tn10d(Tc) insertion in DM2944 was within a previously uncharacterized ORF in E. coli designated yjgF (Fig. 2). Sequencing results demonstrated that the Tn10d(Tc) insertion in DM2946 was in precisely the same location (data not shown).
FIG. 2.
Location of the yjgF2::Tn10d(Tc) insertion. A schematic representation of the 96.8-min region of S. typhimurium (St.) is shown. The yjgF2::Tn10d(Tc) insertion was mapped within the yjgF gene based on homology with E. coli (Ec.). DNA between the yjgF2::Tn10d(Tc) insertion and the pyrB2694::MudA insertion was amplified by PCR. Sequences flanking the yjgF2::Tn10d(Tc) insertion were determined, and the predicted amino acids are shown. Sequence analysis of the region flanking the yjgF1::Tn10d(Tc) insertion placed it at the same location. Amino acids conserved between the two organisms are in boldface, and numbers represent amino acids in E. coli YjgF protein. The inserts contained in the pT7-5YjgF1 and pSU19YjgFa plasmids are indicated by the hatched lines. The mgtA gene is not drawn to scale.
Cloning and sequencing of yjgF.
The DNA between mgtA and pyrI in S. typhimurium was PCR amplified with primers to the C termini of these genes. The resulting 1.4-kb fragment was cloned into the pT7-5 vector, creating pT7-5YjgF1 (Fig. 2). This plasmid complemented the phenotypes (see below) caused by a yjgF mutation. Sequence analysis of the entire 1.4-kb insert in pT7-5YjgF1 indicated that it contained two small ORFs, designated orfA (447 bp) and yjgF (387 bp) (Fig. 2). The gene structure in S. typhimurium shown in Fig. 2 is similar to that of E. coli, the exception being that this region in E. coli does not contain orfA. A BLASTX (1) database search indicated that the predicted OrfA protein showed significant homology (from amino acids 1 to 43) to hypothetical proteins in a number of insertion sequences, including a putative transposase. To confirm that the insertions in yjgF caused the respective phenotypes because of disruption of yjgF (and not polarity on orfA), a single-gene clone of yjgF, pSU19YjgFa (Fig. 2), was constructed as described in Materials and Methods. This clone also complemented the phenotypes caused by the yjgF insertion mutations, indicating that the phenotypes observed were due to loss of the yjgF gene.
The predicted size of the YjgF protein was 128 amino acids (13.5 kDa). A BLASTX database search with S. typhimurium YjgF revealed that this protein belonged to a family of small proteins of approximately 15 kDa designated the YER057c/YjgF family. These proteins are highly conserved across a wide range of organisms. Numerous homologues exist in bacteria, e.g., E. coli hypothetical proteins YjgF and YhaR (5), Haemophilus influenzae hypothetical protein HI0719 (30), Bacillus subtilis hypothetical protein YabJ (52), Helicobacter pylori putative translation initiation inhibitor (62), Lactococcus lactis AldR (35), Aquifex aeolicus hypothetical protein (19), Myxococcus xanthus DfrA, Rhizobium sp. strain NGR234 hypothetical 13.3-kDa protein Y4SK (31), and Azotobacter vinelandii hypothetical protein in the vnfA 5′ region (42). In addition, these proteins are highly conserved in cyanobacteria (Synechocystis strain 6803 hypothetical 13.8-kDa protein) (13), fungi (Saccharomyces cerevisiae chromosome V hypothetical protein YER057c and chromosome IX hypothetical protein YIL051c), and higher eukaryotes (Caenorhabditis elegans hypothetical 19.6-kDa protein, mouse heat-responsive protein 12, rat 14.5-kDa translational inhibitor protein [46, 53], human 14.5-kDa translational inhibitor protein [57], and goat UK114 tumor antigen [12]). Noticeably absent from this list was a representative from the archaea, despite the fact that several genomes have been completely sequenced (9, 44, 59). The homologues listed here ranged from 97% similar (E. coli) to 43% similar (goat). Figure 3 shows the deduced amino acid sequence of S. typhimurium YjgF compared to several of the known homologues. The YER057c/YjgF protein family is defined by a highly conserved signature motif located at the C terminus of these proteins, consisting of the following amino acids: P-[AT]-R-[SA]-X-[LIVMY]-X2-A-X-L-P-X4-[LIVM]-E. The S. typhimurium YjgF protein matches this consensus sequence exactly between amino acids 103 and 120.
FIG. 3.
The YjgF protein is highly conserved. The DNASTAR alignment program was used to align several proteins homologous to YjgF of S. typhimurium. Regions shaded in black represent amino acids that are conserved with respect to YjgF. Proteins in the alignment include E. coli YjgF, B. subtilis YabJ, Synechocystis hypothetical 13.8-kDa protein, S. cerevisiae YER057c, rat 14.5-kDa translational inhibitor protein, and human 14.5-kDa translational inhibitor protein.
Despite the high conservation of the YER057c/YjgF protein family, little is known about the function of these proteins. Studies with two of the eukaryotic proteins (rat and human) have indicated tissue-specific expression of YjgF and upregulation upon cellular differentiation (46, 53, 57). In addition, these two proteins inhibited in vitro translation in a rabbit reticulyte lysate system.
Overexpression of YjgF.
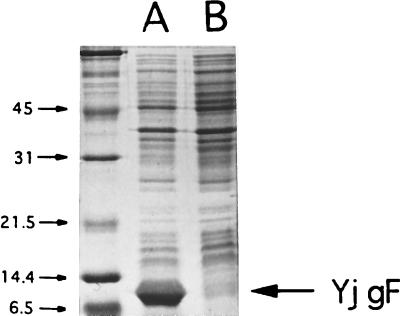
To confirm that the ORF designated yjgF produced a protein of the predicted size, the DNA insert in pSU19YjgFa (Fig. 2) was cloned into the T7 overexpression vectors, pT7-5 and pT7-6, creating pT7-5YjgFa and pT7-6YjgFa, respectively. Restriction enzyme digestion patterns indicated that pT7-6YjgFa contained the yjgF gene in the correct orientation for expression from the T7 promoter. Both plasmids were electroporated into an E. coli strain containing the gene for T7 RNA polymerase on a lambda lysogen (BL21/λDE3), generating strains DM5031 and DM5032, respectively. These strains were subjected to a protocol that induced T7-specific expression as described in Materials and Methods. Proteins of induced crude cell extracts were resolved by SDS-PAGE and visualized by Coomassie blue staining. As shown in Fig. 4, extracts from DM5032 (lane A) contained a prominent band not observed in DM5031 (lane B). Calculations of the molecular mass of the overexpressed protein from diluted samples indicated that the additional band was approximately 12 kDa, which corresponded well with the predicted molecular mass of 13.5 kDa for YjgF.
FIG. 4.
Overexpression of YjgF. Overexpression of YjgF was performed by using the T7 overexpression system as described in Materials and Methods. Proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. Extracts from DM5032 and DM5031, which contain plasmids pT7-6YjgFa and pT7-5YjgFa, respectively, are shown in lanes A and B, respectively. pT7-6YjgFa contains the insert in the proper orientation for YjgF expression from the T7 promoter. Numbers at left indicate molecular mass in kilodaltons.
The YjgF homologues from rats and humans had the unusual property of being soluble in 5% perchloric acid (PCA) and 10% TCA, respectively (46, 53, 57). We therefore tested whether the S. typhimurium YjgF protein was acid soluble. The induced crude cell extract from the overexpressing strain was extracted with 1.25, 2.5, and 5% PCA and 2.5, 5, and 10% TCA. Precipitated proteins were removed by centrifugation, and those remaining in solution were resolved by SDS-PAGE as described above. Staining with Coomassie blue revealed several faint protein bands of high molecular masses in each of the acid extractions, but no significant bands of approximately 14 kDa. This result suggested that the S. typhimurium YjgF protein, unlike the eukaryotic homologues, was not highly soluble in PCA or TCA.
A yjgF mutation suppresses the requirement for the pentose phosphate pathway in thiamine synthesis but not carbon catabolism.
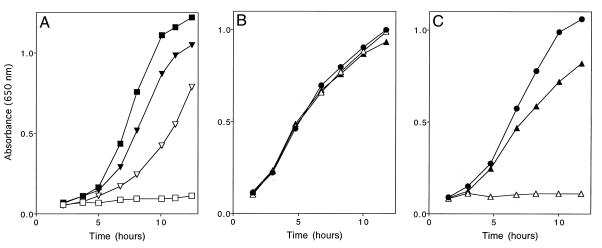
A gnd mutation eliminates function of the APB pathway when strains are grown either on glucose-adenine-pantothenate medium or on a number of nonglucose carbon sources supplemented with adenine, including gluconate (54). We tested whether yjgF mutations alleviated the requirement for Gnd in thiamine synthesis under all conditions. As expected, DM4772 [purF2085 gnd175::Tn10d(Tc) yjgF3::MudJ] grew on glucose-adenine-pantothenate medium while the parent strain, DM574 [purF2085 gnd175::Tn10d(Tc)], did not. When these two strains were grown in gluconate-adenine medium, growth was similarly restored in the yjgF derivative (DM4772) (Fig. 5A). Similar phenotypic tests performed with DM3502 (purF2085 zwf23::MudJ) and DM3503 [purF2085 zwf23::MudJ yjgF1::Tn10d(Tc)] determined that yjgF mutations suppressed the effect of zwf mutations in a similar manner (data not shown). Thus, insertion mutations in yjgF eliminated the requirement for the pentose phosphate pathway in the function of the APB pathway.
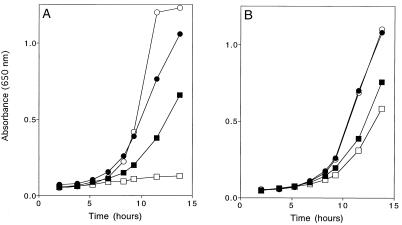
FIG. 5.
Phenotypes caused by a yjgF mutation. Growth curves were obtained from strains grown at 37°C as described in Materials and Methods. (A) Suppression of the requirement for Gnd in thiamine synthesis. Growth of DM574 [purF2085 gnd175::Tn10d(Tc)] in gluconate-adenine medium and gluconate-adenine-thiamine medium is indicated by open and solid squares, respectively. Growth of DM4772 [purF2085 gnd175::Tn10d(Tc) yjgF3::MudJ] in the same medium is indicated by open and solid inverted triangles, respectively. (B and C) Sensitivity to exogenous serine. Shown are growth of LT2 (wild type) (B) and growth of DM3480 (yjgF3::MudJ) (C) in minimal glucose medium (solid circles), glucose medium supplemented with serine (open triangles), and glucose medium supplemented with serine and isoleucine (solid triangles).
To test the simple explanation that yjgF mutations caused their effect by activating a new enzyme(s) that performed the same functions as did Gnd and/or Zwf, we addressed whether yjgF mutations could suppress the requirement for one of these enzymes in carbon catabolism. DM3488 (purF2085 gnd181 pgi::Tn5) was unable to utilize glucose as a sole carbon source because both the pentose phosphate pathway and the glycolytic pathway were blocked. We found that yjgF mutations did not restore the ability of the respective strain [DM3489; purF2085 gnd181 pgi::Tn5 yjgF1::Tn10d(Tc)] to utilize glucose as the sole carbon source. This result indicated that yjgF mutations did not suppress the requirement for the pentose phosphate pathway in carbon catabolism.
It was a formal possibility that yjgF mutations allowed thiamine-independent growth because of a reduction in the cellular TPP requirement, rather than restored synthesis. Conditions that reduce the cellular TPP requirement can be identified based on their ability to allow growth of thi auxotrophs on lower concentrations of thiamine (26). When the yjgF mutations were introduced into known thiamine auxotrophs, they did not alter the amount of thiamine required for growth (data not shown). These results suggested that the yjgF mutation caused its effect by restoring thiamine synthesis.
Mutations in yjgF cause a conditional requirement for isoleucine.
In the course of experiments performed to phenotypically characterize DM1247 [purF2085 apbA1::MudJ gnd181 yjgF1::Tn10d(Tc)], we found that this strain was hypersensitive to the presence of serine when grown on minimal medium-based medium. When 8 μmol of l-serine was spotted on a plate overlaid with DM1247, a large zone of inhibition was observed. Further testing determined that any strain defective in YjgF, regardless of genetic background, exhibited a similar serine sensitivity. The serine sensitivity caused by the yjgF mutation is shown quantitatively in Fig. 5B and C. Growth of DM3480 (yjgF3::MudJ) on minimal glucose medium was completely inhibited in the presence of 4 mM serine (Fig. 5C), while growth of the wild-type strain (LT2) was unaffected (Fig. 5B). Titration experiments determined that serine caused this severe growth defect when present at final concentrations greater than or equal to 500 μM (data not shown).
Compounds that corrected the serine sensitivity of a yjgF mutant were identified by spotting pools containing overlapping compounds on minimal serine plates overlaid with DM3480 (18). Two compounds were found that clearly corrected serine-inhibited growth of DM3480: isoleucine and threonine. Correction of growth by isoleucine is shown quantitatively in Fig. 5C. The sensitivity of DM3480 to serine and correction by isoleucine or threonine (an intermediate in isoleucine synthesis) suggested that the yjgF mutation was exacerbating a condition designated “serine toxicity” that was first observed in E. coli over 40 years ago (2, 15, 16, 55, 56). Wild-type E. coli K-12 undergoes a transient cessation in growth in response to 1.5 mM serine (15, 65); this sensitivity was more severe in different genetic backgrounds (14). In all cases, the serine toxicity was corrected by isoleucine. Serine is thought to induce starvation for isoleucine by negatively affecting isoleucine biosynthesis in several ways. There is evidence that serine inhibits two enzymes required for the synthesis of isoleucine, l-threonine deaminase (55) and homoserine dehydrogenase I (no. 10 and 6, respectively, in Fig. 1B) (36, 37). In addition, serine may lead to increased levels of pyruvate through its catabolism to pyruvate and feedback inhibition of its own synthesis (serine is derived from 3-phosphoglycerate, a precursor to pyruvate) (60). Elevation of pyruvate leads to isoleucine restriction due to the kinetic parameters of the acetohydroxy acid synthase isozymes (4). Increased pyruvate outcompetes α-ketobutyrate for available enzyme (no. 11 and 12 [Fig. 1B]), resulting in valine and leucine synthesis at the expense of isoleucine synthesis. To further characterize the serine sensitivity observed with DM3480, we tested the effects of intermediary metabolites of the isoleucine biosynthetic pathway (Fig. 1B) on serine inhibition. Growth of DM3480 in the presence of serine was restored by isoleucine, α-ketobutyrate, threonine, or homoserine, but not by aspartate (data not shown). These results parallel those observed with E. coli (37) and are consistent with at least one of the causes of isoleucine starvation by serine being inhibition of homoserine dehydrogenase I (no. 6 [Fig. 1B]).
Growth of DM3480 on minimal medium in the presence or absence of serine was examined when a variety of compounds were used as the sole carbon source. Growth of DM3480 was sensitive to serine and corrected by isoleucine on all carbon sources tested, including gluconate, sorbitol, xylose, ribose, pyruvate, glycerol, citrate, succinate, fructose, galactose, and acetate (data not shown). Growth of LT2 was not inhibited by serine under any of these conditions. Significantly, when DM3480 was grown with either pyruvate or glycerol as sole carbon source, the strain showed a significant requirement for isoleucine even in the absence of serine. The fact that yjgF mutants were starving for isoleucine even in the absence of serine suggested that these growth conditions may have introduced increased restrictions on the isoleucine biosynthetic pathway. Levels of pyruvate are likely to be elevated under these two growth conditions and may be contributing to isoleucine restriction as described above.
A yjgF mutation does not cause serine sensitivity by the same mechanism as does a relA mutation.
Mutations exacerbating serine sensitivity have been isolated previously in E. coli (14), the most well-characterized being the relA mutation (64, 65). relA mutants are defective in (p)ppGpp synthetase and consequently are defective in the stringent response (10). A relA mutation seems to cause serine sensitivity by preventing derepression of the ilv genes, a natural way to avoid growth inhibition by serine (64). The serine sensitivity of a relA mutant was suppressed by mutations, including hisT (7, 8), that resulted in constitutive expression of the ilv genes (64). The results of two experiments suggested that the yjgF mutation was not acting in the same manner as a relA mutation. First, we tested if a yjgF mutant was defective in the stringent response by measuring stable RNA accumulation in starved and unstarved cultures as previously described (34). We found that stable RNA synthesis (as measured by 32Pi incorporation into TCA-precipitable material) was dramatically decreased in response to starvation in both DM3480 (yjgF3::MudJ) and LT2 but, as expected, not in DM4440 (relA21::Tn10). 32Pi was incorporated into TCA-precipitable material at rates of 1,608, 1,748, and 1,264 cpm/OD600/min in unstarved cultures of LT2, DM4440, and DM3480, respectively. When these cultures were starved for amino acids, the incorporation rates of LT2 and DM3480 were significantly decreased, to 206 and 217 cpm/OD600/min, respectively. The rate of incorporation into DM4440 remained high, at 1,112 cpm/OD600/min. These results indicated that, unlike a relA mutant, a yjgF mutant was not defective in the stringent response. Second, when we constructed a hisT yjgF double mutant, the resulting strain, DM4442 [yjgF1::Tn10d(Tc) hisT290::Tn5], was as sensitive to serine as the parent strain, DM4053 [yjgF1::Tn10d(Tc)]. This result suggested that the cause of serine sensitivity in a yjgF mutant was not due to an inability to derepress the ilv genes.
Flux through the isoleucine biosynthetic pathway is important for a yjgF mutation to suppress the requirement for Gnd in thiamine synthesis.
As shown in Fig. 5A, a yjgF mutation allowed a purF gnd double mutant to grow in the absence of thiamine. During routine phenotypic experiments, we found that exogenous isoleucine significantly reduced the ability of the yjgF mutation to restore thiamine synthesis in this strain. Figure 6A shows the growth of DM4767 [purF2085 gnd174::MudJ yjgF2::Tn10d(Tc)] in gluconate-adenine and gluconate-adenine-thiamine medium in the presence and absence of isoleucine. Although isoleucine increased the growth of DM4767 slightly when thiamine was included, it inhibited growth in the absence of thiamine. Isoleucine feedback inhibits its own synthesis by inhibiting activity of threonine deaminase (IlvA, no. 10 [Fig. 1B]), which is the first dedicated step to isoleucine biosynthesis (63). Therefore, one possible explanation for the effect of isoleucine described above was that carbon flux through the isoleucine biosynthetic pathway was important for the restoration of thiamine synthesis by the yjgF mutation. To test this possibility, we constructed a derivative of DM4767 that contained a feedback-resistant mutation in ilvA, DM4769 [purF2085 gnd174::MudJ yjgF2::Tn10d(Tc) ilvA219]. The ilvA219 mutation results in a threonine deaminase enzyme that is resistant to feedback inhibition by isoleucine (45). As shown in Fig. 6B, DM4769 was able to grow in the absence of thiamine even when isoleucine was included in the medium. Thus, isoleucine did not interfere with the effect of the yjgF mutation in a strain resistant to feedback inhibition by isoleucine. These results suggested that flux through the isoleucine biosynthetic pathway was important for the yjgF mutation to restore thiamine synthesis.
FIG. 6.
Flux through the isoleucine biosynthetic pathway is important for restoration of thiamine synthesis by a yjgF mutation. Growth curves were obtained from strains grown at 37°C as described in Materials and Methods. Shown are growth of DM4767 [purF2085 gnd174::MudJ yjgF2::Tn10d(Tc)] (A) and growth of DM4769 [purF2085 gnd174::MudJ yjgF2::Tn10d(Tc) ilvA219] (B) in minimal gluconate medium supplemented with adenine (solid squares); adenine and isoleucine (open squares); adenine and thiamine (solid circles); and adenine, thiamine, and isoleucine (open circles).
DISCUSSION
In this study, we describe the isolation and characterization of mutations in a previously uncharacterized ORF, designated yjgF. Mutations in yjgF caused multiple phenotypes. These mutations eliminated the requirement for the oxidative pentose phosphate pathway in PurF-independent thiamine synthesis. In addition, yjgF mutants appeared to be compromised in their ability to synthesize isoleucine, as indicated by their hypersensitivity to conditions that restrict isoleucine biosynthesis, e.g., increased levels of serine and/or pyruvate. This result is in agreement with previous work indicating that growth of the corresponding mutant in L. lactis (aldR) was slowed twofold if isoleucine was omitted from the medium (35).
Data presented here suggest a link between the effect of a yjgF mutation on thiamine synthesis and the apparent defect that this mutation causes in isoleucine synthesis. Results suggest that carbon flux through the isoleucine biosynthetic pathway is important for a yjgF mutation to restore thiamine synthesis. The following general model is proposed to explain these results. We suggest that the yjgF mutation results in the partial block of at least one step in isoleucine biosynthesis (by an as yet undefined mechanism) subsequent to the reaction catalyzed by threonine deaminase (IlvA, no. 10 [Fig. 1B]). This block results in starvation for isoleucine when growth conditions, i.e., increased levels of serine and/or pyruvate, further restrict isoleucine biosynthesis. In addition, we propose that the defect in isoleucine biosynthesis causes accumulation of a compound, likely an isoleucine biosynthetic intermediate or derivative, that is important for restoration of thiamine synthesis in the absence of the oxidative pentose phosphate pathway. When isoleucine is added to the medium, accumulation of the compound (and the corresponding restoration of thiamine synthesis) is prevented by feedback inhibition of the pathway at IlvA. Introduction of an ilvA feedback-resistant mutation thus abolishes isoleucine’s inhibitory effect.
The above model predicts that an isoleucine biosynthetic intermediate, or a derivative, suppresses the requirement of the pentose phosphate pathway in thiamine synthesis. There are several possibilities for how this could occur. Since ribose-5-P also corrects the requirement, the simplest explanation is that the relevant intermediate increases ribose-5-P levels independently of the oxidative pentose phosphate pathway. The compound may be involved in regulating flux through the nonoxidative pathway or another metabolic pathway in which ribose-5-P is a precursor or intermediate. Such scenarios would be consistent with the ability of a yjgF mutation to suppress the requirement for Gnd in thiamine synthesis but not carbon catabolism. Alternatively, the intermediate might allow function of the APB pathway despite low ribose-5-P levels. For example, the relevant metabolite might increase levels of the predicted PRA-forming enzyme or enhance its activity, thus compensating for decreased ribose-5-P. The proposed model for how a YjgF defect affects thiamine synthesis (through the accumulation of an isoleucine biosynthetic intermediate) predicts that isoleucine intermediates added exogenously might also suppress the requirement for the pentose phosphate pathway. Significantly, in a preliminary test of this prediction, none of the intermediates that we tested, including homoserine, threonine, and α-ketobutyrate, restored thiamine synthesis in a purF gnd mutant. Additional genetic and biochemical tests are currently under way to test this proposed model.
This study represents the first detailed phenotypic characterization of mutants defective in the YER057c/YjgF protein family in any organism and provides important clues as to the function of this highly conserved class of proteins. Results presented here suggest that the YjgF protein, or alternatively, the product of the YjgF protein if it functions as an enzyme, is important for appropriate function and/or regulation of the isoleucine biosynthetic pathway. Comparing the levels of isoleucine biosynthetic intermediates and enzymes in wild-type strains and yjgF mutants will be informative in this regard.
ACKNOWLEDGMENTS
We thank Bob LaRossa for supplying the ilvA219 feedback-resistant mutant.
This work was supported by NSF grant MCB 9723830 to D.M.D. J.L.E.-B. was supported by the Pfizer Fellowship in Microbial Physiology.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amos R H, Cohen G N. Amino acid utilization in bacterial growth. J Biochem. 1954;57:338–343. doi: 10.1042/bj0570338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 4.Barak Z, Chipman D M, Gollop N. Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. J Bacteriol. 1987;169:3750–3756. doi: 10.1128/jb.169.8.3750-3756.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Bochner B R, Huang H, Schieven G L, Ames B N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresalier R S, Rizzino A A, Freunlich M. Reduced maximal level of derepression of the isoleucine-valine and leucine enzymes in hisT mutants of Salmonella typhimurium. Nature. 1975;253:279–280. doi: 10.1038/253279a0. [DOI] [PubMed] [Google Scholar]
- 8.Bruni C B, Colantuoni V, Sbordone L, Cortese R, Blasi F. Biochemical and regulatory properties of Escherichia coli K-12 hisT mutant. J Bacteriol. 1977;130:4–10. doi: 10.1128/jb.130.1.4-10.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodeck A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hann M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 11.Castilho B A, Olfson P, Casadaban M J. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceciliani F, Faotto L, Negri A, Colombo I, Berra B, Bartorelli A, Ronchi S. The primary structure of UK114 tumor antigen. FEBS Lett. 1996;393:147–150. doi: 10.1016/0014-5793(96)00850-2. [DOI] [PubMed] [Google Scholar]
- 13.Chavez S, Reyes J C, Chauvat F, Florencio F J, Candau P. The NADP-glutamate dehydrogenase of the cyanobacterium Synechocystis 6803: cloning, transcriptional analysis and disruption of the gdhA gene. Plant Mol Biol. 1995;28:173–188. doi: 10.1007/BF00042048. [DOI] [PubMed] [Google Scholar]
- 14.Cosloy S, McFall E. l-Serine-sensitive mutants of Escherichia coli K-12. J Bacteriol. 1970;103:840–841. doi: 10.1128/jb.103.3.840-841.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosloy S, McFall E. Metabolism of d-serine in Escherichia coli K-12: mechanism of growth inhibition. J Bacteriol. 1973;114:685–694. doi: 10.1128/jb.114.2.685-694.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danchin A, Dondon L. Serine sensitivity of Escherichia coli K 12: partial characterization of a serine resistant mutant that is extremely sensitive to 2-ketobutyrate. Mol Gen Genet. 1980;178:155–164. doi: 10.1007/BF00267224. [DOI] [PubMed] [Google Scholar]
- 17.David S, Estramareix B, Fischer J C, Therisod M. The biosynthesis of thiamine. Synthesis of [1,1,1,5-2H4]-1-deoxy-d-threo-2-pentulose and incorporation of this sugar in biosynthesis of thiazole by Escherichia coli cells. J Chem Soc Perkin Trans. 1982;1:2131–2137. [Google Scholar]
- 18.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 19.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 20.DeMoll E, Shive W. Determination of the metabolic origin of the sulfur atom in thiamine of Escherichia coli by mass spectrometry. Biochem Biophys Res Commun. 1985;132:217–222. doi: 10.1016/0006-291x(85)91010-1. [DOI] [PubMed] [Google Scholar]
- 21.Downs D M. Evidence for a new, oxygen-regulated biosynthetic pathway for the pyrimidine moiety of thiamine in Salmonella typhimurium. J Bacteriol. 1992;174:1515–1521. doi: 10.1128/jb.174.5.1515-1521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downs D M, Petersen L. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1994;176:4858–4864. doi: 10.1128/jb.176.16.4858-4864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downs D M, Roth J R. Synthesis of thiamine in Salmonella typhimurium independent of the purF function. J Bacteriol. 1991;173:6597–6604. doi: 10.1128/jb.173.20.6597-6604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enos-Berlage, J. L., and D. M. Downs. Biosynthesis of the pyrimidine moiety of thiamine independent of the PurF enzyme (phosphoribosylpyrophosphate amidotransferase) in Salmonella typhimurium: incorporation of stable isotope-labeled glycine and formate. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 25.Enos-Berlage J L, Downs D M. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1996;178:1476–1479. doi: 10.1128/jb.178.5.1476-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enos-Berlage J L, Downs D M. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J Bacteriol. 1997;179:3989–3996. doi: 10.1128/jb.179.12.3989-3996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enos-Berlage, J. L., and D. M. Downs. 1996. Unpublished results.
- 28.Estramareix B, David S. Conversion of 5-aminoimidazole ribotide to the pyrimidine of thiamine in enterobacteria: study of the pathway with specifically labeled samples of riboside. Biochim Biophys Acta. 1990;1035:154–160. doi: 10.1016/0304-4165(90)90110-i. [DOI] [PubMed] [Google Scholar]
- 29.Estramareix B, Therisod M. Tyrosine as a factor in biosynthesis of the thiazole moiety of thiamine in Escherichia coli. Biochim Biophys Acta. 1972;273:275–282. [PubMed] [Google Scholar]
- 30.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cottom M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 31.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 32.Frodyma M E, Downs D M. ApbA, the ketopantoate reductase enzyme of Salmonella typhimurium, is required for the synthesis of thiamine via the alternative pyrimidine biosynthetic pathway. J Biol Chem. 1998;273:5572–5576. doi: 10.1074/jbc.273.10.5572. [DOI] [PubMed] [Google Scholar]
- 33.Frodyma M E, Downs D M. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaal T, Gourse R. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goupil-Feuillerat N, Cocaign-Bousquet M, Gocon J-J, Ehrlich S D, Renault P. Dual role of α-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J Bacteriol. 1997;179:6285–6293. doi: 10.1128/jb.179.20.6285-6293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hama H, Kayahara T, Tsuda M, Tsuchiya T. Inhibition of homoserine dehydrogenase I by l-serine in Escherichia coli. J Biochem. 1991;109:604–608. doi: 10.1093/oxfordjournals.jbchem.a123427. [DOI] [PubMed] [Google Scholar]
- 37.Hama H, Sumita Y, Kakutani Y, Tsuda M, Tsuchiya T. Target of serine inhibition in Escherichia coli. Biochem Biophys Res Commun. 1990;168:1211–1216. doi: 10.1016/0006-291x(90)91157-n. [DOI] [PubMed] [Google Scholar]
- 38.Hong J S, Ames B N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci USA. 1971;68:3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes K T, Roth J R. Conditionally transposition-defective derivative of Mu d1 (Amp Lac) J Bacteriol. 1984;159:130–137. doi: 10.1128/jb.159.1.130-137.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes K T, Roth J R. Directed formation of deletions and duplication using Mud (Ap, Lac) Genetics. 1985;109:263–282. doi: 10.1093/genetics/109.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes K T, Roth J R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988;119:9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joerger R D, Jacobson M R, Bishop P E. Two nifA-like genes required for expression of alternative nitrogenases by Azotobacter vinelandii. J Bacteriol. 1989;171:3258–3267. doi: 10.1128/jb.171.6.3258-3267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleckner J, Roth J R, Botstein D. Genetic engineering in vivo using translocatable drug resistant elements. J Mol Biol. 1977;116:125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- 44.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Derlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C, et al. The complete genome sequence of the hyperthermophilic sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 45.LaRossa R, Van Dyk T K, Smulski D R. Toxic accumulation of α-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J Bacteriol. 1987;169:1372–1378. doi: 10.1128/jb.169.4.1372-1378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy-Favatier F, Cuisset L, Nedelec B, Tichonicky L, Kruh J, Delpech M. Characterization, purification, and cDNA cloning of a rat perchloric-acid-soluble 23-kDa protein present only in liver and kidney. Eur J Biochem. 1993;212:665–673. doi: 10.1111/j.1432-1033.1993.tb17704.x. [DOI] [PubMed] [Google Scholar]
- 47.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacAa reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 49.Negre D, Cortay J-C, Donini P, Cozzone A J. Relationship between guanosine tetraphosphate and accuracy of translation in Salmonella typhimurium. Biochemistry. 1989;28:1814–1819. doi: 10.1021/bi00430a058. [DOI] [PubMed] [Google Scholar]
- 50.Newell P C, Tucker R G. Biosynthesis of the pyrimidine moiety of thiamine. Biochem J. 1968;106:279–287. doi: 10.1042/bj1060279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newell P C, Tucker R G. Precursors of the pyrimidine moiety of thiamine. Biochem J. 1968;106:271–277. doi: 10.1042/bj1060271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 53.Oka T, Tsuji H, Noda C, Sakai K, Hong Y-M, Suzuki I, Munoz S, Natori Y. Isolation and characterization of a novel perchloric-acid soluble protein inhibiting cell-free protein synthesis. J Biol Chem. 1995;270:30060–30067. doi: 10.1074/jbc.270.50.30060. [DOI] [PubMed] [Google Scholar]
- 54.Petersen L A, Enos-Berlage J L, Downs D M. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143:37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasko I, Alfoldi L. Biosynthetic l-threonine deaminase as the origin of l-serine sensitivity of Escherichia coli. Eur J Biochem. 1971;21:424–427. doi: 10.1111/j.1432-1033.1971.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 56.Rowley D. Inhibition of E. coli strains by amino-acids. Nature. 1953;171:80–81. doi: 10.1038/171080a0. [DOI] [PubMed] [Google Scholar]
- 57.Schmiedeknecht G, Kerkhoff C, Orso E, Stohr J, Aslanidis C, Nagy G M, Knuechel R, Schmitz G. Isolation and characterization of a 14.5-kDa trichloroacetic-acid-soluble translational inhibitor protein from human monocytes that is upregulated upon cellular differentiation. Eur J Biochem. 1996;242:339–351. doi: 10.1111/j.1432-1033.1996.0339r.x. [DOI] [PubMed] [Google Scholar]
- 58.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 59.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stauffer G V. Biosynthesis of serine, glycine, and one-carbon units. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 506–513. [Google Scholar]
- 61.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel P A, Brent R, Kingston R E, Moore D D, Seidman J C, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 62.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 63.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 442–457. [Google Scholar]
- 64.Uzan M, Danchin A. Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA− mutants. Mol Gen Genet. 1978;165:21–30. doi: 10.1007/BF00270372. [DOI] [PubMed] [Google Scholar]
- 65.Uzan M, Danchin A. A rapid test for the relA mutation in E. coli. Biochem Biophys Res Commun. 1976;69:751–758. doi: 10.1016/0006-291x(76)90939-6. [DOI] [PubMed] [Google Scholar]
- 66.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 67.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 68.Webb E, Febres F, Downs D M. Thiamine pyrophosphate (TPP) negatively regulates transcription of some thi genes of Salmonella typhimurium. J Bacteriol. 1996;178:2533–2538. doi: 10.1128/jb.178.9.2533-2538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]