Abstract
Burkholderia cepacia DBO1 is able to utilize phthalate as the sole source of carbon and energy for growth. Two overlapping cosmid clones containing the genes for phthalate degradation were isolated from this strain. Subcloning and activity analysis localized the genes for phthalate degradation to two separate regions on the cosmid clones. Analysis of the nucleotide sequence of these two regions showed that the genes for phthalate degradation are arranged in at least three transcriptional units. The gene for phthalate dioxygenase reductase (ophA1) is present by itself, while the genes for an inactive transporter (ophD) and 4,5-dihydroxyphthalate decarboxylase (ophC) are linked and the genes for phthalate dioxygenase oxygenase (ophA2) and cis-phthalate dihydrodiol dehydrogenase (ophB) are linked. ophA1 and ophDC are adjacent to each other but are transcribed in opposite directions, while ophA2B is located 4 kb away. The genes for the oxygenase and reductase components of phthalate dioxygenase are located approximately 7 kb away from each other. The gene for the putative phthalate permease contains a frameshift mutation in contrast to genes for other permeases. Strains deleted for ophD are able to transport phthalate into the cell at rates equivalent to that of the wild-type organism, showing that this gene is not required for growth on phthalate.
Phthalates and phthalate esters are widely used in the manufacture of plastics, textiles, papers, insect repellents, pesticides, munitions, and cosmetics (36, 70). Due to the widespread use of phthalates, there has been great concern about their release into the environment (34, 45, 57) and their toxicity to human beings and other organisms (4, 32, 41, 43, 56, 70, 80, 84, 85). Many microorganisms have been isolated from rivers, soil, and even marine regions for their ability to degrade phthalate aerobically or anaerobically (1, 27, 47, 60, 67, 68, 73, 74, 78, 79). Two catabolic pathways have been identified for the aerobic degradation of phthalate. Gram-negative bacteria degrade phthalate via 4,5-dihydroxyphthalate and protocatechuate (47, 60, 65, 73). Gram-positive bacteria also degrade phthalate through protocatechuate, but in this case the pathway proceeds through 3,4-dihydroxyphthalate as an intermediate (26, 65, 74). Protocatechuate, a central metabolite for many aromatic degradation pathways, is further metabolized through either an ortho or meta cleavage pathway.
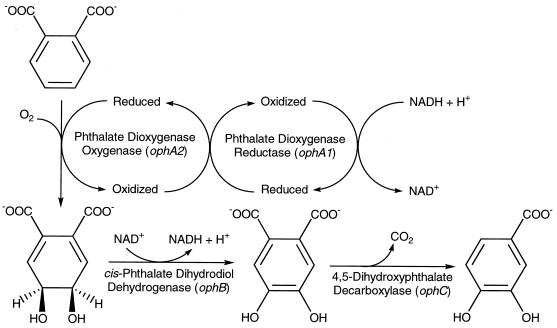
Burkholderia cepacia DBO1 was originally isolated in Florida for its ability to utilize phthalate as the sole carbon and energy source. The strain was first published as Pseudomonas fluorescens PHK (46); it was subsequently known as Pseudomonas putida (12) and was later reclassified as Pseudomonas cepacia on the basis of detailed nutritional studies (6). The P. cepacia species is now associated with the Burkholderia genus (86). The ability of this particular strain to degrade phthalate (see Fig. 1) has been well documented (47, 73, 74). Phthalate is first dihydroxylated by phthalate dioxygenase to give 4,5-dihydro-4,5-dihydroxyphthalate (cis-phthalate dihydrodiol). This two-component enzyme consists of a reductase (phthalate dioxygenase reductase [PDR]) and an oxygenase (phthalate dioxygenase oxygenase [PDO]). Both proteins have been purified to homogeneity (6), and the crystal structure of PDR has been determined at a 2.0-Å resolution (20). The 34-kDa PDR can be subdivided into three parts, a flavin mononucleotide (FMN) binding domain, an NAD+ binding domain, and a plant ferredoxin-type [2Fe-2S] domain, that function to transfer electrons from NADH to PDO. The 48-kDa PDO has one Rieske-type [2Fe-2S] center, which functions to accept electrons from PDR, and a mononuclear iron known to be involved in the actual catalytic addition of oxygen to the aromatic ring. PDO has also been well studied, especially with regard to the Rieske iron sulfur center which has been analyzed by electron nuclear double-resonance spectroscopy (17, 37), pulsed electron paramagnetic resonance spectroscopy (17), resonance Raman spectroscopy (49), and X-ray absorption spectroscopy (81). The ferrous active site has been analyzed by magnetic circular dichroism during substrate binding (33). Phthalate dioxygenase is perhaps the best-studied aromatic dioxygenase from a biophysical point of view.
FIG. 1.
Catabolic pathway for the metabolism of phthalate to protocatechuate by B. cepacia DBO1. The genes for each enzyme have been given the designation oph for o-phthalate degradation.
The second step in the catabolic pathway involves a dehydrogenase that removes two electrons and two hydrogens from cis-phthalate dihydrodiol to form 4,5-dihydroxyphthalate and NADH. One of the two carboxyl groups of the latter compound is removed by 4,5-dihydroxyphthalate decarboxylase (72) to form protocatechuate, a central metabolite in the catabolism of aromatic compounds. In B. cepacia DBO1, protocatechuate then undergoes ortho ring cleavage and is metabolized through the well-known β-ketoadipate pathway to eventually produce tricarboxylic acid cycle intermediates. Protocatechuate 3,4-dioxygenase from B. cepacia DBO1 has been extensively studied at the biochemical (12), genetic (91), and physiological (90) levels.
Although much is known about the initial biochemical mechanisms by which B. cepacia DBO1 metabolizes phthalate, nothing is known about the underlying genetic basis. The goal of the present work was to determine the molecular basis of phthalate degradation by B. cepacia DBO1 to complement the existing biochemical studies and to provide a basis for further investigations on these enzymes. Preliminary reports of this work have been presented elsewhere (14, 15, 54).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
B. cepacia DBO1 is the same strain as the P. putida reported by Bull and Ballou (12) and the P. fluorescens PHK of Ribbons (46). B. cepacia ATCC 29424 is the original PHK strain deposited by Ribbons in the American Type Culture Collection (ATCC). B. cepacia ATCC 17616 (77) is a phthalate-degrading strain isolated by other investigators independently of ATCC 29424. B. cepacia DBO106 is a mutant of DBO1 that accumulates cis-phthalate dihydrodiol (89). Comamonas testosteroni NH1024 is a mutant that accumulates 4,5-dihydroxyphthalate (60). Escherichia coli DH5α [F− φ80dlacZΔ M15 Δ(lacZY-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1] (Gibco-BRL, Gaithersburg, Md.) was used as the recipient strain in the cloning experiments, and E. coli S17-1 (thi pro hsdR hsdM+ recA RP4 tra+) (76) or HB101 (supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1) (9) was used as the recipient strain in the cosmid cloning experiments. The cosmid cloning vectors pMMB34 (29) and pHC79 (39) were used to construct the genomic libraries, and the pGEM series of cloning vectors (Promega, Madison, Wis.), pUC19 (83), pRK415 (44), and pTrc99A (2), were used to construct subclones. Mineral salts basal (MSB) medium (77) was used as minimal medium, and L broth (52) was used as complete medium. Ampicillin and tetracycline were added at 100 and 15 μg/ml, respectively, when needed. Burkholderia strains were grown at 30°C, and E. coli strains were grown at 37°C.
Molecular techniques.
Total genomic DNA from B. cepacia DBO1 was prepared by the method of Olsen et al. (69). Plasmid DNA was isolated by the alkaline-sodium dodecyl sulfate procedure of Birnboim and Doly (8) or purified by the QIAprep spin column procedure (Qiagen, Inc., Santa Clarita, Calif.). Transformation of plasmid DNA into competent E. coli DH5α was performed by the procedure of Hanahan (38) or the calcium chloride-glycerol transformation procedure (75). Restriction digests and ligations of DNA samples were performed as recommended by the supplier (Gibco-BRL). Two genomic libraries were constructed, the first with partially EcoRI-digested total genomic DNA and the cosmid pMMB34 and the other with partially Sau3AI-digested total genomic DNA and the cosmid pHC79, by using procedures described earlier (35, 48). Colony blot and Southern hybridization experiments were performed as described previously (48, 75). Three oligonucleotides corresponding to the amino acid sequences of PDR and PDO were synthesized. PDR-N (5′-CAP-GAP-GAY-GGN-TTY-YT-3′) corresponds to the fourth through ninth amino acids (QEDGFL) from the N-terminal end of PDR (TTPQEDGFL). PDR-C (5′-AC-RCA-NAC-CAT-DAT-YTG-3′) corresponds to amino acids 13 through 18 (QIMVCV) from the C-terminal end of PDR (KGTQIMVCVSRAKSAELVLDL). PDO-N (5′-CAN-CAY-CAP-GAP-AAY-GA-3′) corresponds to the second through the seventh amino acids from the N-terminal end of PDO (LTHQENE). The oligonucleotides were labeled with T4 kinase (Gibco-BRL) for use in Southern hybridization experiments. DNA sequencing was performed with an AmpliTaq DNA polymerase dye-terminator cycle-sequencing kit (Perkin-Elmer, Foster City, Calif.) and resolved on an ABI 373 DNA sequencer (Perkin-Elmer).
Coupled in vitro transcription-translation from purified plasmid DNA was performed with an E. coli S30 extract system as described by the manufacturer (Promega). Tritiated leucine was added to the reaction mix to label the protein product. Aliquots (5 μl) from the reaction mix were analyzed with a denaturing 15% (wt/vol) polyacrylamide gel by using standard procedures (75). The gel was dried, and the signals were enhanced by fluorography and detected by X-ray film exposure. Prestained molecular weight markers (Gibco-BRL) were used to estimate the size of each detected protein.
A knockout mutant of ophD was constructed by gene replacement with an inserted kanamycin resistance gene cassette. A 1.2-kb PCR product containing the ophA1 gene on the left of ophD was PCR amplified by using the primers GCGAATTCTGAGCGAAGCGTAGG and GCGAGCTCCTGGATACCGGCAGG containing EcoRI and SstI restriction sites (underlined), respectively. A 1.2-kb PCR product containing the ophC gene on the right of ophD was PCR amplified by using the primers GCGAGCTCTCAGACAGGAGCAGG and GCTCTAGACCATGCCTTCCTCGC containing SstI and XbaI restriction sites (underlined), respectively. PCR in these cases was performed with Taq polymerase with an initial denaturation step of 1.0 min at 94°C; 30 cycles of 15 s at 94°C (denaturation), 30 s at 50°C (annealing), and 4.0 min at 60°C (extension); followed by 10 min of final extension. The two PCR products were cleaved with the indicated restriction enzymes and cloned into pUC19 cut with EcoRI and XbaI. The resulting plasmid was cut with SstI and an SstI fragment containing a kanamycin resistance cassette from p34S-Km (24) was inserted. The latter construct was electroporated (23) into B. cepacia DBO1 with selection on Lagar containing kanamycin. One strain resulting from a double crossover, designated DBO304, was saved for analysis. This strain is missing the entire ophD gene.
Transformation of phthalate by E. coli strains carrying cloned genes.
Plasmid-containing strains were incubated at 37°C with shaking until the optical density of the culture at 600 nm reached 0.7. The cells were harvested by centrifugation, washed twice with 50 mM sodium/potassium phosphate buffer (pH 7.25), and resuspended in the same buffer supplemented with 20 mM glucose and 10 mM phthalate. After overnight incubation, the cells were removed by centrifugation and the supernatant was collected for analysis by high-performance liquid chromatography (HPLC). A gradient of 0 to 50% methanol in water under acidic (0.1% acetic acid) conditions with a reverse-phase 5-μm C18 column (4.6 by 25 mm) was used to separate the different compounds. Both retention time and UV spectra of the metabolites were compared with those of the standard compounds.
Phthalate transport assay.
Mid-log-phase B. cepacia DBO1 grown on 10 mM phthalate or 10 mM p-hydroxybenzoate and B. cepacia DBO304 grown on 10 mM phthalate were harvested by low-speed centrifugation at room temperature. The cells were washed with an equal volume of 50 mM phosphate buffer (25 mM KH2PO4 and 25 mM Na2HPO4 adjusted to pH 6.8 with KOH) and resuspended in the same buffer containing 10 mM glucose and 10 mM succinate to an optical density of 1.0 at 600 nm. Cell suspensions were gently aerated to prevent oxygen limitation. All assays were done at 25°C. Uptake was initiated by diluting cells into an equal volume of phosphate buffer containing 100 μM [Ring-UL-14C]phthalate (12.6 mCi/mmol; Sigma Chemical Co., St. Louis, Mo.). Samples (0.1 ml) were removed from the reaction mixture at various times (10 s, 30 s, 1 min, 2 min, and 3 min) and filtered through Isopore polycarbonate membranes (0.2-μm pore size; Millipore Corp., Bedford, Mass.). The filters were washed before and after addition of the sample with 2 ml of phosphate buffer. Accumulated phthalate was determined by scintillation counting of the cells retained on the filters. Cell protein was determined by the method of Bradford (10).
Chemicals and biosynthesis of pathway intermediates.
Phthalate and protocatechuate were of the highest quality available (Aldrich Chemical Co., St. Louis, Mo.). cis-Phthalate dihydrodiol and 4,5-dihydroxyphthalate used as standards were synthesized by using mutant strains accumulating each compound. cis-Phthalate dihydrodiol was synthesized by culturing B. cepacia DBO106 in MSB medium supplemented with 10 mM phthalate and 20 mM glucose. The culture supernatant was used directly (without extraction) as an HPLC standard. A single peak was observed at 3.7 min, which is the same retention time as that obtained for cis-phthalate dihydrodiol enzymatically synthesized from phthalate by using purified PDR and PDO (6). Acid-catalyzed dehydration of cis-phthalate dihydrodiol shifted the HPLC peak to 10.7 min, the retention time of authentic 4-hydroxyphthalate (Pfaltz and Bauer, Waterbury, Conn.). 4,5-Dihydroxyphthalate was synthesized by culturing C. testosteroni NH1024 in MSB medium supplemented with 10 mM phthalate, 20 mM glucose, and 20 mM l-glutamate as described previously (60). The culture supernatant yielded a single peak with a retention time of 7.8 min. For comparison, phthalate elutes at 15.8 min and protocatechuate elutes at 11.7 min under the HPLC conditions used.
Nucleotide sequence accession numbers.
The nucleotide sequence has been deposited in the GenBank database under accession no. AF095748.
RESULTS
Cloning and location of the genes for phthalate degradation from B. cepacia DBO1.
A cosmid library was constructed by using partially EcoRI-digested genomic DNA from B. cepacia DBO1, the cosmid cloning vector pMMB34, and the host E. coli strain S17-1. The cosmid clones were transferred by conjugation from E. coli S17-1 to B. cepacia DBO106, a mutant of strain DBO1 previously shown to lack cis-phthalate dihydrodiol dehydrogenase (89), the enzyme responsible for the second step in the catabolic pathway (Fig. 1). One cosmid clone of the 2,000 tested was able to complement the mutation in B. cepacia DBO106. Digestion of this cosmid clone (pGJZ1301) with EcoRI, the cloning enzyme, indicated that at least eight EcoRI fragments totaling approximately 25 kb had been cloned into pMMB34. A subclone (pGJZ1303) containing a 1.6-kb EcoRI fragment from pGJZ1301 allows DBO106 to grow on phthalate.
Although pGJZ1301 contains ophB, it does not contain all of the other genes needed for the phthalate catabolic pathway. Initial biotransformation experiments with E. coli S17-1(pGJZ1301) grown on glucose in the presence of phthalate showed no disappearance of phthalate or appearance of a catabolic intermediate. This suggests that one or both of the genes for the first step in the catabolic pathway is missing or that they are not expressed in E. coli. To explore this further, three oligonucleotides corresponding to the N-terminal and C-terminal sequences of PDR (designated PDR-N and PDR-C) and the N-terminal sequence of PDO (designated PDO-N) were synthesized. In Southern hybridization experiments, both PDR-N and PDR-C hybridize to a 9.2-kb EcoRI fragment and a 3.2-kb SphI fragment of pGJZ1301. The 3.2-kb SphI fragment was subsequently subcloned into pGEM7Zf(−) and designated pGJZ1305. This data places the ophA1 gene at least 5.5 kb away from the ophB gene (Fig. 2). No hybridization was detected between PDO-N and pGJZ1301, suggesting that the ophA2 gene is not present on this cosmid clone.
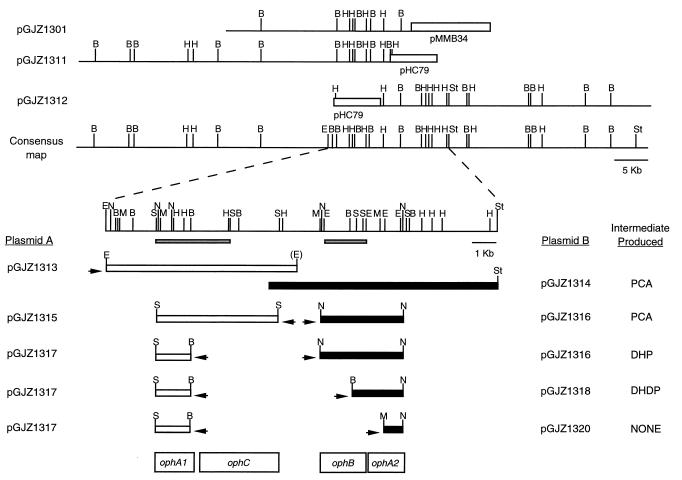
FIG. 2.
Complementation tests were done to determine the locations of the genes for phthalate degradation from B. cepacia DBO1. Abbreviations: PCA, protocatechuate; DHP, 4,5-dihydroxyphthalate; DHDP, cis-phthalate dihydrodiol; B, BamHI; E, EcoRI; H, HindIII; M, MluI; N, NspV; S, SphI; St, SstI. Arrows indicate the direction of transcription of the lac promoter on the vector. The gray bars under the consensus map indicate other regions described in the text: the 3.2-kb SphI fragment on the left is pGJZ1305, and the 1.6-kb EcoRI fragment on the right is pGJZ1303.
A second cosmid library was prepared with partially Sau3A1-digested, size-fractionated total genomic DNA and the cosmid pHC79. By screening the new library with the 3.2-kb SphI fragment from pGJZ1305 containing ophA1, one new clone was obtained, while screening with the 1.6-kb EcoRI fragment from pGJZ1303 containing ophB resulted in three separate clones being obtained. None of these four clones in E. coli were able to transform phthalate. This suggests that either ophA1 and ophA2 are not closely linked or that one or both of these genes are not expressed well in E. coli. Assuming that ophA1 and ophA2 may be distantly linked, two overlapping cosmid clones were examined in more detail. These two clones, pGJZ1311 and pGJZ1312, overlap by 1.9 kb, and each contains approximately 40 kb of cloned DNA; thus, together they represent approximately 80 kb of contiguous DNA from B. cepacia DBO1 (Fig. 2). An 8.2-kb EcoRI fragment from pGJZ1311 was subcloned into pRK415 to represent the left side of the overlapping region (designated pGJZ1313). An SstI deletion was constructed of pGJZ1312 to represent the right side of the overlapping region (designated pGJZ1314). These two compatible plasmids were transformed into the same E. coli DH5α strain and tested for the ability to transform phthalate.
Protocatechuate was detected in the culture supernatant, indicating that these two plasmids together contain all of the genes necessary for the first three steps of phthalate degradation (Fig. 1). To locate the exact positions of the four genes needed for this catabolic transformation, several smaller subclones were prepared and tested. A 5.1-kb (partial) SphI fragment and an internal 1.6-kb SphI-BamHI fragment were subcloned separately from pGJZ1313 into pRK415 and designated pGJZ1315 and pGJZ1317, respectively (Fig. 2). A 3.4-kb NspV, a 1.6-kb BamHI-NspV, and a 1.1-kb MluI-NspV fragment were subcloned separately from pGJZ1312 into pGEM7Zf(−) and designated pGJZ1316, pGJZ1318, and pGJZ1320, respectively (Fig. 2). Different combinations of these five plasmids were inserted into E. coli, and the resulting strains were tested for the ability to transform phthalate to a detectable intermediate. E. coli containing pGJZ1315 and pGJZ1316 is able to transform phthalate to protocatechuate, delimiting the smallest region necessary for the four target genes. E. coli containing the smaller plasmid pGJZ1317 along with pGJZ1316 was only able to transform phthalate to 4,5-dihydroxyphthalate. This indicates that all or part of the gene (ophC) for 4,5-dihydroxyphthalate decarboxylase must be present in the region deleted from pGJZ1315 to form pGJZ1317. Similarly, E. coli containing pGJZ1317 and pGJZ1318 is only able to transform phthalate to cis-phthalate dihydrodiol. This indicates that all or part of the gene (ophB) for cis-phthalate dihydrodiol dehydrogenase must be present in the region deleted from pGJZ1316 to form pGJZ1318. E. coli containing pGJZ1317 and pGJZ1320 is not able to transform phthalate to any detectable products. This indicates that pGJZ1317 and pGJZ1318 must contain the two genes (ophA1 and ophA2) needed for phthalate dioxygenase. It was shown above that ophA1 could be localized to a 3.2-kb SphI fragment by oligonucleotide probing. This places ophA1 on pGJZ1317 and means that ophA2 must be present on pGJZ1318. It is interesting to note that this means that the two genes necessary for the first step in the catabolic pathway are located at least 6.5 kb apart. In addition, there is at least 2.0 kb of space between the ophC and ophB genes that is not needed for the transformation of phthalate to pathway intermediates.
Nucleotide sequence of the genes for phthalate degradation.
To characterize the genes necessary for phthalate degradation in more detail at the molecular level, the two regions necessary for the conversion of phthalate to protocatechuate were sequenced. A diagram of this sequence is shown at the top of Fig. 3. Open reading frames were assigned gene names based on the biochemical data presented above. It is important to note that ophA1 is transcribed in the opposite direction from that of orf1, ophD, and ophC.
FIG. 3.
Different organization of the genes involved in phthalate degradation from B. cepacia DBO1, P. putida NMH102-2 (66), and C. testosteroni M4-1 (51). Designations: ophA1 and pht2, genes coding for phthalate dioxygenase reductase; ophA2 and pht3, genes coding for phthalate dioxygenase; ophB, pht4, and phtC, genes coding for 4,5-dihydro-4,5-dihydroxyphthalate dehydrogenase; ophC, pht5, and phtD, genes coding for 4,5-dihydroxyphthalate decarboxylase; pht3 and phtR, genes coding for putative phthalate transporter; orf1 and ophD, genes coding for products that show homology to the putative phthalate transporter. Abbreviations: B, BamHI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; N, NspV; S, SacI; Sm, SmaI; Sp, SphI. The dashed lines representing phtC and half of phtR indicate that the sequences for these have not been determined.
Analysis of a putative gene for phthalate transport.
A gene (ophD) for a putative phthalate transporter was identified in the nucleotide sequence upstream from ophC, which encodes 4,5-dihydroxyphthalate decarboxylase. Initially this gene was identified through its similarity to other known proteins involved in transport of substrates into the cell and also through its high similarity to a suspected phthalate transporter cloned and sequenced from P. putida (66). However, the gene for this putative transporter in B. cepacia DBO1 is split into two parts due to a frameshift, into an initial, very small, orf1 and the longer ophD (Fig. 2). This frameshift could be due to a sequence or cloning artifact. Two experiments were performed to prove that this was not the case. The region containing the suspected frameshift was PCR amplified from genomic DNA isolated from B. cepacia DBO1, ATCC 29424, and ATCC 17616. (DBO1 and ATCC 29424 are the same strain except that DBO1 passed through the laboratories of a chain of investigators before reaching our lab, while ATCC 29424 was obtained directly from the ATCC.) Each PCR product was then sequenced directly by using the same primers as were used for amplification. The nucleotide sequences of this region in DBO1 and ATCC 29424 are identical, showing that the sequence is correct and not the result of a base change occurring during cloning or subcloning of the genes. This also means that the base change is not due to a recent laboratory-acquired mutation since the DBO1 sequence is identical to the ATCC 29424 sequence. On the other hand, the nucleotide sequence of this region from ATCC 17616 is identical to that from DBO1 except for one additional base: an A found between bases 300 and 301 of orf1 in the DBO1 sequence. The additional base places orf1 in frame with ophD in ATCC 17616. This experiment demonstrates that the missing base is not due to a sequence error since it can be detected in ATCC 17616. This also means that for some reason DBO1, but not ATCC 17616, has lost a single base in this particular gene, splitting it into two halves. This base change does not seem to have an effect on the growth of DBO1 on phthalate since growth curves for DBO1 and ATCC 17616 on phthalate as the sole carbon source are identical (data not shown).
A second experiment was performed to demonstrate that ophD is actually translated (and also to confirm that orf1 and ophD are translationally separate, confirming the missing base). A 1.5-kb BamHI-HindIII fragment containing orf1 and ophD was cloned into the expression vector pTrc99A. An in vitro transcription-translation experiment was then performed, and the products were analyzed by SDS-PAGE (Fig. 4). Two peptides are produced by this clone in contrast to the negative control: one small peptide with a molecular mass of approximately 13 kDa and a large peptide with a molecular mass of approximately 38 kDa. These two molecular masses are consistent with those predicted for the proteins encoded by orf1 and ophD: 12.7 and 37.2 kDa, respectively. However, based on the difference in signal intensities, the gene products are produced in different quantities. Orf1 is produced in a much larger quantity than OphD. This is most likely due to the presence of a ribosome binding site in front of orf1, while translation of ophD is dependent on translational coupling since the putative initiation codon of ophD (ATG) overlaps the stop codon (TGA) of orf1.
FIG. 4.
SDS-PAGE of an in vitro transcription-translation of a clone containing orf1 and ophD. Lane 1, pTRC99A control; lane 2, pGJZ1321. The numbers on the left are the running positions of the molecular weight markers (from top to bottom: ovalbumin, carbonic anhydrase, β-lactoglobulin, and lysozyme; in thousands). The arrows on the right indicate the two translated proteins, Orf1 and OphD.
One question that remains is whether ophD is actually required for growth on phthalate by DBO1. This would provide physiological proof that OphD actually functions as a phthalate transporter. The DBO1 ophD gene region was deleted and replaced with a kanamycin resistance gene cassette by double reciprocal recombination as described in Materials and Methods. The resulting mutant strain (designated DBO304) grows normally on phthalate with a doubling time in liquid culture (MSB medium plus phthalate) equivalent to the wild-type strain. This suggests that ophD is dispensable for growth on phthalate. Phthalate transport assays were performed to prove that ophD is not involved in the transport of phthalate into the cell (Fig. 5). DBO1 grown on phthalate rapidly transports phthalate into the cell (3.2 nmol/min/mg of cells). DBO1 grown on p-hydroxybenzoate does not transport phthalate into the cell at any measurable rate, demonstrating that this is a phthalate-inducible activity. On the other hand, the ophD knockout mutant, DBO304, transports phthalate at a rate identical to that of DBO1. This indicates that ophD, although it is similar to known genes encoding permeases, is not involved in phthalate uptake.
FIG. 5.
Uptake of phthalate by B. cepacia DBO1 (wild type) grown on phthalate (■) and p-hydroxybenzoate (▴) and B. cepacia DBO304 (ophD mutant) grown on phthalate (•). Error bars represent the standard deviation from three independent assays.
DISCUSSION
Many genes for the degradation of aromatic compounds are organized into operons of functional units (3, 25, 30, 66, 82). This does not appear to be the case for the B. cepacia DBO1 genes for phthalate degradation described here. The five structural genes coding for the conversion of phthalate to protocatechuate and a nonfunctional permease are arranged in at least three transcriptional units based on orientation: ophA1, ophDC, and ophA2B. In fact, the two genes (ophA1 and ophA2) encoding the oxygenase and reductase components of phthalate dioxygenase, the first step in the pathway, are located at opposite ends of the sequenced region, approximately 7.0 kb apart. This gene organization is not a unique feature of DBO1 as another B. cepacia strain, ATCC 17616, which also degrades phthalate, has an identical restriction fragment length polymorphism pattern when probed with the cloned genes described here (13). The genes for phthalate degradation from P. putida NMH102-2 have also been cloned and sequenced (Fig. 3). The genes in this case were identified simply by sequence analysis, and their functions were predicted by homology to genes encoding other proteins. The P. putida genes are clustered and transcribed in the same direction in the order pht1 (coding for permease), pht2 (phthalate oxygenase reductase), pht3 (phthalate oxygenase), pht4 (cis-phthalate dihydrodiol dehydrogenase), and pht5 (4,5-dihydroxyphthalate decarboxylase). The order of the genes is thus the same as the order in the catabolic pathway. Three different transcripts were identified by Northern blotting, and the authors’ analysis of the nucleotide sequence suggested promoters in front of pht1, pht3, and pht5. However, although the genes for phthalate degradation in P. putida NMH102-2 and B. cepacia DBO1 are closely related based on their nucleotide sequence (see below), one cannot postulate a simple mechanism for the evolutionary rearrangement of the genes going from one strain to another. It is interesting to note that the genes for phthalate degradation are present on a plasmid in P. putida NMH102-2 (64) and in the chromosome of B. cepacia DBO1 and ATCC 17616 (16, 87). The gene for 4,5-dihydroxyphthalate decarboxylase from C. testosteroni M4-1 has also been cloned and sequenced (51). In this case, the authors suggested that a putative transport gene is located upstream (based on a partial nucleotide sequence) and a gene for cis-phthalate dihydrodiol dehydrogenase is located over 2 kb away (based on functional assays). Although the genes for the oxygenase and the reductase were not located and the complete nucleotide sequences of two of the three genes located are not available, the gene organization postulated for C. testosteroni M4-1 is similar to that identified here for B. cepacia DBO1.
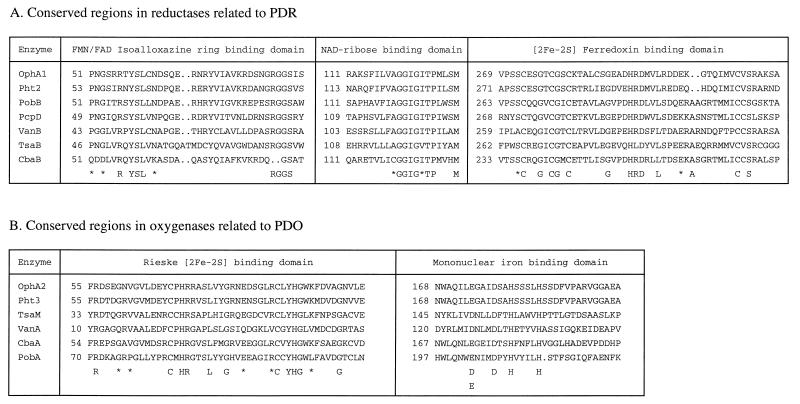
The two components of B. cepacia DBO1 phthalate dioxygenase, PDO and PDR, have been well studied (see the introduction). The corresponding genes have been identified in the present work by activity assays of subclones and by correlation with N- and C-terminal sequences of the purified proteins. The crystal structure of PDR has been solved at a 2.0-Å resolution (20) by using a partial amino acid sequence obtained through protein sequencing of the purified protein. The deduced amino acid sequence presented here will enable a more defined examination of the structure of the reductase. Since phthalate dioxygenase belongs to a family of oxygenases comprised of two components (Fig. 6), it should be possible to extend by analogy what is known about PDR and PDO to other members of this family. For instance, the crystal structure of PDR reveals an FMN binding domain in the N terminus, an NAD binding domain in the center, and a plant-type ferredoxin [2Fe-2S] domain in the C terminus. One arginine, one tyrosine, one serine, and one leucine are conserved in this reductase family for binding the FMN-isoalloxazine ring (Fig. 7). The PDR crystal structure shows that Tyr-58 contacts the si face of the flavin while Phe-226 stacks against the re side of the flavin ring (20). The consensus sequence XRGGS (where X is G or S) for binding the phosphate of FMN is conserved in all of the reductases but CbaB (Fig. 7). Arg-81 in PDR binds intimately to the FMN phosphate group and contributes to the specificity for FMN over flavin adenine dinucleotide. The fingerprint sequence GXGXXP for NADH or NADPH binding can also be detected, but in the case of this family, the consensus is the larger sequence XGGIGZTP (where X is A or C and Z is I or V) (Fig. 7). Pro-126 in this sequence contacts the bound nicotinamide (20). Finally, there are a number of conserved amino acids found around the conserved [2Fe-2S] chloroplast-type ferredoxin binding site (Fig. 7).
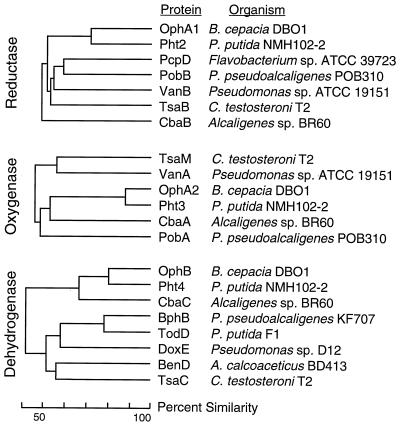
FIG. 6.
Dendrograms showing the relationship of OphA1, OphA2, and OphB to similar proteins. Reference strains include B. cepacia DBO1 (this work), P. putida NMH102-2 (66), Flavobacterium sp. strain ATCC 39823 (50), P. pseudoalcaligenes POB310 (22), Pseudomonas sp. strain ATCC 19151 (11), C. testosteroni T2 (42), Alcaligenes sp. strain BR60 (58), P. pseudoalcaligenes KF707 (31), P. putida F1 (88), Pseudomonas sp. strain D12 (25), and A. calcoaceticus BD413 (61).
FIG. 7.
Conserved regions in PDR, PDO, and related proteins. The consensus shown below the aligned sequences is where all sequences are identical. An asterisk indicates those positions where all but one sequence is identical. A period indicates a gap in the aligned sequences. The number before each sequence is the distance from the N-terminal end of the protein.
The oxygenase components of this two-component dioxygenase family are also related, at approximately the same level of similarity as seen with the reductase components (Fig. 6). All of the dioxygenases have the consensus sequence (CXHX16–17CXXH) which has been proposed for binding a Rieske-type [2Fe-2S] center (55, 62). Figure 7 shows this region for sequences in the same family as PDO. In addition to the Rieske-type [2Fe-2S] center, the oxygenase components also contain a mononuclear iron. In many oxygenases, one glutamate, one aspartate, one tyrosine, and two histidines are conserved for binding mononuclear iron (40). However, in phthalate dioxygenase and related enzymes, the glutamate and the tyrosine are not conserved (Fig. 7). The glutamate in some cases is conservatively replaced with an aspartate. The conserved amino acids Glu-214, Asp-219, Tyr-221, His-222, and His-228 of TodC1 (the α subunit of the terminal oxygenase component of toluene dioxygenase) from P. putida F1 were independently replaced with alanine residues by site-directed mutagenesis (40). Toluene dioxygenase with mutations at Glu-214, Asp-219, His-222, or His-228 completely lost activity. However, TodC1 with an alanine substitution at Tyr-221 retained 42% activity. These laboratory-generated data are consistent with the observation provided by nature in the amino acid sequence alignments shown in Fig. 7: the tyrosine is dispensable.
OphB is quite dissimilar from most other cis-dihydrodiol dehydrogenases involved in aromatic compound degradation (Fig. 6). OphB groups with the similar enzyme Pht4 from P. putida NMH102-2 (66) and CbaC from Alcaligenes sp. strain BR60 (58). However, OphB is 50 amino acids shorter than Pht4. That these three proteins cluster together into a new family of dihydrodiol dehydrogenases is probably due to the fact that their substrate is a compound with the dihydrodiol moiety opposite a carboxyl on the aromatic ring. On the other hand, OphB shows only 41 to 46% similarity to dihydrodiol dehydrogenases that act on neutral aromatic substrates such as BphB (biphenyl cis-dihydrodiol dehydrogenase) from Pseudomonas pseudoalcaligenes KF707 (31) and TodD (toluene cis-dihydrodiol dehydrogenase) from P. putida F1 (88) and that act on acidic substrates such as BenD (benzoate cis-dihydrodiol dehydrogenase) from Acinetobacter calcoaceticus BD413 (61) and TsaC (p-toluenesulfonate cis-dihydrodiol dehydrogenase) from C. testosteroni T2 (42).
The enzyme responsible for the third step of the catabolic pathway, 4,5-dihydroxyphthalate decarboxylase, has been purified from both B. cepacia DBO1 and C. testosteroni NH1000 (59, 72). The C. testosteroni enzyme has a molecular mass of 38 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and a molecular weight of 150,000 as determined by gel filtration through Sephadex G-200 (59). However, the same enzyme purified from B. cepacia DBO1 has a molecular mass of 66 kDa as determined by SDS-PAGE and a molecular weight of 420,000 as determined by gel filtration through Sepharose 6B (72). The calculated molecular mass for 4,5-dihydroxyphthalate decarboxylase (ophC gene product) based on the nucleotide sequence presented above is 37.4 kDa. This is in excellent agreement with that calculated (37.2 kDa) for the decarboxylase based on the nucleotide sequence of phtD from C. testosteroni M4-1 (51). It is also in agreement with that calculated (37.8 kDa) for the decarboxylase based on the corrected sequence of pht5 from P. putida NMH102-2. (We suspect that the NMH102-2 nucleotide sequence is missing a base immediately after position 6387 based on a comparison with the sequences from B. cepacia DBO1 and C. testosteroni M4-1.) These data correlate well with those derived from the purified enzyme from C. testosteroni NH1000, a 37- to 38-kDa monomer. However, the molecular mass based on the data derived from the purified decarboxylase from DBO1 is almost twice that derived from the nucleotide sequence. Further investigation is required to resolve this discrepancy.
The gene for a putative transporter (ophD) was identified by the similarity of the deduced protein to known or suspected aromatic acid transporters such as PcaK for protocatechuate and p-hydroxybenzoate (63), BenK for benzoate (19), VanK for vanillate (18), TfdK for 2,4-dichlorophenoxyacetate (53), HppK (5) and MhpT (28) for m-hydroxyphenylpropionate, HpaX for p-hydroxyphenylacetate (71), and Pht1 for phthalate (66). However, it is obvious through comparisons with these other related gene products that a frameshift mutation caused the production of two peptides (Orf1 and OphD). This frameshift is present in B. cepacia DBO1 and B. cepacia ATCC 29424 (ostensibly identical strains) but not in B. cepacia 17616. Both Orf1 and OphD are produced, as evidenced by in vitro transcription-translation, with a noticeable difference in the levels of protein produced (Fig. 5). Although this frameshift separates the leader sequence from the main body of the protein, the split occurs after two putative membrane-spanning regions of the protein. Since phthalate is a negatively charged compound at neutral pH, it is highly likely that it must be transported into the cell and thus a transport protein would be required. It has been shown that a signal sequence is not required for lactose permease to insert itself into the cell membrane and to function normally (7), and this may also be the case for phthalate permease in DBO1. However, a knockout mutation of ophD results in no detectable loss of the ability to transport phthalate into the cell (Fig. 5), demonstrating that OphD is not involved in phthalate transport in DBO1. One possibility is that a related aromatic acid transporter can take its place, as has been shown in Acinetobacter for the transport of aromatic acids (18, 21). However, no phthalate uptake was detected when DBO1 was grown on p-hydroxybenzoate (the phthalate and p-hydroxybenzoate catabolic pathways intersect at protocatechuate), demonstrating not only that phthalate transport is a phthalate-inducible phenotype but also that related aromatic acid transporters are not involved.
ACKNOWLEDGMENTS
We thank Teruko Nakazawa, David Ribbons, and Thomas Lessie for supplying strains; Chris Batie, Carl Correll, David Ballou, and Martha Ludwig for providing N- and C-terminal sequences of purified PDO and PDR; and Ruilong Liu, Ravindra Ramadhar, and Michael Murillo for technical assistance.
This work was supported by a National Science Foundation Young Investigator Award to G.J.Z. and cooperative agreement CR822634 from the U.S. Environmental Protection Agency Gulf Breeze Environmental Research Laboratory.
REFERENCES
- 1.Aftring R P, Taylor B F. Aerobic and anaerobic catabolism of phthalic acid by a nitrate respiring bacteria. Arch Microbiol. 1981;130:101–104. [Google Scholar]
- 2.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 3.Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- 4.Autian J. Toxicity and health threats of phthalate esters: a review of the literature. Environ Health Perspect. 1973;4:3–26. doi: 10.1289/ehp.73043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes M R, Duetz W A, Williams P A. A 3-(3-hydroxyphenyl)propionic acid catabolic pathway in Rhodococcus globerulus PWD1: cloning and characterization of the hpp operon. J Bacteriol. 1997;179:6145–6153. doi: 10.1128/jb.179.19.6145-6153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 7.Bibi E, Stearns S M, Kaback H R. The N-terminal 22 amino acid residues in the lactose permease of Escherichia coli are not obligatory for membrane insertion or transport activity. Proc Natl Acad Sci USA. 1992;89:3180–3184. doi: 10.1073/pnas.89.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol. 1988;170:4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bull C, Ballou D P. Purification and properties of protocatechuate 3,4-dioxygenase from Pseudomonas putida. A new iron to subunit stoichiometry. J Biol Chem. 1981;256:12673–12680. [PubMed] [Google Scholar]
- 13.Chang H-K. Ph.D. thesis. New Brunswick, N.J: Rutgers University; 1997. [Google Scholar]
- 14.Chang H-K, Zylstra G J. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Molecular analysis of phthalate degradation by P. cepacia DBO1, abstr. Q-223; p. 440. [Google Scholar]
- 15.Chang H-K, Zylstra G J. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Novel organization of the genes for phthalate degradation from Pseudomonas cepacia DBO1, abstr. Q-167; p. 414. [Google Scholar]
- 16.Cheng H P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cline J F, Hoffman B M, Mims W B, LaHaie E, Ballou D P, Fee J A. Evidence for N coordination to Fe in the [2Fe-2S] clusters of Thermus Rieske protein and phthalate dioxygenase from Pseudomonas. J Biol Chem. 1985;260:3251–3254. [PubMed] [Google Scholar]
- 18.Coco W M, Ornston L N. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Acinetobacter PcaK is a multispecificity active facilitator of aromatic acid transport, abstr. K-81; p. 355. [Google Scholar]
- 19.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain Adp1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Correll C C, Batie C J, Ballou D P, Ludwig M L. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 21.d’Argenio D A, Segyra A, Bunz P V, Coco W M, Ornston L N. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Genetic and physiological interactions associated with transport of aromatic compounds in Acinetobacter, abstr. K-79; p. 255. [Google Scholar]
- 22.Dehmel U, Engesser K-H, Timmis K N, Dwyer D F. Cloning, nucleotide sequence, and expression of the genes encoding a novel dioxygenase involved in metabolism of carboxydiphenyl ethers in Pseudomonas pseudoalcaligenes POB310. Arch Microbiol. 1995;163:35–41. doi: 10.1007/BF00262201. [DOI] [PubMed] [Google Scholar]
- 23.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Methods in molecular biology. 47. Electroporation protocols for microorganisms. Totowa, N.J: Humana Press Inc.; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 24.Dennis, J. J., and G. J. Zylstra. Unpublished data.
- 25.Denome S A, Stanley D C, Olson E S, Young K D. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J Bacteriol. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eaton R W, Ribbons D W. Metabolism of dibutylphthalate and phthalate by Micrococcus sp. strain 12B. J Bacteriol. 1982;151:48–57. doi: 10.1128/jb.151.1.48-57.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelhardt G, Wallnöfer P R, Rast H G. Metabolism of o-phthalic acid by different gram-negative and gram-positive soil bacteria. Arch Microbiol. 1976;109:109–114. [Google Scholar]
- 28.Ferrandez A, Garcia J L, Diaz E. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J Bacteriol. 1997;179:2573–2581. doi: 10.1128/jb.179.8.2573-2581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frey J, Bagdasarian M, Feiss D, Franklin F C H, Deshusses J. Stable cosmid vectors that enable the introduction of cloned fragments into a wide range of gram-negative bacteria. Gene. 1983;24:299–308. doi: 10.1016/0378-1119(83)90090-2. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation. 1995;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa K, Arimura N, Miyazaki T. Nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene of Pseudomonas pseudoalcaligenes. J Bacteriol. 1987;169:427–429. doi: 10.1128/jb.169.1.427-429.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganning A E, Brunk U, Dallner G. Phthalate esters and their effect on the liver. Hepatology. 1984;4:541–547. doi: 10.1002/hep.1840040331. [DOI] [PubMed] [Google Scholar]
- 33.Gassner G T, Ballou D P, Landrum G A, Whittaker J W. Magnetic circular dichroism studies on the mononuclear ferrous active site of phthalate dioxygenase from Pseudomonas cepacia show a change of ligation state on substrate binding. Biochemistry. 1993;32:4820–4825. doi: 10.1021/bi00069a017. [DOI] [PubMed] [Google Scholar]
- 34.Giam C S, Chan H S, Neff G S, Atlas E L. Phthalate ester plasticizers: a new class of marine pollutant. Science. 1978;199:419–420. [PubMed] [Google Scholar]
- 35.Goyal A K, Zylstra G J. Molecular cloning of novel genes for polycyclic aromatic hydrocarbon degradation from Comamonas testosteroni GZ39. Appl Environ Microbiol. 1996;62:230–236. doi: 10.1128/aem.62.1.230-236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham P R. Phthalate ester plasticizers—why and how they are used. Environ Health Perspect. 1973;3:3–12. doi: 10.1289/ehp.73033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurbiel R J, Batie C J, Sivaraja M, True A E, Fee J A, Hoffman B M, Ballou D P. Electron-nuclear double resonance spectroscopy of 15N-enriched phthalate dioxygenase from Pseudomonas cepacia proves that two histidines are coordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989;28:4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D. Studies on transformation of E. coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 39.Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 40.Jiang H, Parales R E, Lynch N A, Gibson D T. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178:3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jobling S, Reynolds T, White R, Parker M G, Sumpter J P. A variety of environmentally persistent chemicals, including some phthalate plasticizers, are weakly estrogenic. Environ Health Perspect. 1995;103:582–587. doi: 10.1289/ehp.95103582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junker F, Kiewitz R, Cook A M. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroni T-2. J Bacteriol. 1997;179:919–927. doi: 10.1128/jb.179.3.919-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaul A F, Souney P F, Osathanondh R. A review of possible toxicity of diethylhexylphthalate (DEHP) in plastic intravenous containers: effects on reproduction. Drug Intell Clin Pharm. 1982;16:689–692. doi: 10.1177/106002808201600908. [DOI] [PubMed] [Google Scholar]
- 44.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 45.Keith L M, Telliard W A. Priority pollutants. I. A perspective view. Environ Sci Technol. 1979;13:416–423. [Google Scholar]
- 46.Keyser P. Aerobic metabolism of the phthalates by selected pseudomonads. M.S. dissertation. Miami, Fla: University of Miami; 1974. [Google Scholar]
- 47.Keyser P, Pujar R W, Eaton R W, Ribbons D W. Biodegradation of phthalates and their esters by bacteria. Environ Health Perspect. 1976;18:159–166. doi: 10.1289/ehp.7618159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim E, Zylstra G J. Molecular and biochemical characterization of two meta-cleavage dioxygenases involved in biphenyl and m-xylene degradation by Beijerinckia sp. strain B1. J Bacteriol. 1995;177:3095–3103. doi: 10.1128/jb.177.11.3095-3103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuila D, Fee J A, Schoonover J R, Woodruff W H. Resonance Raman spectra of the [2Fe-2S] clusters of the Rieske protein from Thermus and phthalate dioxygenase from Pseudomonas. J Am Chem Soc. 1987;109:1559–1561. [Google Scholar]
- 50.Lange C C. Ph.D. thesis. Moscow: University of Idaho; 1994. [Google Scholar]
- 51.Lee J-H, Omori T, Kodama T. Identification of the metabolic intermediates of phthalate by Tn5 mutants of Pseudomonas testosteroni and analysis of the 4,5-dihydroxyphthalate decarboxylase gene. J Ferment Bioeng. 1994;77:583–590. [Google Scholar]
- 52.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 53.Leveau J H, Zehnder A J, van der Meer J R. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1998;180:2237–2243. doi: 10.1128/jb.180.8.2237-2243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu R, Olsen R H, Zylstra G J. Abstracts of the 92nd General Meeting of the American Society for Microbiology 1992. Washington, D.C: American Society for Microbiology; 1992. Cloning and characterization of the genes for phthalate degradation from Pseudomonas cepacia DBO1, abstr. K-37; p. 262. [Google Scholar]
- 55.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 56.Mayer F L J, Sanders H O. Toxicity of phthalic acid esters in aquatic organisms. Environ Health Perspect. 1973;3:153–157. doi: 10.1289/ehp.7303153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayer F L J, Stalling D L, Johnson J L. Phthalate esters as environmental contaminants. Nature (London) 1972;238:411–413. doi: 10.1038/238411a0. [DOI] [PubMed] [Google Scholar]
- 58.Nakatsu C H, Wyndham R C. Cloning and expression of the transposable chlorobenzoate-3,4-dioxygenase genes of Alcaligenes sp. strain BR60. Appl Environ Microbiol. 1993;59:3625–3633. doi: 10.1128/aem.59.11.3625-3633.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakazawa T, Hayashi E. Phthalate and 4-hydroxyphthalate metabolism in Pseudomonas testosteroni: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl Environ Microbiol. 1978;36:264–269. doi: 10.1128/aem.36.2.264-269.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakazawa T, Hayashi E. Phthalate metabolism in Pseudomonas testosteroni: accumulation of 4,5-dihydroxyphthalate by a mutant strain. J Bacteriol. 1977;131:42–48. doi: 10.1128/jb.131.1.42-48.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neidle E, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Cis-diol dehydrogenases encoded by the TOL pWWO plasmid xylL gene and the Acinetobacter calcoaceticus chromosomal benD gene are members of the short-chain alcohol dehydrogenase superfamily. Eur J Biochem. 1992;204:113–120. doi: 10.1111/j.1432-1033.1992.tb16612.x. [DOI] [PubMed] [Google Scholar]
- 62.Neidle E L, Harnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequence of the Acinectobacter calcoaceticus benABC genes for benzoate-1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nichols N N, Harwood C S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nomura Y, Harashima S, Oshima Y. PHT, a transmissible plasmid responsible for phthalate degradation in Pseudomonas putida. J Ferment Bioeng. 1990;70:295–300. [Google Scholar]
- 65.Nomura Y, Harashima S, Oshima Y. A simple method for detection of enzyme activities involved in the initial step of phthalate degradation in microorganisms. J Ferment Bioeng. 1989;67:291–296. [Google Scholar]
- 66.Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng. 1992;74:333–344. [Google Scholar]
- 67.Nomura Y, Takada N, Oshima Y. Isolation and identification of phthalate-utilization bacteria. J Ferment Bioeng. 1989;67:297–299. [Google Scholar]
- 68.Nozawa T, Maruyama Y. Denitrification by a soil bacteria with phthalate and other aromatic compounds as substrates. J Bacteriol. 1988;170:2501–2505. doi: 10.1128/jb.170.6.2501-2505.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peakall D B. Phthalate esters: occurrence and biological effects. Residue Rev. 1975;54:1–41. doi: 10.1007/978-1-4612-9857-1_1. [DOI] [PubMed] [Google Scholar]
- 71.Prieto M A, Diaz E, Garcia J L. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J Bacteriol. 1996;178:111–120. doi: 10.1128/jb.178.1.111-120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pujar B G, Ribbons D W. Phthalate metabolism in Pseudomonas fluorescens PHK: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl Environ Microbiol. 1985;49:374–376. doi: 10.1128/aem.49.2.374-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ribbons D W, Evans W C. Oxidative metabolism of phthalic acid by soil pseudomonads. Biochem J. 1960;76:310–317. doi: 10.1042/bj0760310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ribbons D W, Keyser P, Kunz D A, Taylor B F, Eaton R W, Anderson B N. Microbial degradation of phthalate. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker Inc.; 1984. pp. 371–397. [Google Scholar]
- 75.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 76.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 77.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 78.Taylor B F. An Na+-independent decarboxylation in a marine bacterium: ion-controlled biotransformations with intact cells. FEMS Microbiol Lett. 1985;29:279–283. [Google Scholar]
- 79.Taylor B F, Curry R W, Corcoran E F. Potential for biodegradation of phthalic acid esters in marine regions. Appl Environ Microbiol. 1981;42:590–595. doi: 10.1128/aem.42.4.590-595.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas J A, Wienckowski D B, Gillies B A, Thomas M J, J Y E. Effects of phthalic acid esters (PAES) on the neonate and aspects of teratogenic actions. Environ Health Perspect. 1986;65:243–248. doi: 10.1289/ehp.8665243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsang H T, Batie C J, Ballou D P, Penner-Hahn J E. X-ray absorption spectroscopy of the [2Fe-2S] Rieske cluster in Pseudomonas cepacia phthalate dioxygenase. Determination of core dimensions and iron ligation. Biochemistry. 1989;28:7233–7240. doi: 10.1021/bi00444a015. [DOI] [PubMed] [Google Scholar]
- 82.van der Meer J R, De Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 84.Wams T J. Diethylhexylphthalate as an environmental contaminant—a review. Sci Total Environ. 1987;66:1–16. doi: 10.1016/0048-9697(87)90072-6. [DOI] [PubMed] [Google Scholar]
- 85.Woodward K N. Phthalate esters, cystic kidney disease in animals and possible effects on human health: a review. Hum Exp Toxicol. 1990;9:397–401. doi: 10.1177/096032719000900607. [DOI] [PubMed] [Google Scholar]
- 86.Yakabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes, 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 87.Zylstra, G. J. Unpublished data.
- 88.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 89.Zylstra, G. J., and R. H. Olsen. Unpublished data.
- 90.Zylstra G J, Olsen R H, Ballou D P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989;171:5907–5914. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zylstra G J, Olsen R H, Ballou D P. Genetic organization and sequence of the Pseudomonas cepacia genes for the alpha and beta subunits of protocatechuate 3,4-dioxygenase. J Bacteriol. 1989;171:5915–5921. doi: 10.1128/jb.171.11.5915-5921.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]