Abstract
Understanding plant-microbe interactions requires examination of root exudation under nutrient stress using standardized and reproducible experimental systems. We grew Brachypodium distachyon hydroponically in fabricated ecosystem devices (EcoFAB 2.0) under three inorganic nitrogen forms (nitrate, ammonium, and ammonium nitrate), followed by nitrogen starvation. Analyses of exudates with liquid chromatography–tandem mass spectrometry, biomass, medium pH, and nitrogen uptake showed EcoFAB 2.0’s low intratreatment data variability. Furthermore, the three inorganic nitrogen forms caused differential exudation, generalized by abundant amino acids–peptides and alkaloids. Comparatively, nitrogen deficiency decreased nitrogen-containing compounds but increased shikimates-phenylpropanoids. Subsequent bioassays with two shikimates-phenylpropanoids (shikimic and p-coumaric acids) on soil bacteria or Brachypodium seedlings revealed their distinct capacity to regulate both bacterial and plant growth. Our results suggest that (i) Brachypodium alters exudation in response to nitrogen status, which can affect rhizobacterial growth, and (ii) EcoFAB 2.0 is a valuable standardized plant research tool.
EcoFAB 2.0, a fabricated ecosystem device, has low data variability in studies of plant traits.
INTRODUCTION
A substantial portion of the total fixed C from the plant photosynthate may be exuded from roots in the form of sugars, amino acids, organic acids, and secondary metabolites (1–3). These chemical cocktails are dynamic, complex, and associated with diverse functions in the rhizosphere (4, 5). Some exudate metabolites, particularly cinnamic acid derivatives (e.g., p-coumaric, caffeic, or ferulic acids), were shown to have allelopathic effects in plants (6, 7). In contrast, some exogenous metabolites can promote root growth, significantly contributing to plant C budget (8). Exudates can also inhibit the activity of nitrifying bacteria, while other compounds support N2-fixing symbiotic microorganisms (9). Since these compounds generate a nutrient-rich region in soils, they are a primary driver for microbial community formation that ideally benefit the plant (10). For example, specific root exudates from the wild oat Avena barbata, including several aromatic acids (nicotinic, shikimic, salicylic, cinnamic, and indole-3-acetic), were shown to be preferentially consumed by rhizosphere bacteria (5). Plant exudation can also alter the cycling of soil organic matter to bioavailable N in soil (11). In particular, oxalic acid can cause the microbe-independent destabilization of mineral-associated organic N, while glucose can activate microbial-mediated N mining (12, 13). The optimized N cycling via plant-microbe interactions can be a key to the increased production of bioenergy grasses on marginal N-limited soils (14, 15).
Nitrogen is an essential and often plant growth-limiting macronutrient that plants uptake through roots primarily as ammonium (NH4+) and nitrate (NO3−) and to a lesser extent as organic N (8, 16, 17). Previous studies showed that inorganic nitrogen (iN) form affects plant development. For example, the supply of NH4+ or NO3− alters the phenotypes and cell wall composition of the model grass plant Brachypodium distachyon (18). On the other hand, N deficiency causes changes in the transcriptome, chlorosis due to chlorophyll degradation, a decrease in plant foliar N content, and changes in auxin levels resulting in initial root foraging (16, 19–21). Multiple studies showed exudate changes by N supply in several plant grass species. For example, a recent paper showed the increase in aromatic and other organic acids in marginal N-depleted soils of the switchgrass rhizosphere, while added nitrogen (+N) and nitrogen phosphate (+NP) fertilizer increased the abundance of amino acids (22). A study on rice (Oryza sativa) found that roots actively release many metabolites in response to N and P deficiency (23). Another study showed that axenically grown maize under N-, P-, Fe-, or K-deficient conditions changed the abundance of specific exuded amino acids, sugars, and organic acids (24).
The annual grass B. distachyon, an accepted model for many bioenergy and grain crops (23), has many favorable traits for laboratory studies, including a relatively small diploid genome, small size, and a short reproduction cycle (25). Despite the importance of B. distachyon to understanding fundamental processes associated with grass growth and genetics, currently, there is a lack of studies systematically exploring the effects of inorganic N (NH4NO3, NO3−, or NH4+) starvation on root exudation in B. distachyon. Given that changes in response to the form of iN may be subtle, we require methods for achieving highly reproducible and sterile plant growth, exudate collection, and phenotyping across hundreds of plants. We have previously described “EcoFAB” devices that were shown to support reproducible B. distachyon growth and phenotyping across four different laboratories (26). However, these EcoFABs were made by “hand” molding polydimethylsiloxane rubber in three-dimensional (3D) printed molds, followed by chemically bonding them to microscope slides, thus limiting the scale of studies they can support (27).
Here, we aim to increase the understanding of B. distachyon exudation dynamics under varying N supply. We hypothesize that iN form and starvation will change the plant exudate profile and that iN starvation–induced exudates have diverse functions in the rhizosphere that will alter the growth of microbes and seedlings. To test this hypothesis, we developed and used an improved EcoFAB device (EcoFAB 2.0), made using a high-throughput fabrication. The combination of different inorganic N sufficient (iN+) treatments (NH4NO3, NO3−, or NH4+) with N starvation (iN−) in a hydroponic environment of EcoFAB 2.0 devices allowed us to assess the modulation of root exudation in B. distachyon Bd21-3 via liquid chromatography–tandem mass spectrometry (LC-MS/MS)–based metabolomics analysis, followed by spectral matching using an online database and feature-based molecular networking. We combined the LC-MS/MS analysis with the Global Natural Products Social Molecular Networking (GNPS) tool, which provides us with feature annotations based on similarities to spectral libraries (28). We also used NPClassifier, a deep-neural network tool that classifies compounds based on structural characteristics (29). Metabolomics was followed by plant and bacterial bioassays to access the functions of selected exudate compounds.
RESULTS
EcoFAB 2.0 capabilities
The EcoFAB (https://eco-fab.org/) platform aims to create standardized systems to study environmental microbiomes and plant exudation (30). Initial devices were made using polydimethylsiloxane rubber and 3D printed molds (27). At the same time, we found that the resulting devices had high cross-laboratory reproducibility of plant morphology and exometabolome (26). EcoFAB 2.0 is another sterile, self-contained system suitable for sterile plant growth and imaging (U.S. patent application no: 16/876,415). Version 2.0 is larger to allow for studying small plants such as B. distachyon over their entire life cycle or large plants but only at early growth stages. Critically, EcoFAB 2.0 is made using injection molding, which enables high-throughput fabrication. The EcoFAB 2.0 comprises three injection-molded polycarbonate parts, a silicone gasket, a glass slide, and eight screws. It has a ~10-ml root chamber that slightly slopes downward away from the plant port, while the device is sitting flat and has two sampling ports and a 264-ml shoot chamber (height × length × width = 50 mm × 66 mm × 80 mm) with four luer-compatible gas vent ports (fig. S1A). The root zone used in this study was ~3 mm in thickness. However, this thickness is determined by the gasket thickness and is, therefore, adjustable. All parts can be sterilized by autoclaving or ethylene oxide (31). The base dimensions of the EcoFAB 2.0 are the same as a standard multiwell plate (SBS format), making them compatible with liquid handling equipment (fig. S1A).
EcoFABs 2.0 are compatible with plant hydroponic and soil experiments (fig. S1B) and have been used to grow not only B. distachyon but also Arabidopsis thaliana, Lotus japonicus, and Camelina suneson for at least 4 weeks. Because the parts are made of injection-molded polycarbonate, dyes can be added to the EcoFAB 2.0 base to shield the roots chamber from specific wavelengths or all visible light for light-sensitive rhizosphere experiments (fig. S1B). During development, we have also tested visualization of root architecture by scanning with flatbed scanners, inverted microscopy on roots, sampling of microbes for determination of root microbiome structure by 16S sequencing, metabolomics on spent medium, and gas analysis in the shoot top chamber (fig. S1C). While large glass slides of standard thickness were used for this study, the backing plate in the EcoFAB 2.0 can be reversed to accommodate thinner slides (i.e., 0.55-mm gorilla glass). In our hydroponic experiments, we tested the sterility of EcoFAB 2.0 at the end of weeks 3 and 5 by incubating the spent plant growth medium from each replicate on Luria-Bertani agar plates for several days at 27°C in the dark; this revealed 100% sterility of the EcoFAB 2.0 system over the experimental period of 5 weeks.
Form of iN changes plant biomass
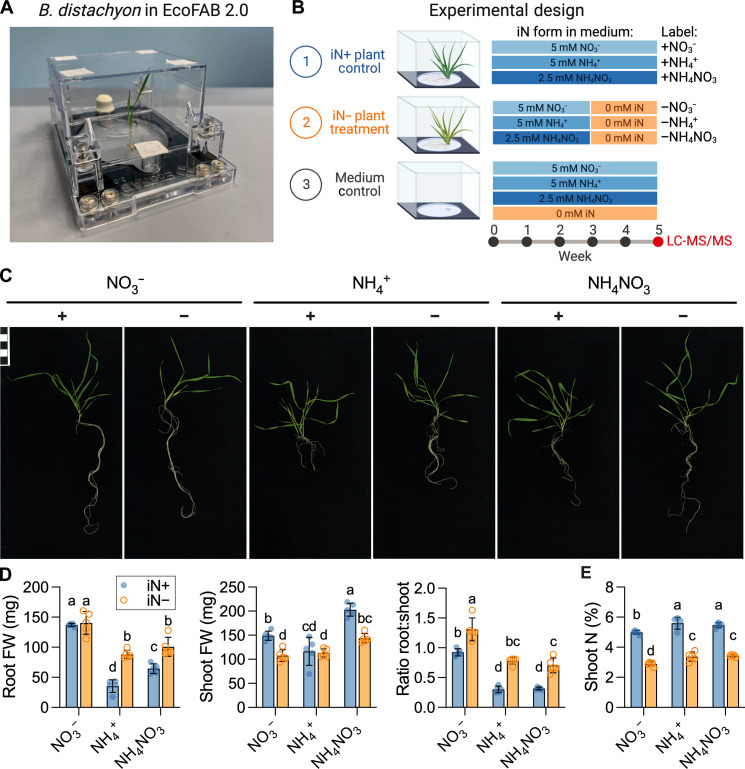
We grew B. distachyon in a hydroponic EcoFAB 2.0 setup and analyzed plant biomass after 5 weeks (Fig. 1, A and B). The type of iN treatment was correlated with phenotypical differences in the B. distachyon Bd21-3 plants (Fig. 1C). After 5 weeks, all plants were at the tillering/main stem elongation stage (Fig. 1C). The 2 weeks on iN− resulted in mild chlorosis on the bottom leaves (Fig. 1C). The iN form strongly affected the plant fresh biomass (Fig. 1D) with +NO3− yielding the highest root weight, followed by +NH4NO3, while the +NH4+ treatments showed the smallest root weights. The +NH4NO3 supply resulted in the highest shoot weight, followed by the +NO3− and + NH4+ treatments. The transfer to iN− conditions significantly increased the root:shoot fresh biomass ratio relative to the iN+ treatments by an average of 42% for NO3−, 123% for NH4NO3, and 158% for NH4+ (Fig. 1D). The number of tillers remained consistent across all treatments, averaging at three per plant (fig. S2A). Nitrogen deficiency did not affect leaf number, but the type of nitrogen source had a significant impact, with plants supplied with +NH4NO3 and +NH4+ exhibiting the highest leaf count, while those supplied with +NO3− had the lowest leaf count (fig. S2B). The iN form significantly affected the maximum root length with +NO3− resulting in longest roots, followed by shorter roots in +NH4NO3 medium and the shortest roots in +NH4+. The iN starvation caused significant root length increase relative to the iN sufficient treatments (fig. S2C).
Fig. 1. Analysis of plant biomass grown in EcoFAB 2.0.
(A) An example of EcoFAB 2.0 containing hydroponically grown B. distachyon BD21-3 seedling. (B) Design of the plant growth experiment. B. distachyon Bd21-3 grew in EcoFAB 2.0 devices for 5 weeks with weekly medium changes. Control iN+ plants (blue) received 5 mM iN as nitrate (+NO3−), ammonium (+NH4−), or ammonium nitrate (NH4NO3), while the plants in the iN− treatment group (orange) were transferred to iN− medium (−NO3−, −NH4+, or −NH4NO3) after 3 weeks. The EcoFABs 2.0 filled with growth medium without plants were used as technical controls. Each iN treatment had five biological replicates. We sampled plant spent medium for exudate analysis by LC-MS/MS at week 5. The figure was created with BioRender.com. (C) The phenotypes of 5-week-old B. distachyon grown on different iN forms. The iN+ controls are marked with plus signs (+), and the iN− treatments with minus signs (−). The scale bar in the top left indicates 1 cm × 1 cm. All phenotype pictures are available at https://doi.org/10.6084/m9.figshare.21376017. (D) Fresh weight (FW) of roots, shoots, and their ratio. (E) Total nitrogen in dry shoot weight (%, w/w). Different letters indicate statistically significant differences [two-way analysis of variance (ANOVA) with Tukey’s post hoc test; n = 5; P ≤ 0.05]. Blue-shaded circles show control iN+ plants, while orange circles indicate iN− plants.
The isotope ratio MS analysis was primarily used to obtain total N content (%, w/w) from small shoot samples. Consequently, the shoot total C content and stable isotope values (δ13C and δ15N) served as additional variables obtained from the technique. The iN− resulted in significantly lower shoot total N content at <3.4% (w/w) than iN+ conditions at >5.0% (w/w), thus showing that the N-limited conditions caused changes in plant nutritional status (Fig. 1E). Shoot total C content was statistically identical across iN treatments at means between 47.2 and 49% (w/w) (fig. S2D). When comparing the δ13Cshoot values, the +NH4NO3 plants were the most 13C-enriched, pointing to increased photosynthetic rates (fig. S2E). The δ15Nshoot values were generally not affected by iN starvation, but differences were observed between plants grown on different nitrogen forms (fig. S2F) with general 15N depletion in shoots relative to the iN sources from the grown medium (fig. S2G).
iN form modulates nitrogen uptake and rhizosphere pH
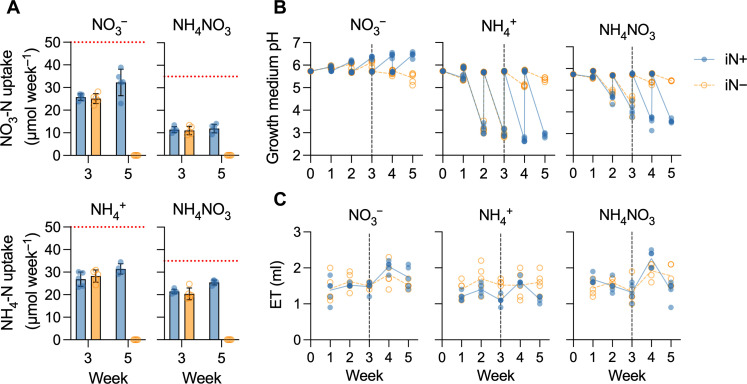
We measured the iN concentration in the spent plant growth medium from the EcoFAB 2.0 root zones in the medium controls and plant treatments at the end of weeks 3 and 5. The NH4+ and NO3− technical controls each contained, on average, 50 μmol of iN per 10 ml of medium in EcoFAB 2.0, which equals the intended 5 mM iN. However, the NH4NO3 technical control contained, on average, 35 μmol of both NH4-N and NO3-N per 10 ml of medium, totaling 7 mM iN (40% higher than the expected concentration of 5 mM). Plants did not take up all available iN from the growth medium, leaving behind more than 36% bioavailable iN each week (Fig. 2A). The iN plant uptake rates of solely supplied NO3− or NH4+ were comparable at 25 or 28 μmol of iN week−1 at week 3 and increased with plant age to 32 or 31 μmol iN week−1 at week 5, respectively. However, B. distachyon supplied with NH4NO3 preferred NH4+ over NO3− in an approximate ratio of 2:1, uptaking on average of 20 μmol of NH4-N week−1 and 11 μmol of NO3-N week−1 at week 3 and increasing to 25 μmol of NH4-N week−1 and 12 μmol of NO3-N week−1 at week 5 (Fig. 2A).
Fig. 2. EcoFAB 2.0 growth medium characterization.
(A) Plant nitrate-N and ammonium-N uptake (in micromoles per week) from the measured average concentration in the growth medium (dotted red line) for plants supplied with NO3−, NH4+, or NH4NO3. Bars show means ± SD, n = 5; +NH4+ treatment, n = 4. (B) Growth medium weekly pH shift from the average baseline value of 5.8. (C) Evapotranspiration (ET) expresses weekly medium volume loss from EcoFAB 2.0 containing B. distachyon. The dashed vertical line at the end of week 3 indicates the transition to an iN− medium for the iN− treatment plants (empty orange circles and dashed lines), while the iN+ control plants received iN for 5 weeks (full blue circles and lines). The lines in (B) and (C) connect weekly means. Media (V = 10 ml) were changed completely weekly.
Each week, we measured the pH of the spent medium and replaced it with fresh medium pH of 5.8. Despite the presence of the MES buffer in the growth medium, plants changed the medium pH depending on the iN source, increasing the weekly pH amplitudes with age (Fig. 2B). The NO3− uptake increased medium alkalinity to a pH of 6.5 at week 5, while NH4+ acidified the medium to a pH of 2.9 at week 5. NH4NO3 supply caused medium acidification to a pH of 3.6 at week 5 (Fig. 2B), correlated with a higher proportion of NH4+ to NO3− uptake (Fig. 2A). Plants in the iN-free medium maintained the initial pH of 5.8 regardless of the previous iN supply (Fig. 2B). While performing the medium replacement, we measured its volume, enabling us to determine water loss due to evapotranspiration. The EcoFAB 2.0 exhibited an average evapotranspiration of 1.6 ± 0.2 ml week−1, equivalent to 16% of the initial medium volume (Fig. 2C). This underscores the need for weekly water replenishment in the EcoFAB 2.0.
Root exudation changes with nitrogen supply
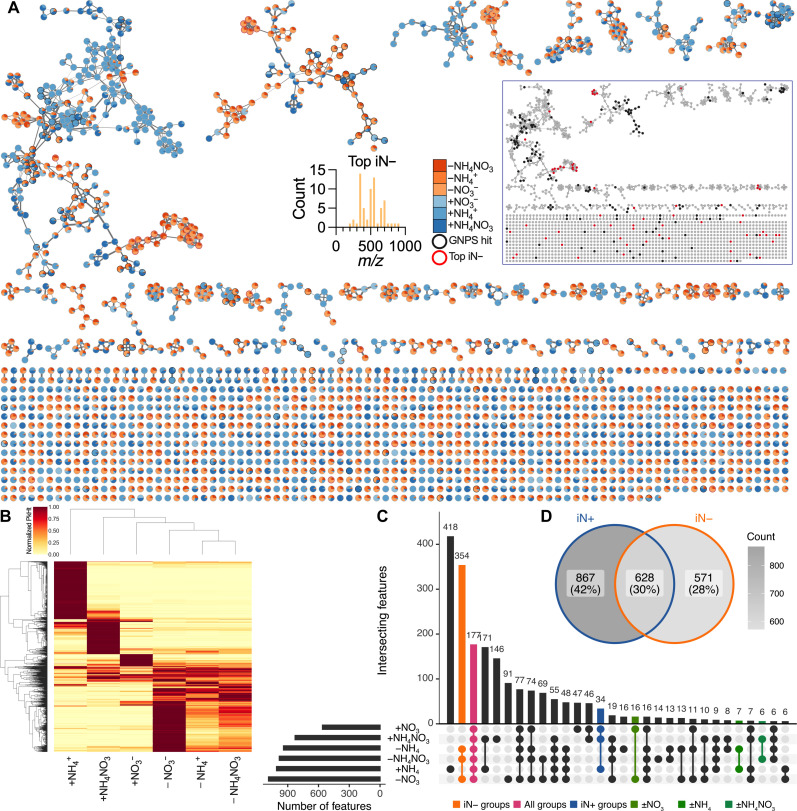
We analyzed the root exudate composition by LC-MS/MS (table S1). The analysis was filtered down to 2065 features across positive and negative polarities, followed by the creation of molecular networks (Fig. 3A and table S2). The network shows that the compositions of plant exudates are modulated by iN source with evident clusters of features differentially responding to iN supply (Fig. 3A). There were, in total, 155 features (7.8% of all features) with robust annotation, defined by MQScore (cosine score) of >0.7, to a library reference compound in GNPS across all samples/conditions (Fig. 3A). Many of these 155 features also had secondary GNPS annotations, most of which shared the same NPClassifier biosynthetic pathway as the top annotation (table S3); this allowed for high confidence in accurate feature classifications. We show that 65 features were >1000-fold statistically higher (t test, P ≤ 0.05) in the iN− (n = 14) than in iN+ (n = 15) plant treatments (Fig. 3A). These top iN− features had no annotations in GNPS. Most top iN− features had relatively high mass-to-charge ratio (m/z) values of >300. Notably, the top iN− features are clustered separately from the annotated features, thus prohibiting identification by mass shift (Fig. 3A). The evaluation of whether some features were identified twice in both positive and negative modes revealed 94 pairs of corresponding features in the 2065 filtered features set and 6 pairs in the 155 annotated features set (table S4), which represents feature duplication of less than 5%; thus, these features were kept in the datasets for downstream analyses.
Fig. 3. Filtered features detected by polar metabolomics in B. distachyon exudates under varying iN supply in EcoFAB 2.0.
(A) Feature-based molecular networking (merged polarities, n = 2065). Edge widths increase with the cosine score. Pie charts show the mean peak heights of iN+ (blue gradient) and iN− (orange gradient) treatments. The black bordered nodes signify GNPS-annotated features (MQScore > 0.7), while the red bordered nodes show 65 features that increased >1000-fold in N deficiency (top iN−). Most top iN− features have m/z > 300. Note no GNPS annotations for the top iN− features. The inset network shows low proximity of the top iN− features (red nodes) to GNPS features (black nodes), thus prohibiting their mass-shift identification. The high-resolution interactive network is available via Network Data Exchange (NDEx) at https://doi.org/10.18119/N9FW3X. (B) The heatmap shows hierarchical clustering of the filtered features normalized to the maximum average peak intensity across N treatments. (C) The UpSet plot shows the number of shared filtered features across the iN treatments among the first 30 largest intersecting sets. The second largest intersection of 354 features was across iN− treatments (orange), 177 features were shared across all iN groups (pink), and 34 features were shared within the iN+ group (blue), while a relatively low number of features were shared between the individual iN+ treatments and their iN− counterparts (greens). (D) Venn diagram shows feature intersections between grouped iN+ versus iN− treatments.
We summarized the normalized relative feature peak height in a heat plot (Fig. 3B), revealing distinct clustering between iN− and iN+ treatments. In the iN− treatments, the −NO3− treatment showed high intensities for more features than the −NH4+ or −NH4NO3 treatments. Conversely, in the iN+ treatments, both the +NH4+ and +NH4NO3 plant exudates displayed more high intensity features than the +NO3− treatment. In addition, the nonmetric multidimensional scaling ordination plot provided further evidence of separate clustering between the iN− and iN+ treatments based on raw peak heights (fig. S3).
When we compared shared features across our six individual treatments, we observed minimal overlaps between the three iN+ treatments versus their iN− counterparts (16 features for +NO3 versus −NO3, 7 features for +NH4 versus −NH4, and 6 features +NH4NO3 versus −NH4NO3), while the three iN− treatments (−NO3 versus −NH4 versus −NH4NO3) shared a large proportion (354) of features (Fig. 3C). This suggests that the exposure of plants to iN− conditions causes change of exudation regardless of the previous iN form. The three iN+ treatments (+NO3, +NH4, and +NH4NO3) shared only 34 features, suggesting that different N forms cause different exudate profiles. The UpSet plot showed 177 features intersecting across all groups, thus potentially comprising a core B. distachyon exometabolome under these growth conditions. In addition, the number of features uniquely present only in one of the treatments (418 in +NH4+, 146 in +NH4NO3, 47 in +NO3−, 91 in −NO3−, 14 in −NH4NO3, and 16 in −NH4+) (Fig. 3C) corresponds with the highest peak intensity patterns in the heat plot (Fig. 3B).
The iN+ treatments collectively contained the highest number of features at 867 features (42% of all features). The iN+ and iN− treatments shared 628 features (31% of all features), while 571 features (27% of all features) were specific only to iN− condition (Fig. 3D), indicating the highest cumulative feature diversity of polar metabolites under a sufficient iN supply.
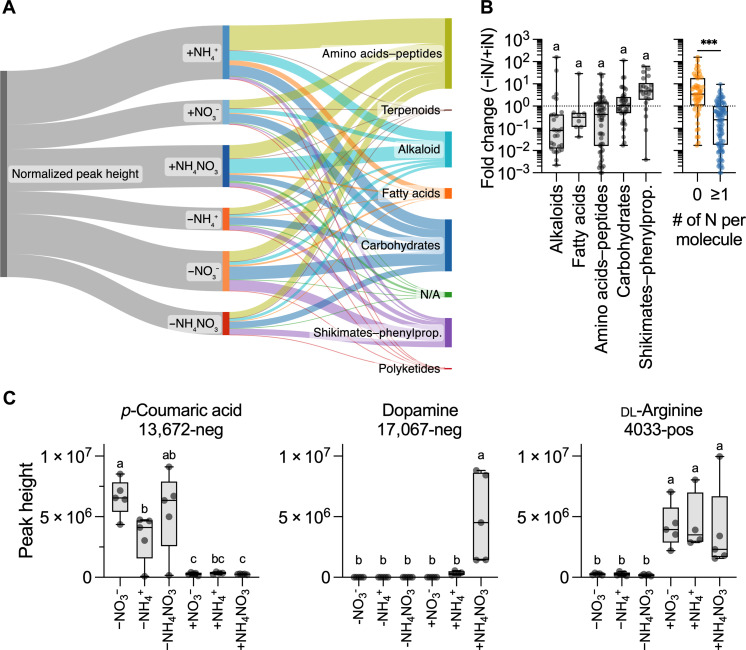
The 155 annotated features belonged to diverse biosynthetic pathways according to the NPClassifier: amino acids–peptides (53), carbohydrates (39), alkaloids (27), shikimates-phenylpropanoids (22), fatty acids (8), no classification (4), terpenoid (1), and polyketides (1) (fig. S4). These 155 annotations matched 110 unique metabolites (table S2). There was a tendency (not statistically significant) for differential production of compound classes based on nitrogen supply (Fig. 4, A and B). For example, signals for putatively annotated sugars were equally present across N treatments, and amino acid–annotated features were high under N sufficiency. However, features annotated to shikimates-phenylpropanoids had higher peak intensities under N-depleted conditions (Fig. 4B). In general, under iN− conditions, the exudation of N-containing compounds was decreased relative to iN+ conditions, whereas the exudation of N-free compounds was increased relative to iN+ conditions (Fig. 4B). Specifically, iN− conditions increased the abundance of features with annotation to aromatic acids and their precursors (fig. S5).
Fig. 4. GNPS annotated features with MQScore of >0.7 (n = 155) in the polar metabolomic analysis of B. distachyon exudates.
(A) Sankey plot showing the distribution of compound classes for GNPS-annotated features, which are classified by biosynthetic pathway and iN treatment. Widths represent the summed values of the normalized peak heights (relative to the maximum value for each feature). The plot was made with SankeyMATIC (sankeymatic.com). (B) Fold change (Fc) peak heights in iN− versus iN+ treatments for annotated features grouped by biosynthetic pathways (excluded singleton and unclassified groups). Different letters indicate statistically significant differences (ANOVA with Tukey’s post hoc test, all n.s., P ≤ 0.05). There was a significant decrease in metabolites containing ≥1 N atom (blue) relative to N-free molecules (orange) under iN− conditions (t test, ***P < 0.001). (C) Annotated features that showed differential patterns of abundance based on iN supply and were verified against library reference standards. The exudation of p-coumaric acid was increased in iN−, and dopamine was +NH4NO3 specific, while dl-arginine increased only under iN+ conditions. Different letters above box plots indicate statistically significant differences (two-way ANOVA with Tukey’s post hoc test; n = 5; +NH4+, n = 4; P ≤ 0.01). All box plots show all points, hinges extend from the 25th to 75th percentiles, the middle line indicates the median, and the whiskers extend to minimum and maximum values.
The supply of iN+ in the form of +NH4+ and +NH4NO3 resulted in the production of the largest number of unique features compared to +NO3−, whereas for iN−, the −NO3− condition resulted in the largest number of features compared with −NH4+ and −NH4NO3 (Fig. 3C); these patterns were also present across the annotated features for relative normalized peak heights (Fig. 4A). In particular, the +NH4+ led to higher relative peak intensities for amino acids–peptides, while alkaloid biosynthesis was the highest under +NH4NO3 (Fig. 4A). In general, the transition of plants from iN+ to iN− conditions caused a reduction in total normalized peak intensities, except for the ±NO3− treatments; however, relative intensities of some molecular classes went up (shikimates in −NO3− and +NH4NO3 and carbohydrates in −NO3− treatment).
Statistical comparison [analysis of variance (ANOVA) with Tukey’s post hoc test, P ≤ 0.01] between peak heights of filtered features within iN− treatments, iN+ controls, and individual ∓iN pairs confirmed that exudation is modulated by iN source (table S5). The next aim was to verify some of the most interesting putative GNPS annotations. The statistical analysis combined with UpSet grouping (Fig. 3C) of annotated features determined 6 features unique to iN− conditions, 2 in iN+, 10 specific to +NH4+, 4 in +NH4NO3, and 1 in +NO3− (table S6). These differentially produced features were matched against an in-house library of standard reference compounds analyzed using the same LC-MS/MS methods, resulting in four matches, of which three aligned to m/z (corresponding adduct), retention time, and MS/MS spectra of the reference (table S6). Dopamine (feature 17,067-neg) classified in the alkaloid pathway was uniquely present under the supply of +NH4NO3 (Fig. 4C). The p-coumaric acid (feature 13,672-neg), classified as shikimates-phenylpropanoids, was significantly higher in iN− than iN+, while an opposing pattern was present for dl-arginine (4033-pos), which significantly increased under iN+ conditions.
To predict the identification of the 65 top iN− features without GNPS annotation (Fig. 3A), we used two computational tools: CANOPUS (class assignment and ontology prediction using MS) and GNPS analog search. CANOPUS predicted formula and NPClassifier pathways for 33 features (table S7), mostly associated with amino acids–peptides or alkaloids. Approximately 91% of the predicted formulas contained N atoms, some of which also contain S and/or P atoms. GNPS analog search revealed 16 features with robust analog hits to known metabolites (table S8). Seven analog compounds containing N and S atoms, similar to the MES buffer in the growth medium, were classified as alkaloids and were not found naturally in plants (fig. S6). On the other hand, eight analog compounds naturally occurred in plants and belonged to shikimates-phenylpropanoids, without N and S atoms (fig. S6). Together, these tools suggest the presence of NS compounds, potentially originating from plant-mediated reactions of the MES buffer under N-free conditions, alongside natural plant exudate compounds.
Shikimic and p-coumaric acids affect the growth of bacteria and plants
Given the observed increase in features annotated as shikimates-phenylpropanoids under N deficiency, we sought to assess their biological effects. We selected two representative compounds, p-coumaric and shikimic acids. In addition, we used oxalic acid and glucose because they are common in root exudates (13, 32). We tested the individual effects of these four exogenous metabolites on model rhizosphere bacteria and B. distachyon Bd21-3 seedlings.
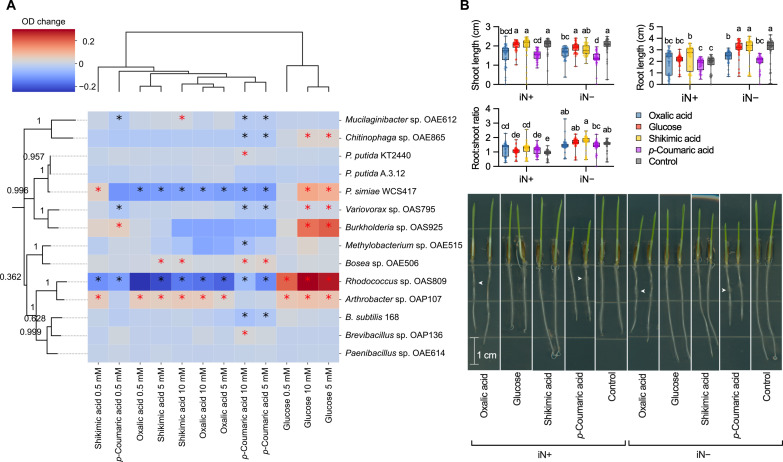
To test whether the exogenous compounds can support improved bacterial growth, we grew 14 diverse bacteria individually for 96 hours in culture medium supplemented with each exometabolite at three concentrations and measured optical density at 600 nm (OD600) (fig. S7). The effect of the exometabolites on bacterial growth was strain specific (Fig. 5A). Glucose had a significant dose-dependent positive effect on growth of six strains. Oxalic acid and shikimic acid had similar effects, both promoting Arthrobacter growth, while inhibiting Pseudomonas simiae and Rhodococcus. Moreover, shikimic acid also stimulated growth of Bosea and Mucilaginibacter, particularly at the highest dose. Notably, p-coumaric acid caused divergent dose-dependent growth modulation, promoting four strains while inhibiting seven strains.
Fig. 5. Exometabolite bioassays showing microbial growth and plant development on oxalic acid, glucose, shikimic acid, and p-coumaric acid.
(A) Heatmap showing the bacterial growth differences (OD change) within 96 hours in medium [0.1× R2A broth ± 5% dimethyl sulfoxide (DMSO)] supplemented with a metabolite (0.5, 5, or 10 mM) compared to compound-free control. Red asterisks indicate significant increases compared to both inoculated and uninoculated medium, while black asterisks denote significant decreases (Tukey’s post hoc test, P < 0.05). Treatments cluster via unweighted pair group method with arithmetic mean (UPGMA) algorithm based on Euclidean distances, rows by phylogenetic distances of 16S ribosomal RNA gene sequences (average length, 1500 base pairs; bootstrap values). (B) Influence of exometabolites (0.5 or 0 mM for control) on root and shoot length of 3-day-old B. distachyon seedlings supplemented with 2.5 or 0 mM NH4NO3 (iN+ or iN−, respectively). Distinct letters indicate significant differences (two-way ANOVA, Tukey’s post hoc test; n = 48; P ≤ 0.05). Representative seedling phenotypes on day 3 after germination are shown in the bottom panel. Arrows pinpoint root areas with concentrated root hair density under oxalic and p-coumaric acid treatments. High-resolution seedling images are available at https://doi.org/10.6084/m9.figshare.21433065.
It is known that exometabolites can alter root growth (8); therefore, we also tested whether these compounds at 0.5 mM affect seedling development under iN+ or iN− conditions (2.5 or 0 mM NH4NO3, respectively). The assay revealed that B. distachyon seedlings under the iN+ conditions statistically increased root length on shikimic acid to 2.36 cm relative to metabolite-free control at 1.80 cm. However, the iN+ condition did not follow this pattern (Fig. 5B). The p-coumaric acid supply significantly reduced mean shoot length relative to control from 1.99 to 1.51 cm (24% reduction) under the iN+ condition and from 2.02 to 1.38 cm (32% reduction) under the iN− condition. However, the mean root length suppression by p-coumaric acid relative to the control was significant only under the iN− condition from 3.11 to 1.98 cm (37% reduction) but insignificant under the iN+ condition (Fig. 5B). The oxalic acid biomass reduction relative to control followed the same pattern as p-coumaric acid by decreasing significantly shoot length under both iN+ and iN− conditions to 1.56 cm (22% reduction) and 1.67 cm (18% reduction), respectively, in addition to significant root length reduction to 2.38 cm (23% lower) that was exclusive to iN− conditions. The oxalic and p-coumaric acids caused patchy root hair density by visual inspection (Fig. 5B).
Reproducibility of B. distachyon measurements in EcoFAB 2.0 experiments
Several measurements were made during the experiment as described above, including plant shoot and root biomass measurements, tiller and shoot counts, maximum root length, biomass elemental compositions, growth medium NH4+ and NO3− levels after 3 and 5 weeks of growth, pH and evapotranspiration, and untargeted LC-MS/MS feature comparisons. To assess reproducibility within this experiment, the coefficient of variation (CV; %) was calculated for each of these measurements under each experimental treatment and is detailed in (table S9). Briefly, for 7 of 12 of the biomass measurements, the CVs were below 10%, all 12 were below 20%, and all N and C biomass composition measurements were below 10%. Specific phenotypes varied more and depended on the treatments. Leaf numbers were the most reproducible with all but the +NH4+ treatment under 15%. Tiller numbers were the least reproducible with all CVs over 15% and the +NH4+ treatment at ~49%. Maximum root length variation ranged from 6 to 28% with an average of ~17%. The flow injection analysis (FIA) measurements used to determine NH4+ and NO3− concentrations after 3 and 5 weeks of growth had CV percentages almost exclusively below 20% except for the week 5 “−NO3−” treatment with a CV of ~31%. All pH measurements, i.e., six conditions and 5 weeks, had CVs below 10%, and while evapotranspiration measurements varied more overall, none exceeded 30%.
As mentioned, after filtering on the basis of intensity and statistical relevance, 2065 LC-MS features were used to compare B. distachyon exudate profiles across different nutrient growth media. Between 596 and 1159 of these features were detected under each experimental condition with an average peak height intensity greater than 5 × 105 (table S9). Except for the “−NH4+” condition, where one of the five replicates was removed, 22.6 to 31.2% of those features had a CV % of peak height below 30%.
DISCUSSION
EcoFAB 2.0 is suitable for studying nitrogen nutrition impact on plant traits
EcoFAB 2.0 experiments provide a plant habitat that is amenable to a wide variety of plant measurements while maintaining realistic growth conditions. In the following section, we describe the reproducible plant phenotypes obtained through the EcoFAB 2.0 experiments. We observed generally slower plant development manifested by the lack of flowering (Fig. 1C); this was due to the relatively short photoperiod of 12 hours, which previously prohibited flowering in 35-day-old Brachypodium Bd21-3 (33, 34). Keeping plants at the vegetative stage is advantageous because of EcoFAB’s compact size (Fig. 1A). Furthermore, the developmental stage is essential when comparing exudate data between studies; for example, the exudate profiles of another grass plant A. barbata showed a shift with plant development (5).
The plants supplied exclusively with NH4+ developed typical symptoms of ammonium toxicity characterized by stunted growth (Fig. 1C) and the onset of leaf chlorosis and necrosis. These ammonium toxicity symptoms have previously been observed for ammonium-grown Brachypodium (18, 35). Ammonium toxicity is caused by ionic imbalances, disturbance of pH gradients across cell membranes, or oxidative stress (36). Our observed prohibition of NH4+ toxicity by cosupply of NO3− in the +NH4NO3 treatment (Fig. 1, C and D) can be explained by NO3− ability to induce root signals that determine NH4+ tolerance, alongside alkalinization of rhizosphere during NO3− uptake, which we also have observed (Fig. 2B) (37). In contrast, nitrate did not cause toxicity (Fig. 1), as excess is stored in plant vacuoles and used for biosynthesis when needed (38). The NO3− fed plants developed bigger root biomass (Fig. 1D) and longer roots (fig. S2C), perhaps via NO3−-driven regulation of auxin levels in roots (19, 20). However, more research must be completed to determine the causality between nitrate uptake, root development, and hormone homeostasis in Brachypodium.
Our iN− plants showed only minor chlorosis (Fig. 1C) combined with root foraging (Fig. 1D and fig. S2C), suggesting mild N deficiency (16). For our plants, the shoot N content decreased in the N-free medium to <3% (Fig. 1E). According to data from closely related wheat, values <3.4% at the late tillering stage constitute N deficiency (39). On the basis of the reported critical N values in wheat and a general increase in root:shoot biomass under iN− conditions (Fig. 1D), our Brachypodium was N-deficient. However, other studies observed lower N contents of ~1% for chronically undernourished (0.1 mM NO3−) 35-day-old Brachypodium (17). A previous study measured Brachypodium shoot N contents at the tillering stage at ~5% when supplied with sufficient NO3− and ~6% for NH4+ or NH4NO3 (18). These values align with our results for N-supplied plants (Fig. 1E).
Nitrogen form determines rhizosphere pH that can influence root exudation
Factors influencing root exudation in response to nitrogen treatments include the interplay between nitrogen forms and resulting rhizosphere pH changes, which contribute to the variations observed in exudate profiles and have implications for understanding plant-microbe interactions. Consistent with previous findings (40), our study confirms the higher uptake of NH4 relative to NO3 for Brachypodium (Fig. 2A). The iN form drove bidirectional pH change in our spent medium (Fig. 2B). Plant uptake of ammonium decreases pH in the rhizosphere, while nitrate causes an increase in pH due to their root-mediated efflux of H+ or OH−, respectively (41). We could not separate the pH and iN form effects in our experiments. However, this iN form–associated pH change is a natural process. Plants can alter the pH in a sharp gradient from the root surface (2 to 3 mm), and the size of this layer increases with lower soil buffering capacity. This interface is a hotspot for exudation and plant-microbe interactions (42). Our observations are, therefore, relevant from the perspective of natural rhizosphere processes.
The acidification during NH4+ and NH4NO3 treatments (Fig. 2B) might cause a high number of unique exudate features (Fig. 3C) in comparison to the alkalinized rhizosphere under NO3− supply (Fig. 2B). It is well established that proton gradients across the membranes facilitate the transport of compounds into and from the rhizosphere (43). For example, the exudation of terpenoid 3-epi-brachialactone in grass Brachiaria humidicola was promoted during low rhizosphere pH and NH4+ nutrition due to changes in proton motive force (44).
Nitrogen sufficiency and deficiency in grasses produce conserved patterns of root exudation
Metabolomic analyses revealed marked changes in exudate composition in response to nitrogen supply (Figs. 3 and 4 and fig. S5), with many of the detected annotations (fig. S4 and table S2) aligning with organic, aromatic, or amino acids previously reported in root exudates of grasses (5, 32, 45, 46). This alignment with existing literature is crucial for validating our observed root exudation patterns in EcoFAB 2.0, enhancing our understanding of the associated metabolic processes linked to plant nutrition.
Our data showed that B. distachyon supplied with iN+ increased abundance of N-containing compounds, including alkaloids and amino acid annotations; this was in contrast to iN− treatments that decreased the abundance of these compounds (Fig. 4, A and B). Similar findings were observed in soil-grown switchgrass that showed an increased amino acid exudation pattern under +N and +NP fertilization relative to unfertilized controls (22). In addition, axenic hydroponically grown maize under N deficiency lowered the amino acid amounts in root exudates (24). These consistent results underscore the significance of nitrogen in shaping amino acid exudation patterns across various grass species.
Levels of other metabolites can be indicative of stress response related to plant nutritional status. Here, we found that B. distachyon produced dopamine in the root exudates only in the +NH4NO3 treatment (Fig. 4C). The number of studies showing dopamine production in plant tissues is limited (47). Dopamine has a range of physiological and biochemical functions in plants, including tolerance against abiotic stresses, such as drought, salt, and nutrient stress (47). A recent study showed that exogenous dopamine could mediate nitrogen uptake and metabolism regulation at low ammonium levels in a tree Malus hupehensis (48). Our study contributes valuable insights into the limited body of literature about dopamine exudation in grasses. However, the production of another neurotransmitter, serotonin, was documented to be also N dependent in the rhizosphere of switchgrass (22). Combined, this indicates that the root exudation of neurotransmitters in grasses can depend on optimal iN supply.
N deficiency generally reduced the abundance of many N-containing compounds and increased the abundance of N-free shikimates-phenylpropanoids (Fig. 4 and fig. S5). The shikimate-phenylpropanoid pathway comprises shikimic acid precursors (e.g., quinic acid) and derivatives, aromatic acids (e.g., cinnamic acid derivatives), coumarins, hydroxybenzoic acids, flavonoids, lignans, or lignins (29, 49). For instance, recent research showed an increase in several organic and aromatic acids in the rhizosphere of switchgrass grown in a greenhouse under marginal N-limited soil (22), and another study showed that the root exudation of p-coumaric acid in hydroponically grown rice increased by 1.95-fold under N-deficient conditions compared to N-sufficient conditions (23). Therefore, our results from B. distachyon combined with the previous result suggest conserved patterns of increased production of compounds from the shikimate-phenylpropanoid pathway in root exudates of grasses under N deficiency (22, 23).
Shikimic and p-coumaric acids have diverse functions in the rhizosphere
We observed in vitro growth-modulating effects of shikimic acid on a number of soil bacteria (Fig. 5A). Other study showed that rhizosphere bacteria prefer to consume shikimic acid alongside other organic acids (nicotinic, salicylic, cinnamic, and indole-3-acetic) exuded by roots of grass A. barbata (5). Moreover, shikimic acid was shown to be a highly consumed substrate by diverse phylogeny of soil isolates in the recently developed Northen Lab Defined Medium (50). The shikimic acid promoted the growth of the roots only under iN+ conditions relative to a control (Fig. 5B). The root growth–promoting effects of 0.5 mM shikimic acid in the presence of 0.5 mM NH4NO3 were also shown in 10-day-old Arabidopsis seedlings (8). Synthesis of previous observations and our exometabolomics, microbial, and root growth data (Figs. 3 to 5) suggest that exogenous shikimic acid can promote root growth, serve as substrates to support the growth of selected soil bacteria and thus shape microbiome assembly in the rhizosphere (5, 8, 22, 50).
We found negative effects of increasing concentration of p-coumaric acid on a half of studied soil bacteria (Fig. 5A). This is consistent with a recent study that showed that coumaric acid is generally not a preferred microbial substrate by soil bacterial isolates in vitro (51). Our data suggest that concentrations of 0.5 mM and higher can have phytotoxic activity on Brachypodium seedlings and may negatively affect crop productivity (Fig. 5B). In particular, p-coumaric acid strongly inhibited root growth exclusively under iN limitation (Fig. 5B). It was shown that N-limiting conditions induce root foraging via the regulation of auxin transport (16, 19, 20). Therefore, we propose that p-coumaric acid in Brachypodium might alter root hormone homeostasis, likely via stimulation of indole-3-acetic acid oxidase activity shown for other aromatic compounds (7, 52). In addition, the inhibition of shoot growth was independent of N supply (Fig. 5B). The shoot reduction might be related to photosynthesis alterations via peroxidase-mediated chlorophyll degradation shown for p-coumaric acid previously (53).
Limitations and future work
The current experimental design compromises sufficient growth conditions and exudate signals. In our setup (EcoFAB 2.0), some compounds might not accumulate in detectable levels in our exudate samples due to the need for weekly medium changes to maintain adequate N supply and acceptable pH (Fig. 2). Growing B. distachyon in a larger medium volume than 10 ml to avoid pH shifts would dilute the exudates concentration. Moreover, metabolite extraction from the greater volume of spent medium would result in higher ion suppression due to high salts and MES buffer concentrations. While EcoFAB 2.0 can have an opaque base to shield the roots, this study used a transparent base; thus, the light exposure could have affected the exudate profiles due to stress.
The compact EcoFAB size limits the number of plant species to small plants. Notably, recent research demonstrated that the previous EcoFAB 1.0 design did not impose space constraints on Brachypodium biomass and root microbiome structure within a 14-day growth period compared to traditional pots (54). Grasses larger than B. distachyon, such as wheat, rice, maize, sorghum, or switchgrass, can be grown only during the early seedling developmental stages to avoid crowding inside the EcoFAB 2.0 device. The future research will aim at developing an alternative EcoFAB design to accommodate larger plant species and investigate ecological relevance of EcoFAB-derived results for field-grown plants.
As mentioned above, the interconnected effects of the N source and rhizosphere pH (Fig. 2) have combined effects on the exudate composition (Fig. 3). From this perspective, it would be interesting to assess the sole effect of pH gradient on exudate composition in plants grown with one nitrogen source (i.e., NH4NO3). A controlled study establishing causality between pH and root exudate composition still needs to be conducted in grasses. Furthermore, future work elucidating dopamine pathways and exudation in grasses would benefit our understanding of grassland function under N limitation and stress.
The typical GNPS annotation rate is 5 to 6% (55). Thus, expectedly, the top fold-increasing iN− features were unannotated (Fig. 3C). To supplement these unknown features, we subsequently used the GNPS analog search tool as well as SIRIUS and CANOPUS to infer molecular formulas and putative compounds classifications for these datasets (28, 56, 57). However, expansion of annotated reference libraries is needed to gain further insight into these “dark matter” molecules (58).
We conclude that the use of EcoFAB 2.0 devices result in reproducible plant growth, which enabled us to determine that the iN form (NO3−, NH4+, or NH4NO3) and starvation are significant factors modulating root exudation in the grass species B. distachyon. Relative to the +NH4+ and +NO3− treatments, the +NH4NO3 supply produces large shoots, robust root exudate profiles (signal intensity and feature diversity) that include dopamine, balanced rhizosphere pH changes, and preferential uptake of NH4+ relative to NO3−. Transient iN-limiting conditions for 2 weeks remodulate root exudation relative to the previous iN forms. The root exudate profiles of plants supplied with NO3−, NH4+, or NH4NO3 differed from each other and were collectively more diverse than the exudates of the iN-starved plants. The N-deficient plants increased the abundance of compounds in the shikimate-phenylpropanoid pathway, e.g., p-coumaric acid, while decreasing the abundance of N-containing compounds. We demonstrate on the example of shikimic and p-coumaric acids that the exometabolites increasing in the rhizosphere of N-stressed plants can serve diverse functions from microbial substrates to allelochemicals. Our work increases the understanding of nitrogen nutrition and root exudation in the model plant B. distachyon, which can be useful for studies of economically important grass crops. Furthermore, the EcoFAB 2.0 device proved to be a valuable standardized tool in plant research and has the potential to enable scientists to work on shared systems and facilitate replicability in environmental microbiome research.
MATERIALS AND METHODS
Plant growth and harvest
Seeds of B. distachyon Bd21-3 with removed lemma were surface-sterilized with 70% (v/v) ethanol for 30 s, followed by 5 min in 6% (w/v) sodium hypochlorite and washed five times with Milli-Q water. Seeds were then stratified on plates composed of 1.5% (w/v) Phytoagar and incubated at 4°C for 3 days in the dark. Germination was done by placing plates vertically in a growth chamber (CU36L4, Percival Scientific Inc., Perry, IA, USA) set to 22°C, photosynthetic photon flux density at 150 μmol m−2 s−1 and a 12-hour photoperiod. We transferred 3-day-old seedlings aseptically into fabricated ecosystem (EcoFAB 2.0) devices (Fig. 1A). EcoFAB 2.0 devices were assembled and sterilized following the instructions described in the in the assembly protocol available online at Protocols.io (dx.doi.org/10.17504/protocols.io.q26g7p693gwz/v1).
We supplied each EcoFABs with 10 ml of plant growth medium modified from Glazovska et al. (18). The basal salt medium (BSM) consisted of 0.06 μM MoNa2O4, 0.3 μM CuSO4, 0.8 μM ZnSO4, 9 μM MnCl2, 50 μM H3BO4, 50 uM Fe(III)-EDTA-Na, 0.5 mM KH2PO4, 1 mM MgSO4, and 5 mM MES buffer at pH 6. From the BSM, we created four types of media with varying iN composition: (i) The NO3− medium contained 2 mM Ca(NO3)2 and 1 mM KNO3; (ii) the NH4+ medium contained 2.5 mM (NH4)2SO4; (iii) the NH4+/NO3− medium contained 2.5 mM NH4NO3; and (iv) iN− medium consisted of BSM without additional ingredients. All iN+ media contained iN at 5 mM. All media were filtered via 0.22-μm polyethersulfone (PES) membrane and stored at 4°C.
The iN− treatment, iN+ plant control, and medium control received their respective iN media (Fig. 1B). The EcoFABs 2.0 were placed into the growth chamber, continuing the settings during germination. We used five biological replicates for each of the 10 treatments groups (3× iN+ plant treatments: +NO3−, +NH4+, and +NH4NO3; 3× iN− plant treatments: −NO3−, −NH4+, and −NH4NO3; and 4× technical control treatments with: NO3−, NH4+, NH4NO3, and N-free medium), resulting in a total of 50 EcoFABs 2.0 (Fig. 1B). The plant growth medium was entirely changed weekly in a sterile environment of a biosafety cabinet by opening EcoFAB 2.0 sampling ports and using 10-ml sterile serological pipettes. Treatment group plants received iN+ medium for 3 weeks before being transferred to iN− medium for 2 weeks. We checked the sterility of all EcoFABs 2.0 by incubating a drop of spent medium from weeks 3 and 5 on Luria-Bertani agar plates for 3 days at 30°C.
Plant harvest occurred at week 5 by dismantling EcoFAB 2.0 and gently pulling plants out of the root chamber opening. Autoclaved wipers (Texwipe TX604 TechniCloth) removed residual medium from the roots, and the whole plants were scanned (HP Scanjet G4050). The determination of Brachypodium developmental stages, tiller number, and leaf number followed the work by Hong et al. (33). The roots and shoots were separated, and fresh biomass was immediately weighed. Roots were flash-frozen in liquid N2, while shoots were frozen by placing them at −80°C. Frozen tissues were stored at −80°C. Lyophilization (Labconoco FreeZone 2.5) removed water from the frozen shoots before homogenization using a beat mill (MM 400, Retsch GmbH, Haan, Germany) set to 30 Hz for 10 min. The plant scans were used to quantify maximum root length using RootNav 1.8.1 software (59).
Spent medium sampling and analyses of pH and iN
We sampled the spent medium from EcoFAB 2.0 weekly and quantified evapotranspiration as a medium volume loss. We adjusted the medium samples to the initial 10 ml with Milli-Q water and then sterile-filtered using Acrodisc Syringe Filters 25 mm with 0.2-μm Supor PES membrane (Pall Corporation, NY, USA) attached to a 10-ml plastic syringe. We split each medium sample into three aliquots: 1 ml for pH measurement, 1 ml for FIA, and 8 ml for LC-MS. The pH was measured weekly using HALO Wireless pH Meter (HI10832, Hanna Instruments). The FIA was done on the samples from weeks 3 and 5, while LC-MS/MS was done on samples from week 5. One of the plant control +NH4+ medium samples (EcoFAB #22) at week 5 was lost during sampling; thus, it was excluded from the downstream analyses resulting in n = 4 for this treatment.
The analytical facility at UC Davis conducted the FIA analysis. The method involved the quantification of ammonium (NH4-N), nitrate (NO3-N), and nitrite (NO2-N) in aqueous samples. The spent medium sample was diluted 10× from 1 ml to 10 ml with Milli-Q water and used for the FIA (Lachat Quikchem 8500 Series 2). The detection limit for the 10× dilution was 0.5 mg liter−1 for all analytes. The reported nitrate values include nitrite because nitrite is typically an insignificant fraction of the N in a sample. We calculated plant iN uptake (Nuptake) (in micromoles) (Fig. 2A) from 10 ml of the growth medium (V) using the measured initial iN concentration (in milligrams per liter) in the fresh medium (ci) and the final concentration in spent medium (cf) and molar mass of nitrogen (MN = 14.0067 g mol−1) according to Eq. 1
| (1) |
Analysis of shoot elemental and stable isotope compositions
We conducted shoot C and N elemental (% dry weight) and isotopic composition (δ13C and δ15N values, respectively) analysis at the Center for Stable Isotope Biogeochemistry, University of California, Berkeley, CA, USA. The analytical system consisted of a CHNOS elemental analyzer (vario ISOTOPE cube, Elementar, Hanau, Germany) coupled to a continuous flow stable isotope ratio MS (IsoPrime 100 mass spectrometer, Isoprime Ltd., Cheadle, UK). The dried powdered shoots and iN sources from media were packed into tin foils and analyzed. We report the stable isotope abundances as delta (δ) notation in parts per thousand (‰) according to Eq. 2
| (2) |
where Rsample and Rstandard are the ratios of the heavy to light isotope (i.e., 15N/14N or 13C/12C) in the sample versus an international standard, respectively. The international standards are atmospheric nitrogen (air) for δ15N values and Vienna PeeDee Belemnite for δ13C values. Raw stable isotope data were corrected for drift and linearity and normalized to the international stable isotope reference scale. The normalization used three laboratory reference materials with different carbon and nitrogen delta values. These laboratory reference materials are calibrated annually against IAEA (International Atomic Energy Agency, Vienna, Austria)–certified reference materials. We used National Institute of Standards and Technology (Gaithersburg, MD, USA) SMR 1577c (bovine liver) previously calibrated against IAEA-certified reference materials for quality control. The long-term external precision for C and N isotope analyses was ±0.1 and ±0.2‰, respectively.
LC-MS analysis of Brachypodium exudates
The root exudate samples from week 5 (8 ml) were frozen at −80°C and then freeze-dried (Labconco Freeze-Zone). The dried material was resuspended by rinsing tubes in 700, 500, and then 300 μl of LC-MS grade methanol (Sigma-Aldrich); rinse volumes were combined in 2-ml tubes and dried in a Thermo Speed-Vac concentrator. We resuspended the dried material in 150 μl of LC-MS grade methanol containing internal standards (table S1). The solution was vortexed 2 × 10 s, bath-sonicated in ice water for 15 min, and centrifuged (10,000g for 5 min at 10°C) to pellet insoluble material, and then supernatants were filtered using 0.22-μm polyvinylidene difluoride microcentrifuge filtration devices (Pall) (10,000g for 5 min at 10°C); filtrates were transferred to 384-well plates and sealed with foil before analysis. Metabolites were separated using hydrophilic interaction chromatography for polar metabolomics; eluted compounds were detected using a Thermo Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer. LC-MS/MS parameters are in table S1.
Untargeted metabolomics and molecular networking
The GNPS and MZmine2 version 2.39 workflow generated molecular networking files for positive (12,109 features) (MZmine2 parameters in file S1) and negative (4770 features) (MZmine2 parameters in file S2) polarities (28). First, a baseline filter accepted features with an retention time (RT) of >0.6 min (after solvent front) and a maximum peak height of >1 × 106 and >10× the peak height maximum in extraction (n = 10) and technical controls (n = 5). We added +1 to the numerator and denominator in the fold change calculations to avoid errors when dividing by 0. In addition, we applied a statistical filter (ANOVA) accepting only features significantly higher (P ≤ 0.01) in plant samples than extraction and technical controls. We selected the top iN− features based on the statistically significant and >1000 fold increase in iN− (n = 14) versus iN+ (n = 15) treatments in exudates at week 5 (P ≤ 0.05, t test). We accepted top GNPS annotations with cosine scores of MS/MS spectral mirror match (MQScore) to library reference of >0.7 (155 features) (table S3). NPClassifier determined metabolite classifications based on annotated features’ biosynthetic pathways (29). For each feature, we calculated the fold change of average peak height in iN− treatments versus iN+ in exudates at week 5. We determined in which treatment groups (six treatment groups = UpSet or two treatment groups = Venn) for the filtered features were present on the basis of their maximum peak height in the group exceeding a threshold of 1 × 106 (table S2). For filtered features, we compared the peak height between plant treatments for statistical differences [ANOVA with Tukey’s post hoc test, P ≤ 0.01 using the “pairwise_tukeyhsd” function from the statsmodels (v. 0.11.1) Python package] (table S5). For annotated features that showed differential abundances between iN treatments (based on UpSet grouping and statistical differences), we verified the feature’s top annotation against library reference in the metabolite atlas by matching RT (maximum of Δ0.6 min), adduct, m/z integer, and MS/MS spectra of the reference (table S6) (60).
Features detected and putative identifications are limited to metabolites that are soluble in the extraction solvent, retain on the chromatography used, and ionize under the conditions set in the mass spectrometer. In addition, the availability of standards in the spectral database limits our ability to putatively identify metabolites based on MS/MS matching to known reference standards. Identifications using GNPS are considered level 2 per the Metabolomics Standards Initiative (61). Most stereoisomers and some positional isomers are not resolvable; thus, the sample may contain a different or a mixture of isomers similar to the GNPS match. There may be other metabolites in the samples that we cannot extract, measure, or identify on the methods used.
Using Cytoscape software (62), we conducted feature-based molecular networking by merging the filtered features from positive and negative polarities into a single molecular network (2065 nodes) and following a step-by-step protocol (55). The Cytoscape network merging in our figures did not create edges between features across polarities but provided a concise data representation. Alternative polarity merging in GNPS (using a delta RT of 0.1 min, mass tolerance of 30 parts per million, and delta m/z of 2H) created edges between polarities and allowed assessment of potentially identical structures (table S4) (28).
To shed light on the top iN− features that did not receive GNPS annotation, we used two in silico tools. The first one was CANOPUS, which is a deep neural network–based tool integrated in SIRIUS software and designed to predict compound classes and formulas from fragmentation spectra (56, 57). The second tool, GNPS analog search, is based on modified cosine correlations against a GNPS reference library and considers fragment ions in the alignment that differ by the same mass delta as the parent ions (28, 55). In this study, the analog search setting was a 200 dalton maximum mass difference and a minimum analog cosine score (Analog_MQScore) of 0.7.
Growth of bacteria on selected exudate compounds
We selected representative compounds from shikimate (shikimic acid) and phenylpropanoid (p-coumaric acid) pathways alongside two common root exudate compounds (glucose and oxalic acid) to test their impact on the growth of 14 different root-colonizing soil bacteria: Pseudomonas putida KT2440 (ATCC 47054), P. putida A.3.12 (ATCC 12633), P. simiae WCS417 (obtained from Benjamin Cole, Joint Genome Institute), Bacillus subtilis 168 (ATCC 27370), Bosea sp. OAE506 (DSM113628), Methylobacterium sp. OAE515 (DSM113602), Mucilaginibacter sp. OAE612 (DSM113562), Paenibacillus sp. OAE614 (DSM113526), Chitinophaga sp. OAE865 (DSM113563), Arthrobacter sp. OAP107 (DSM113524), Brevibacillus sp. OAP136 (DSM113625), Variovorax sp. OAS795 (DSM113622), Rhodococcus sp. OAS809 (DSM113518), and Burkholderia sp. OAS925 (DSM113627) (63–67). We resuspended each bacterium to an OD600 (1 cm in pathlength) of 0.01 in 0.1× R2A (Himedia M1687) liquid medium supplemented with a phosphate buffer [Na2HPO4 (6 g liter−1) and KH2PO4 (3 g liter−1)] at pH 7 and containing either oxalic acid, glucose, shikimic acid, or p-coumaric acid at 0.5, 5, or 10 mM (n = 6). The pH of the growth medium after the compound addition was 7. The p-coumaric acid treatments also contained 5% dimethyl sulfoxide (DMSO), which was used as a solvent. Our controls were blank medium with and without 5% DMSO and bacteria-inoculated medium with and without 5% DMSO (n = 6). The bacteria were grown in 0.2-ml aliquots in a 96-well microtiter plate covered with Breathe-easy sealing membrane (Sigma-Aldrich, Z380059) incubated at 28°C without shaking. The bacterial growth was measured through periodic measurements in OD600 at 0, 12, 24, 48, 72, and 92 hours using a Synergy H1 (BioTech Instruments) reader. For subsequent data analysis, we calculated net absorbance difference between the maximum OD600 and the initial (t0) reading. Then, we determined the change in the net OD600 readings for each bacterium-inoculated metabolite treatment relative to inoculated metabolite–free control. The phylogenetic tree was constructed using partial bacterial 16S ribosomal RNA gene sequences as described previously (50).
Growth of B. distachyon seedlings on selected exudate compounds
Dehusked and surface-sterilized seeds of B. distachyon BD21-3 were placed on 10-cm2 plates containing N-free ½ MS salts in 0.75% (w/v) Phytoagar containing no metabolite (control) or 0.5 mM oxalic acid, glucose, shikimic acid, or p-coumaric acid and either 0 or 2.5 mM NH4NO3. The p-coumaric acid treatments also contained 0.1% ethanol as a solvent. Each treatment was done in four replicate plates, each containing 12 seeds; this resulted in a total of 48 seedlings per treatment. The plates with seeds were stratified for 3 days at 4°C in the dark and then germinated vertically for 3 days in the growth chamber using the previous Brachypodium growth settings listed in the previous section. The root and shoot length for each plant were analyzed using ImageJ 1.53k software by setting the scale of each image to 1 cm and using the segmented line function to trace the center of the tissue from the embryo pole to the tip of the primary root and from the embryo pole to the tip of the first leaf, respectively.
Statistical analyses and data visualization
We conducted feature filtering and t tests between iN− versus iN+ treatments in Microsoft Excel version 16.66.1. Data visualization and statistics were done in Prism 9 (GraphPad Software LLC) version 9.3.0 unless indicated otherwise. We checked the data for normality and followed ANOVA with Tukey’s post hoc test when performing ANOVA analyses. Rstudio generated UpSet and nonmetric multidimensional scaling plots using the UpSetR and vegan packages, respectively (68, 69). The heatmap was generated using the Seaborn package (0.10.1) in Python (3.8.3). The statistical analysis of peak height data across plant treatments was also done in Python using ANOVA with Tukey’s post hoc test using the pairwise_tukeyhsd function within Statsmodels (0.11.1) (table S5). The Sankey plot was generated with the online tool SankeyMATIC (sankeymatic.com). The statistically significant difference was assumed at P ≤ 0.05 except for ANOVA of peak height data, which was performed at a threshold of P ≤ 0.01 following false discovery rate correction (70). To compare the variation of quantitative traits measured in this study, we used the CV that was calculated as the SD of replicate samples divided by their mean and then converted to percentage (71).
Acknowledgments
The Analytical Laboratory at UC Davis conducted the FIA analysis. We thank T. Dawson and S. Mambelli from the Center for Stable Isotope Biogeochemistry at UC Berkeley for elemental and stable isotope analysis and S. Glazovska from the Department of Plant and Environmental Sciences at the University of Copenhagen for providing excellent advice for hydroponic medium formulation.
Funding: We gratefully acknowledge funding from the U.S. Department of Energy (DOE) Office of Science, Office of Biological and Environmental Research. The research described was funded under contract DE-AC02-05CH11231 to Lawrence Berkeley National Laboratory as part of a project led by UC San Diego (DE-SC0021234). The EcoFAB 2.0 was developed as part of the project Trial Ecosystem Advancement for Microbiome Science (TEAMS) under contract DE-AC02-05CH11231 to Lawrence Berkeley National Laboratory. Data analysis used resources of the National Energy Research Scientific Computing Center, a Department of Energy Office of Science User Facility operated under contract number DE-AC02-05CH11231.
Author contributions: V.N. designed the research with input from P.F.A., T.R.N., K.Z., B.P.B., and S.M.K. P.F.A. developed the EcoFAB 2.0 device and assembly protocol. V.N., P.F.A., Y.D., and A.N.G. performed the research. V.N., P.F.A., B.P.B., T.V.H., M.C.M.v.W., T.R.N., and S.M.K. analyzed, collected, and interpreted data. V.N. wrote the first draft of the manuscript with input from P.F.A., C.T., and T.R.N. K.S.H., T.V.H., and M.C.M.v.W. contributed with writing of the revised manuscript. All authors edited the paper.
Competing interests: P.F.A. and T.R.N. are inventors on patent US11510376B2 held by University of California that covers Ecosystem device for determining plant-microbe interactions. T.R.N. has an equity interest in Brightseed Bio, which uses artificial intelligence to find bioactive compounds for human health. All other authors declare that they have no competing interests.
Data and materials availability: GNPS-negative and -positive modes for the polar metabolite analysis (HILIC) are available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=7e8210b68fd049198579c2eff3068876 and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=15c14ac5a13c466db52068de85bc8894. The interactive feature–based molecular network and node metadata can be accessed via Network Data Exchange (NDEx) at https://doi.org/10.18119/N9FW3X under the UUID: 0e3c4384-35fa-11ed-ac45-0ac135e8bacf. Phenotypes of plants grown under varying iN supplies are available at https://doi.org/10.6084/m9.figshare.21376017. Phenotypes of seedlings grown on selected root exudate compounds are available at https://doi.org/10.6084/m9.figshare.21433065. All raw data used in this paper are available at https://doi.org/10.6084/m9.figshare.21440748. The MS data in mzML format are available as a MassIVE dataset at https://massive.ucsd.edu/ under the ID number MSV000092053. The authors deposited an earlier version of this manuscript in bioRxiv: https://doi.org/10.1101/2023.01.18.524647. The EcoFAB 2.0 assembly protocol is available at dx.doi.org/10.17504/protocols.io.q26g7p693gwz/v1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S9
Legends for files S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S9
Files S1 and S2
REFERENCES AND NOTES
- 1.Lynch J. M., Whipps J. M., Substrate flow in the rhizosphere. Plant and Soil 129, 1–10 (1990). [Google Scholar]
- 2.Jones D. L., Nguyen C., Finlay R. D., Carbon flow in the rhizosphere: Carbon trading at the soil–root interface. Plant and Soil 321, 5–33 (2009). [Google Scholar]
- 3.Pausch J., Kuzyakov Y., Carbon input by roots into the soil: Quantification of rhizodeposition from root to ecosystem scale. Glob. Chang. Biol. 24, 1–12 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Badri D. V., Vivanco J. M., Regulation and function of root exudates. Plant Cell Environ. 32, 666–681 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Zhalnina K., Louie K. B., Hao Z., Mansoori N., da Rocha U. N., Shi S., Cho H., Karaoz U., Loqué D., Bowen B. P., Firestone M. K., Northen T. R., Brodie E. L., Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Li Z.-H., Wang Q., Ruan X., Pan C.-D., Jiang D.-A., Phenolics and plant allelopathy. Molecules 15, 8933–8952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng F., Cheng Z., Research progress on the use of plant allelopathy in agriculture and the physiological and ecological mechanisms of allelopathy. Front. Plant Sci. 6, 1020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. Hu, P. F. Andeer, Q. Zheng, S. M. Kosina, K. J. Jardine, Y. Ding, L. Z. Han, Y. Gao, K. Zengler, B. P. Bowen, J. C. Mortimer, J. P. Vogel, T. R. Northen, Extensive plant use of exometabolites. bioRxiv 2022.07.29.496484 [Preprint] (30 July 2022). 10.1101/2022.07.29.496484. [DOI]
- 9.Coskun D., Britto D. T., Shi W., Kronzucker H. J., How plant root exudates shape the nitrogen cycle. Trends Plant Sci. 22, 661–673 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Kuzyakov Y., Blagodatskaya E., Microbial hotspots and hot moments in soil: Concept & review. Soil Biol. Biochem. 83, 184–199 (2015). [Google Scholar]
- 11.Daly A. B., Jilling A., Bowles T. M., Buchkowski R. W., Frey S. D., Kallenbach C. M., Keiluweit M., Mooshammer M., Schimel J. P., Grandy A. S., A holistic framework integrating plant-microbe-mineral regulation of soil bioavailable nitrogen. Biogeochemistry 154, 211–229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiluweit M., Bougoure J. J., Nico P. S., Pett-Ridge J., Weber P. K., Kleber M., Mineral protection of soil carbon counteracted by root exudates. Nat. Clim. Chang. 5, 588–595 (2015). [Google Scholar]
- 13.Jilling A., Keiluweit M., Contosta A. R., Frey S., Schimel J., Schnecker J., Smith R. G., Tiemann L., Grandy A. S., Minerals in the rhizosphere: Overlooked mediators of soil nitrogen availability to plants and microbes. Biogeochemistry 139, 103–122 (2018). [Google Scholar]
- 14.Weyens N., van der Lelie D., Taghavi S., Newman L., Vangronsveld J., Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 27, 591–598 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Liu Y.-X., Zhang N., Hu B., Jin T., Xu H., Qin Y., Yan P., Zhang X., Guo X., Hui J., Cao S., Wang X., Wang C., Wang H., Qu B., Fan G., Yuan L., Garrido-Oter R., Chu C., Bai Y., NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684 (2019). [DOI] [PubMed] [Google Scholar]
- 16.de Bang T. C., Husted S., Laursen K. H., Persson D. P., Schjoerring J. K., The molecular-physiological functions of mineral macronutrients and their consequences for deficiency symptoms in plants. New Phytol. 229, 2446–2469 (2021). [DOI] [PubMed] [Google Scholar]
- 17.David L. C., Girin T., Fleurisson E., Phommabouth E., Mahfoudhi A., Citerne S., Berquin P., Daniel-Vedele F., Krapp A., Ferrario-Méry S., Developmental and physiological responses of Brachypodium distachyon to fluctuating nitrogen availability. Sci. Rep. 9, 3824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Głazowska S., Baldwin L., Mravec J., Bukh C., Fangel J. U., Willats W. G., Schjoerring J. K., The source of inorganic nitrogen has distinct effects on cell wall composition in Brachypodium distachyon. J. Exp. Bot. 70, 6461–6473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacheco-Villalobos D., Díaz-Moreno S. M., van der Schuren A., Tamaki T., Kang Y. H., Gujas B., Novak O., Jaspert N., Li Z., Wolf S., Oecking C., Ljung K., Bulone V., Hardtke C. S., The effects of high steady state auxin levels on root cell elongation in brachypodium. Plant Cell 28, 1009–1024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ötvös K., Marconi M., Vega A., O’Brien J., Johnson A., Abualia R., Antonielli L., Montesinos J. C., Zhang Y., Tan S., Cuesta C., Artner C., Bouguyon E., Gojon A., Friml J., Gutiérrez R. A., Wabnik K., Benková E., Modulation of plant root growth by nitrogen source-defined regulation of polar auxin transport. EMBO J. 40, e106862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.V. Römheld, Diagnosis of deficiency and toxicity of nutrients, in Marschner’s Mineral Nutrition of Higher Plants (Elsevier, 2012), pp. 299–312. [Google Scholar]
- 22.N. R. Baker, K. Zhalnina, M. Yuan, D. Herman, J. Ceja-Navarro, J. Sasse, J. S. Jordan, B. P. Bowen, L. Wu, C. Fossum, A. Chew, Y. Fu, M. Saha, J. Zhou, J. Pett-Ridge, T. Northen, M. Firestone, Nutrient and moisture limitation reveal keystone metabolites that link switchgrass rhizosphere metabolome and microbiome dynamics. bioRxiv 2022.06.20.496911 [Preprint] (21 June 2022). 10.1101/2022.06.20.496911. [DOI]
- 23.Tawaraya K., Horie R., Wagatsuma T., Saito K., Oikawa A., Metabolite profiling of shoot extract, root extract, and root exudate of rice under nitrogen and phosphorus deficiency. Soil Sci. Plant Nutr. 64, 312–322 (2018). [Google Scholar]
- 24.Carvalhais L. C., Dennis P. G., Fedoseyenko D., Hajirezaei M.-R., Borriss R., von Wirén N., Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J. Plant. Nutr. Soil Sci. 174, 3–11 (2011). [Google Scholar]
- 25.International Brachypodium Initiative , Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Sasse J., Kant J., Cole B. J., Klein A. P., Arsova B., Schlaepfer P., Gao J., Lewald K., Zhalnina K., Kosina S., Bowen B. P., Treen D., Vogel J., Visel A., Watt M., Dangl J. L., Northen T. R., Multilab EcoFAB study shows highly reproducible physiology and depletion of soil metabolites by a model grass. New Phytol. 222, 1149–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Sasse J., Lewald K. M., Zhalnina K., Cornmesser L. T., Duncombe T. A., Yoshikuni Y., Vogel J. P., Firestone M. K., Northen T. R., Ecosystem fabrication (EcoFAB) protocols for the construction of laboratory ecosystems designed to study plant-microbe interactions. J. Vis. Exp. e57170, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., Nguyen D. D., Watrous J., Kapono C. A., Luzzatto-Knaan T., Porto C., Bouslimani A., Melnik A. V., Meehan M. J., Liu W.-T., Crüsemann M., Boudreau P. D., Esquenazi E., Sandoval-Calderón M., Kersten R. D., Bandeira N., Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H. W., Wang M., Leber C. A., Nothias L.-F., Reher R., Kang K. B., van der Hooft J. J. J., Dorrestein P. C., Gerwick W. H., Cottrell G. W., NPClassifier: A deep neural network-based structural classification tool for natural products. J. Nat. Prod. 84, 2795–2807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zengler K., Hofmockel K., Baliga N. S., Behie S. W., Bernstein H. C., Brown J. B., Dinneny J. R., Floge S. A., Forry S. P., Hess M., Jackson S. A., Jansson C., Lindemann S. R., Pett-Ridge J., Maranas C., Venturelli O. S., Wallenstein M. D., Shank E. A., Northen T. R., EcoFABs: Advancing microbiome science through standardized fabricated ecosystems. Nat. Methods 16, 567–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. Ranjan, S. Sahni, B. D. Prasad, R. R. Kumar, K. Rajani, V. K. Jha, V. Sharma, M. Kumar, V. Kumar, Sterilization Technique (Apple Academic Press, 2017). [Google Scholar]
- 32.Kawasaki A., Donn S., Ryan P. R., Mathesius U., Devilla R., Jones A., Watt M., Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLOS ONE 11, e0164533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong S. Y., Park J. H., Cho S. H., Yang M. S., Park C. M., Phenological growth stages of Brachypodium distachyon: Codification and description. Weed Res. 51, 612–620 (2011). [Google Scholar]
- 34.Ream T. S., Woods D. P., Schwartz C. J., Sanabria C. P., Mahoy J. A., Walters E. M., Kaeppler H. F., Amasino R. M., Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 164, 694–709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britto D. T., Kronzucker H. J., NH4+ toxicity in higher plants: A critical review. J. Plant Physiol. 159, 567–584 (2002). [Google Scholar]
- 36.Liu Y., von Wirén N., Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 68, 2581–2592 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Esteban R., Ariz I., Cruz C., Moran J. F., Review: Mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248, 92–101 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Tegeder M., Masclaux-Daubresse C., Source and sink mechanisms of nitrogen transport and use. New Phytol. 217, 35–53 (2018). [DOI] [PubMed] [Google Scholar]
- 39.D. Reuter, J. B. Robinson, Eds., Plant Analysis: An Interpretation Manual (CSIRO Publishing, 1997). [Google Scholar]
- 40.Kuang W., Sanow S., Kelm J. M., Müller Linow M., Andeer P., Kohlheyer D., Northen T., Vogel J. P., Watt M., Arsova B., N-dependent dynamics of root growth and nitrate and ammonium uptake are altered by the bacterium Herbaspirillum seropedicae in the cereal model Brachypodium distachyon. J. Exp. Bot. 73, 5306–5321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinsinger P., Plassard C., Tang C., Jaillard B., Origins of root-mediated pH changes in the rhizosphere and their responses to environmental constraints: A review. Plant and Soil 248, 43–59 (2003). [Google Scholar]
- 42.Kuzyakov Y., Razavi B. S., Rhizosphere size and shape: Temporal dynamics and spatial stationarity. Soil Biol. Biochem. 135, 343–360 (2019). [Google Scholar]
- 43.Sasse J., Martinoia E., Northen T., Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Egenolf K., Verma S., Schöne J., Klaiber I., Arango J., Cadisch G., Neumann G., Rasche F., Rhizosphere pH and cation-anion balance determine the exudation of nitrification inhibitor 3-epi-brachialactone suggesting release via secondary transport. Physiol. Plant. 172, 116–123 (2021). [DOI] [PubMed] [Google Scholar]
- 45.McLaughlin S., Zhalnina K., Kosina S., Northen T. R., Sasse J., The core metabolome and root exudation dynamics of three phylogenetically distinct plant species. Nat. Commun. 14, 1649 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasse J., Kosina S. M., de Raad M., Jordan J. S., Whiting K., Zhalnina K., Northen T. R., Root morphology and exudate availability are shaped by particle size and chemistry in Brachypodium distachyon. Plant Direct 4, e00207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q., Gao T., Liu W., Liu Y., Zhao Y., Liu Y., Li W., Ding K., Ma F., Li C., Functions of dopamine in plants: A review. Plant Signal. Behav. 15, 1827782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du P., Yin B., Zhou S., Li Z., Zhang X., Cao Y., Han R., Shi C., Liang B., Xu J., Melatonin and dopamine mediate the regulation of nitrogen uptake and metabolism at low ammonium levels in Malus hupehensis. Plant Physiol. Biochem. 171, 182–190 (2022). [DOI] [PubMed] [Google Scholar]
- 49.N. Francenia Santos-Sánchez, R. Salas-Coronado, B. Hernández-Carlos, C. Villanueva-Cañongo, Shikimic acid pathway in biosynthesis of phenolic compounds, in Plant Physiological Aspects of Phenolic Compounds (IntechOpen, 2019), pp. 1–15. [Google Scholar]
- 50.de Raad M., Li Y. V., Kuehl J. V., Andeer P. F., Kosina S. M., Hendrickson A., Saichek N. R., Golini A. N., Han L. Z., Wang Y., Bowen B. P., Deutschbauer A. M., Arkin A. P., Chakraborty R., Northen T. R., A defined medium for cultivation and exometabolite profiling of soil bacteria. Front. Microbiol. 13, 855331 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Wilhelm R. C., Swenson T. L., Silver A., Andeer P. F., Golini A., Kosina S. M., Bowen B. P., Buckley D. H., Northen T. R., Substrate utilization and competitive interactions among soil bacteria vary with life-history strategies. Front. Microbiol. 13, 914472 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salvador V. H., Lima R. B., dos Santos W. D., Soares A. R., Böhm P. A. F., Marchiosi R., Ferrarese M. d. L. L., Ferrarese-Filho O., Cinnamic acid increases lignin production and inhibits soybean root growth. PLOS ONE 8, e69105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamauchi N., Funamoto Y., Shigyo M., Peroxidase-mediated chlorophyll degradation in horticultural crops. Phytochem Rev. 3, 221–228 (2004). [Google Scholar]
- 54.Acharya S. M., Yee M. O., Diamond S., Andeer P. F., Baig N. F., Aladesanmi O. T., Northen T. R., Banfield J. F., Chakraborty R., Fine scale sampling reveals early differentiation of rhizosphere microbiome from bulk soil in young Brachypodium plant roots. ISME Commun. 3, 54 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aron A. T., Gentry E. C., McPhail K. L., Nothias L.-F., Nothias-Esposito M., Bouslimani A., Petras D., Gauglitz J. M., Sikora N., Vargas F., van der Hooft J. J. J., Ernst M., Kang K. B., Aceves C. M., Caraballo-Rodríguez A. M., Koester I., Weldon K. C., Bertrand S., Roullier C., Sun K., Dorrestein P. C., Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 15, 1954–1991 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Dührkop K., Fleischauer M., Ludwig M., Aksenov A. A., Melnik A. V., Meusel M., Dorrestein P. C., Rousu J., Böcker S., SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Dührkop K., Nothias L.-F., Fleischauer M., Reher R., Ludwig M., Hoffmann M. A., Petras D., Gerwick W. H., Rousu J., Dorrestein P. C., Böcker S., Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 39, 462–471 (2021). [DOI] [PubMed] [Google Scholar]
- 58.da Silva R. R., Dorrestein P. C., Quinn R. A., Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. U.S.A. 112, 12549–12550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pound M. P., French A. P., Atkinson J. A., Wells D. M., Bennett M. J., Pridmore T., RootNav: Navigating images of complex root architectures. Plant Physiol. 162, 1802–1814 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao Y., Sun T., Wang T., Ruebel O., Northen T., Bowen B. P., Analysis of metabolomics datasets with high-performance computing and metabolite atlases. Metabolites 5, 431–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., Fan T. W.-M., Fiehn O., Goodacre R., Griffin J. L., Hankemeier T., Hardy N., Harnly J., Higashi R., Kopka J., Lane A. N., Lindon J. C., Marriott P., Nicholls A. W., Reily M. D., Viant M. R., Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molina L., Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32, 315–321 (2000). [Google Scholar]
- 64.Cole B. J., Feltcher M. E., Waters R. J., Wetmore K. M., Mucyn T. S., Ryan E. M., Wang G., Ul-Hasan S., McDonald M., Yoshikuni Y., Malmstrom R. R., Deutschbauer A. M., Dangl J. L., Visel A., Genome-wide identification of bacterial plant colonization genes. PLOS Biol. 15, e2002860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coker J., Zhalnina K., Marotz C., Thiruppathy D., Tjuanta M., D’Elia G., Hailu R., Mahosky T., Rowan M., Northen T. R., Zengler K., A reproducible and tunable synthetic soil microbial community provides new insights into microbial ecology. mSystems 7, e0095122 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liffourrena A. S., Lucchesi G. I., Alginate-perlite encapsulated Pseudomonas putida A (ATCC 12633) cells: Preparation, characterization and potential use as plant inoculants. J. Biotechnol. 278, 28–33 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Gallegos-Monterrosa R., Mhatre E., Kovács Á. T., Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology 162, 1922–1932 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Conway J. R., Lex A., Gehlenborg N., UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., Szoecs E., Wagner. H., vegan: Community Ecology Package. R Package Version 2, 5–7 (2020). https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 70.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. Methodol. 57, 289–300 (1995). [Google Scholar]
- 71.Pélabon C., Hilde C. H., Einum S., Gamelon M., On the use of the coefficient of variation to quantify and compare trait variation. Evol. Lett. 4, 180–188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao J., Liu P., Hu Y., Liu T., Guo Y., Sun P., Zheng J., Ren Z., Wang Y., Antiviral activities of Artemisia vulgaris L. extract against herpes simplex virus. Chin. Med. 18, 21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goursot J.-F., A collaborative work on quantitative HPLC methods for the routine determination of atranol and chloroatranol in moss extracts. Flavour Fragr. J. 34, 28–35 (2019). [Google Scholar]
- 74.Fraga M. F., Uriol E., Diego L. B., Berdasco M., Esteller M., Cañal M. J., Rodríguez R., High-performance capillary electrophoretic method for the quantification of 5-methyl 2'-deoxycytidine in genomic DNA: Application to plant, animal and human cancer tissues. Electrophoresis 23, 1677–1681 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Carcache-Blanco E. J., Kang Y.-H., Park E. J., Su B.-N., Kardono L. B. S., Riswan S., Fong H. H. S., Pezzuto J. M., Kinghorn A. D., Constituents of the stem bark of Pongamia pinnata with the potential to induce quinone reductase. J. Nat. Prod. 66, 1197–1202 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Miyake Y., Hiramitsu M., Isolation and extraction of antimicrobial substances against oral bacteria from lemon peel. J. Food Sci. Technol. 48, 635–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Villarino M., Sandín-España P., Melgarejo P., De Cal A., High chlorogenic and neochlorogenic acid levels in immature peaches reduce Monilinia laxa infection by interfering with fungal melanin biosynthesis. J. Agric. Food Chem. 59, 3205–3213 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Niwa M., Liu G.-Q., Tatematsu H., Hirata Y., Chamaechromone, a novel rearranged biflavonoid from Stellera chamaejasme L. Tetrahedron Lett. 25, 3735–3738 (1984). [Google Scholar]
- 79.Topal M., Gocer H., Topal F., Kalin P., Köse L. P., Gülçin İ., Çakmak K. C., Küçük M., Durmaz L., Gören A. C., Alwasel S. H., Antioxidant, antiradical, and anticholinergic properties of cynarin purified from the Illyrian thistle (Onopordum illyricum L.). J. Enzyme Inhib. Med. Chem. 31, 266–275 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S9
Legends for files S1 and S2
References
Tables S1 to S9
Files S1 and S2