Abstract
Defects in adipocyte lipolysis drive multiple aspects of cardiometabolic disease, but the transcriptional framework controlling this process has not been established. To address this, we performed a targeted perturbation screen in primary human adipocytes. Our analyses identified 37 transcriptional regulators of lipid mobilization, which we classified as (i) transcription factors, (ii) histone chaperones, and (iii) mRNA processing proteins. On the basis of its strong relationship with multiple readouts of lipolysis in patient samples, we performed mechanistic studies on one hit, ZNF189, which encodes the zinc finger protein 189. Using mass spectrometry and chromatin profiling techniques, we show that ZNF189 interacts with the tripartite motif family member TRIM28 and represses the transcription of an adipocyte-specific isoform of phosphodiesterase 1B (PDE1B2). The regulation of lipid mobilization by ZNF189 requires PDE1B2, and the overexpression of PDE1B2 is sufficient to attenuate hormone-stimulated lipolysis. Thus, our work identifies the ZNF189-PDE1B2 axis as a determinant of human adipocyte lipolysis and highlights a link between chromatin architecture and lipid mobilization.
Perturbation screening of adipocytes uncovers ZNF189 as a transcriptional regulator of lipolysis through PDE1B2 repression.
INTRODUCTION
White adipose tissue (WAT) stores triglycerides by lipogenesis and mobilizes these lipids via lipolysis. The latter is a tightly controlled process involving stepwise hydrolysis reactions catalyzed by adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and monoglyceride lipase (1). While insulin is the major anti-lipolytic signal, there are multiple factors, e.g., catecholamines and natriuretic peptides, that induce lipolysis through distinct receptor-mediated proximal signaling steps, which converge on lipase activation (2). Blunted WAT responsiveness to these pro-lipolytic signals is a feature of insulin resistance and type 2 diabetes (3), highlighting the importance of retained ability to mobilize lipids.
Sophisticated transcriptomic mapping in brown adipocytes has recently underscored the complex cross-talk between lipolysis and gene transcription (4). While perturbed WAT lipase expression can drive metabolic disease, this only accounts for a small part of the individual variation in lipolytic responsiveness (5), emphasizing the existence of other factors controlling lipolysis. To identify these factors and the transcriptional mechanisms controlling their expression, we performed a targeted perturbation screen of lipid mobilization in primary human adipocytes. Our systematic approach identified multiple hits affecting lipolysis including the transcription factor zinc finger protein 189 (ZNF189), which we show increases hormone-stimulated lipolysis by repressing PDE1B expression at an adipocyte-specific promoter. Together, this work provides a resource for future studies of lipolytic regulators in fat cells.
RESULTS
Perturbation screening identifies distinct classes of lipolytic regulators
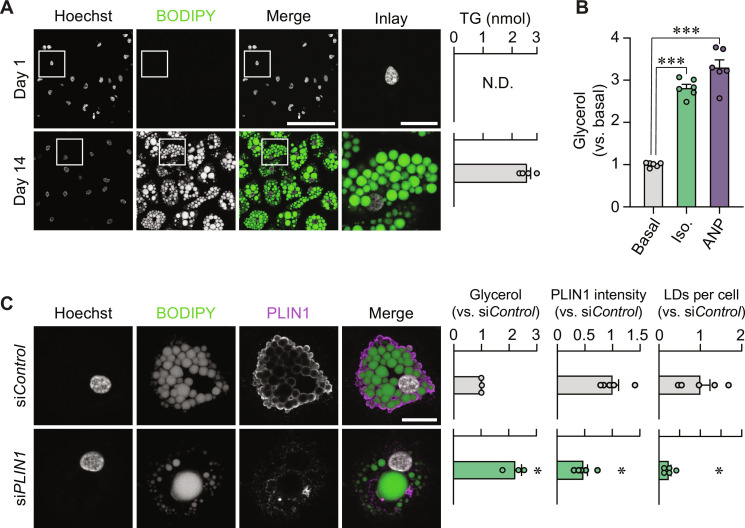
To identify transcriptional determinants of lipid mobilization, we performed a targeted perturbation screen in human white adipocytes. For this, we obtained stromal cells from subcutaneous abdominal liposuctions, and after careful optimizations, we generated progenitor-derived adipocytes that (i) mobilize lipids upon isoprenaline or atrial natriuretic peptide (ANP) incubations and (ii) can be specifically depleted of target proteins following transfections with small interfering RNAs (siRNAs) (Fig. 1, A to C, and fig. S1A). Perilipin 1 (PLIN1), a lipid droplet coating protein that regulates lipid turnover (6), was used as a positive control (Fig. 1C), and we reproduced its depletion effects on glycerol release using siRNA oligonucleotides from two additional vendors (fig. S1B).
Fig. 1. Setting up a human adipocyte screening system.
(A) Representative images of cells stained with Hoechst (nuclei) and BODIPY (lipid droplets) at days 1 (top) and 14 (bottom) after adipogenic induction. Scale bars, 100 μm (merge) and 20 μm (inlay). Triglyceride (TG) quantification from the same time points is shown to the right. N.D., not detectable. (B) Isoprenaline (Iso.) and ANP responsiveness of adipocytes. Results are displayed as glycerol fold change versus unstimulated cells (basal). ***P < 0.001 from Tukey’s post hoc test after a one-way analysis of variance (ANOVA). (C) Representative immunofluorescence images of adipocytes transfected with nontargeting siControl or PLIN1-targeting siRNA (siPLIN1). Cells were stained with Hoechst, BODIPY, and PLIN1 antibody. Scale bar, 20 μm. Glycerol release, PLIN1 intensity, and lipid droplets (LDs) per cell are presented to the right versus siControl. *P < 0.05 from a Welch’s two-tailed t test. Replicates are highlighted by dots and are based on at least three independent experiments. Bar charts are presented as means ± SEM.
For the screen, we used a commercially available RNA interference (RNAi) library targeting 1530 genes encoding transcriptional regulators. To ensure that we only targeted genes expressed in adipocytes, we mined the FANTOM5 database (7), which contains an atlas of promoters and enhancers active in the majority of mammalian cell types, including the primary progenitor-derived adipocytes used in the present work. On the basis of these single molecule sequencing results, we found that 700 of the 1530 genes were expressed (≥10 tags per million) in our cell model (Fig. 2A). We therefore selected these target genes to screen for their effects on spontaneous (basal) glycerol release 72 hours after gene silencing. The screen was performed in triplicate reactions using ~30 million cells isolated from three individuals undergoing surgery for nonmalignant disease (Fig. 2B). Before selecting hits, we assessed the quality of the screen by replicate correlations and found our data to be highly reproducible (Fig. 2C and table S1). By performing statistical analyses, we identified 38 hits that upon silencing either repressed or stimulated basal lipid mobilization (Fig. 2D and table S2), results that were generally reproducible in cells from a fourth donor (fig. S1C). To rule out cytotoxic effects, we measured lactate dehydrogenase levels in parallel reactions. Our data demonstrated that only one hit, UBB (encoding ubiquitin B), altered this readout, and we therefore excluded it from further analyses (fig. S1D).
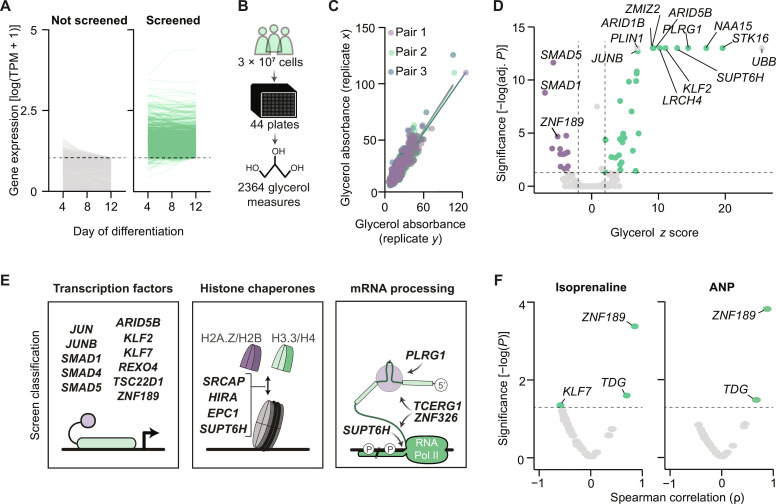
Fig. 2. Perturbation screening identifies 37 regulators of lipolysis.
(A) Expression of genes from Dharmacon’s transcriptional regulator siRNA library measured at three time points during adipogenesis. Genes with tags per million (TPM) > 10 at day 12 of differentiation were selected for screening. (B) Schematic representation of the lipolysis screen. (C) Pearson correlations for replicate pairs of glycerol measures from the lipolysis screen. (D) Thirty-seven hits from the lipolysis screen are highlighted in purple (stimulators) and green (suppressors). P values reflect Dunnet’s post hoc test after a one-way ANOVA. (E) Classification of lipolysis screen hits from (D). (F) Volcano plots displaying Spearman’s correlations between gene expression of screen hits and isoprenaline and ANP-stimulated lipolysis. Measures were performed in subcutaneous WAT samples (n = 12).
To mine these hits for function, we performed protein-protein interactor queries using the STRING database and literature searches to identify clusters regulating lipolysis. As displayed in Fig. 2E, we found three major functional classes: (i) transcription factors with experimentally determined DNA binding sequences (ARID5B, JUN, JUNB, KLF2, KLF7, SMAD1, SMAD4, SMAD5, REXO4, TSC22D1, and ZNF189), (ii) histone variant chaperones (EPC1, HIRA, SRCAP, and SUPT6H), and (iii) mRNA processing proteins (PLRG1, SUPT6H, TCERG1, and ZNF326). Emphasizing the validity of this screen as a resource of lipolysis regulators, JUN and SMAD protein family members have been previously linked to lipolytic suppression (8, 9). Therefore, this screen uncovers both known and unexplored lipolytic regulators in human adipocytes. To select screen hits associated with stimulated lipolysis, we isolated subcutaneous abdominal adipocytes from human subjects with a range of body fat percentages (fig. S2A) and measured isoprenaline- and ANP-induced lipolysis. In paired WAT samples, we determined gene expression levels of the hits for comparison to the lipolytic response. Our data revealed that of all measured genes, ZNF189 expression displayed the strongest association with isoprenaline- and ANP-stimulated lipolysis (Fig. 2F). To further assess the relationship between ZNF189 mRNA levels and metabolic parameters, we performed a meta-analysis of its expression in subcutaneous WAT samples across nine published clinical cohorts (10, 11). This revealed an inverse correlation between ZNF189 mRNA and two measures of metabolic health: waist-to-hip ratio and homeostasis model assessment insulin resistance (fig. S2B). Further studies were therefore focused on this transcription factor.
ZNF189 regulates hormone-stimulated lipolysis in white adipocytes
To uncover how ZNF189 regulates lipolysis, we performed in-depth analyses in human adipocytes differentiated from CD55+/DPP4+ progenitor cells (10, 12). We found ZNF189 to be highly enriched in the nuclear fraction, and this pool was efficiently depleted upon gene knockdown without effects on adipogenesis (Fig. 3, A to D, and fig. S3, A and B). In concordance with its relationship to hormone-stimulated lipolysis in human isolated adipocytes (Fig. 2F), ZNF189 depletion reduced both isoprenaline- and ANP-induced lipolysis (Fig. 3E), effects that were replicated using siRNAs from two additional vendors (fig. S3C). In contrast to the screen, ZNF189 silencing did not affect basal lipolysis in these short-term experiments. This discrepancy is likely due to the difficulties in detecting reductions in basal glycerol release following incubations for only 3 hours.
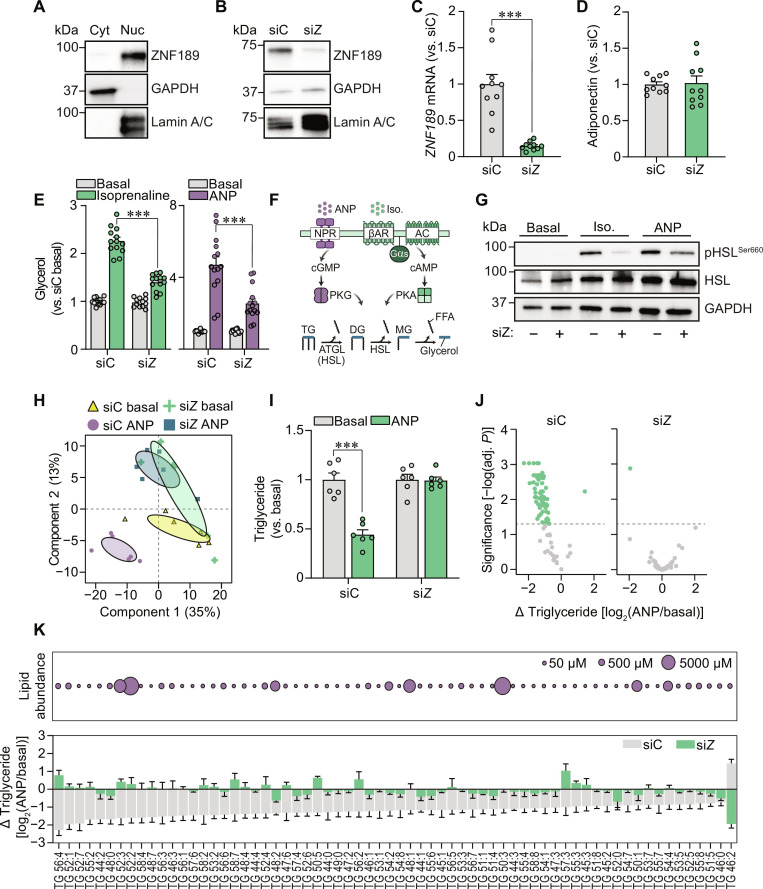
Fig. 3. ZNF189 regulates hormone-stimulated lipolysis.
(A and B) Representative Western blots of ZNF189, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (cytosolic), and Lamin A/C (nuclear) in cytosolic (Cyt) and nuclear (Nuc) adipocyte fractions (A) and nuclear fractions from adipocytes electroporated with siRNAs targeting ZNF189 (siZ) or nontargeting control (siC) (B). (C to E) Gene expression (C), adiponectin secretion (D), and glycerol levels (E) of siC and siZ adipocytes. Glycerol was measured in conditioned media following a 3-hour incubation under basal or stimulated conditions using either isoprenaline or ANP. (F) Overview of the ANP and isoprenaline signaling pathways. NPR, natriuretic peptide receptor; βAR, beta-adrenergic receptor; Gαs, Gs alpha subunit, AC = adenylate cyclase, PKG/PKA, protein kinase G and A; DG, diglyceride; MG, monoglyceride. (G) Representative Western blots for HSL phosphorylated at serine-660 (activating phospho-site; pHSLSer660), total HSL (HSL), and GAPDH from basal, isoprenaline- and ANP-treated siC and siZ cells. (H) Principal components analysis of 307 lipid species measured by shotgun lipidomics of basal and ANP-stimulated siC and siZ adipocytes. Confidence ellipses display 95% confidence intervals. (I) Total triglycerides for the samples in (H) presented as fold change versus same siRNA basal companion. (J and K) Individual triglyceride species comparing siC versus siZ adipocytes. Volcano plots for ANP responsiveness of all triglycerides are shown in (J), and lipid abundance and ANP responsiveness are highlighted for the species with altered responsiveness following ZNF189 depletion in (K). In (J), adjusted P reflects FDR correction on multiple comparisons analyses performed using limma. Bar charts are presented as means ± SEM. In (C) to (E) and (I), replicates are highlighted by dots. For targeted analyses, data are based on at least three independent experiments. ***P < 0.001 for Welch’s t test [(C) and (I)] or Tukey’s multiple comparisons test (E).
Because isoprenaline and ANP act through distinct receptor-kinase signaling pathways that converge on lipase activation (Fig. 3F), we hypothesized that the attenuated lipolytic responsiveness in ZNF189 depleted cells is regulated at a level shared by these two signaling cascades. To probe lipase activity, we measured activated [at protein kinase phosphorylation site (13)] and total protein levels of HSL in ZNF189 silenced (siZNF189) versus control (siC) adipocytes following hormonal stimulation. We found that both isoprenaline- and ANP-induced HSL phosphorylation were attenuated in ZNF189-depleted cells, with no change in total protein levels (Fig. 3G). Furthermore, as displayed in fig. S3D, there were no changes in mRNA levels of key lipolytic regulators PLIN1, PNPLA2 (encoding ATGL), or ADRB2 (encoding the major β-adrenergic receptor in our cell model) comparing siZNF189 and siC cells. These results indicate that ZNF189 depletion leads to impaired lipase activation without effects on the expression of established key lipolytic regulators.
To extend these observations, we also determined the effects of ZNF189 silencing on triglyceride levels (the substrate for the first hydrolysis step in lipolysis). For this, we performed shotgun lipidomics of ANP-stimulated siC and siZNF189 adipocytes. Our principal components analyses of 307 lipid species revealed that while lipolytic stimulation in control cells resulted in a major shift along component #1, this response was abrogated in adipocytes depleted of ZNF189 (Fig. 3H). At the individual lipid level, we confirmed that many (n = 66) triglyceride species were reduced in control cells following ANP stimulation, an effect that was almost entirely absent in siZNF189 adipocytes (Fig. 3, I to K). Thus, our results suggest that ZNF189 regulates lipolysis through a step affecting lipase activation that is shared by the isoprenaline and ANP signaling cascades.
ZNF189 is a putative transcriptional repressor
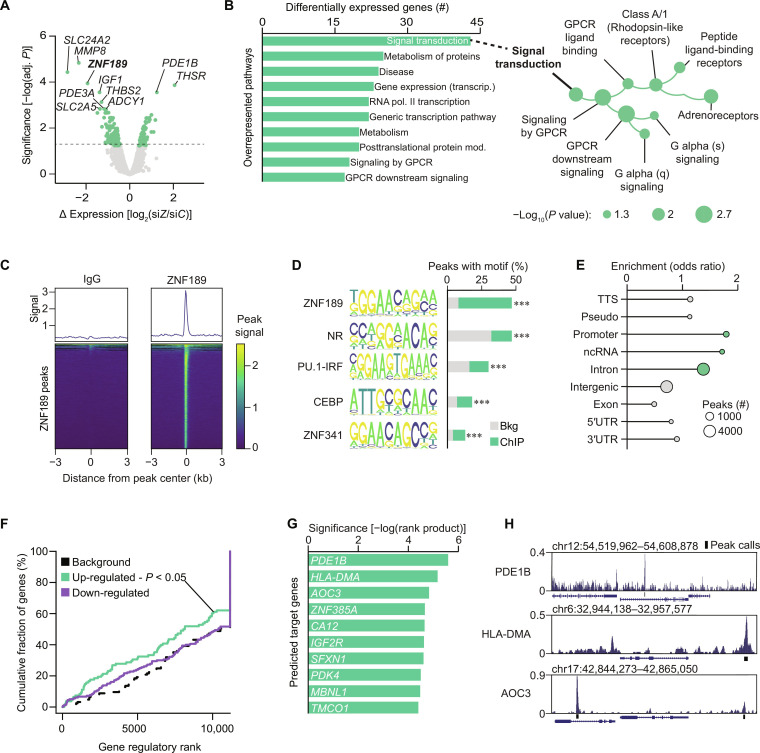
Having established through orthogonal approaches the level at which ZNF189 affects lipolysis, we next set out to identify the target genes governing these effects. For this, we transcriptionally profiled ZNF189-depleted adipocytes and performed chromatin immunoprecipitation of ZNF189 followed by deep sequencing (ChIPseq). We found that 186 genes were either up- or down-regulated in siZNF189 versus siC adipocytes and overrepresentation analyses revealed these genes to be enriched for signal transduction pathways including signaling downstream of G protein–coupled receptors (Fig. 4, A and B). Our ZNF189 ChIPseq identified 9276 peaks and showed that the ZNF189 binding motif, previously reported in human embryonic kidney (HEK) 293T cells (14), was top ranked and present in 47% of target sequences (Fig. 4, C and D). Moreover, ZNF189 binding was enriched at gene promoters (Fig. 4E), which supported the validity of our results as this has been shown previously (14). Using Binding and Expression Target Analysis (BETA) (15) integrative analyses of our transcriptome and ChIPseq results, we predicted that ZNF189 primarily functions as a transcriptional repressor (Fig. 4F). We also used BETA to rank target genes and inspection of our ChIP data surrounding these top genes showed good quality peak signals (Fig. 4, G and H). The highest scoring and most up-regulated gene among these target genes was PDE1B, which encodes the PDE1 family member phosphodiesterase 1B (Fig. 4G). PDE1 proteins degrade cyclic nucleotides with dual sensitivity to cyclic adenosine 3′,5′-monophosphate (cAMP) and cyclic guanosine 3′,5′-monophosphate (cGMP), making this protein a potential regulator of both isoprenaline- (signals through cAMP) and ANP- (signals through cGMP) stimulated lipolysis (16).
Fig. 4. ZNF189 is a sequence-specific transcriptional repressor.
(A) Volcano plot for transcriptomic analysis of ZNF189-depleted adipocytes (siZ) versus control cells (siC). Adj. P reflects FDR correction on multiple comparisons analyses performed by limma. (B) Overrepresentation analysis for results from (A). (C) Peak density plots of average peak values shown atop heatmaps of all ZNF189 peaks in immunoglobulin G (IgG) (left) and ZNF189 (right) chromatin immunoprecipitation followed by sequencing (ChIPseq). Scores are peak signal relative to IgG. (D) Top five enriched known motifs by homer analysis of ChIPseq peaks identified in (C). ***P < 0.001. (E) Binding distribution of ChIPseq peaks from (C). Enrichment per genomic region was assessed with hypergeometric testing and regions enriched with P < 0.05 are shown in green. (F) Activating or repressing function prediction by BETA. P value reflects gene activation (up-regulated) or repression (down-regulated) prediction for ZNF189 by a Kolmogorov-Smirnov test. (G) Direct gene target prediction by BETA. Values are −log(rank product), which reflect a P value for the combined score for binding potential. (H) ChIP signal at the top three BETA-predicted target genes. TTS, transcription termination site; 3′UTR, 3′ untranslated region; ncRNA, noncoding RNA; GPCR, G protein–coupled receptor.
ZNF189 regulates chromatin architecture at the PDE1B locus
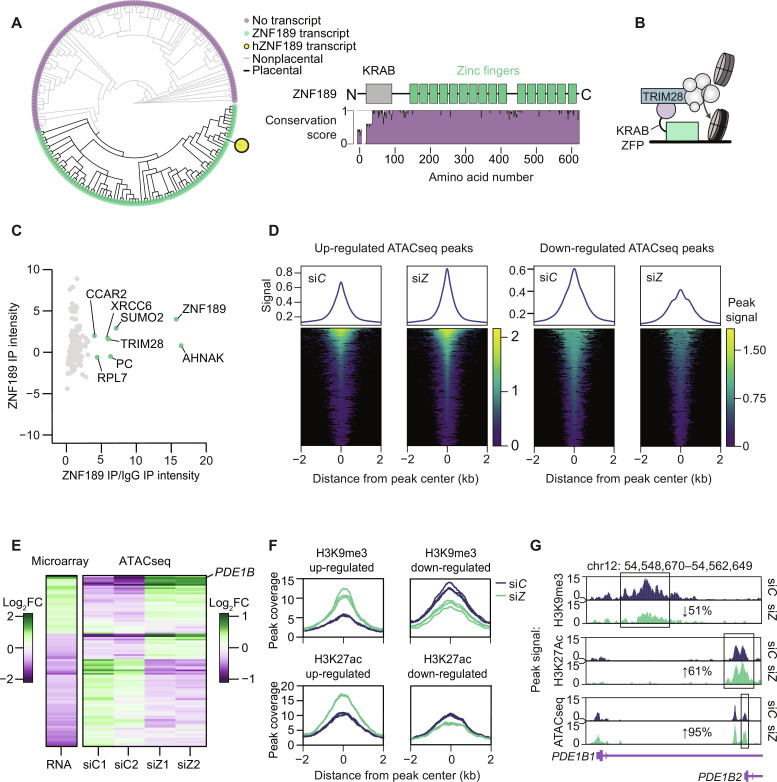
To understand how ZNF189 exerts its repressive effects on transcription, we first investigated the structure and conservation of this gene/protein across multiple species. We found ZNF189 homologs in all placentals in the Ensembl database and identified 16 highly conserved C2H2 zinc fingers and a Krüppel-associated box (KRAB) (Fig. 5A). The latter is a strong repressive domain present in one-third of all eukaryotic zinc finger proteins (17), which regulate transcription by interacting with tripartite motif protein 28 (TRIM28; also known as KRAB-associated protein 1), a scaffold protein that recruits mediators of chromatin accessibility (Fig. 5B) (18). As not all KRAB domain proteins interact with TRIM28 (19), we next immunoprecipitated ZNF189 from adipocytes and measured protein-protein interactors using mass spectrometry (MS). This confirmed that ZNF189 interacts with TRIM28 and other chromatin-associated proteins (e.g., CCAR2, ZMYM4, and GTF2I) in fat cells (Fig. 5C and table S3). These data support our functional genomics analyses and suggests that ZNF189 represses gene transcription, possibly via TRIM28-mediated regulation of chromatin accessibility.
Fig. 5. ZNF189 contains a KRAB domain and interacts with TRIM28.
(A) Left: Phylogenetic tree showing gain of ZNF189 gene in placentals (eutheria). Right: ZNF189 protein conservation (Valdar scores from 10 homologs) aligned with ZNF189 protein domains (Uniprot O75820). (B) Schematic overview of the KRAB-TRIM28 protein complex showing ZNF189 interacting with DNA and the TRIM28 complex. TRIM28 is depicted maintaining closed chromatin via a protein complex. (C) Normalized protein intensities from MS on ZNF189 immunoprecipitation (IP) samples (n = 4). (D) Peak density plots and heatmaps for increased and decreased ATACseq peaks in control (siC) and siZNF189 (siZ) cells (n = 2). (E) Fold change [log2(FC)] from the microarray and ATACseq for genes with significant regulation by both assays. RNA data reflects fold changes versus siC and ATACseq data reflects fold changes versus the mean of the comparison condition. (F) Peak coverage plots for regulated H3K9me3 and H3K27ac peaks by CUT&Tag (n = 3). (G) Average peak signal for chromatin profiling at the PDE1B locus, which contains two promoters encoding PDE1B1 and PDE1B2.
To directly assess ZNF189 regulation of chromatin accessibility, we performed assay for transposase-accessible chromatin using sequencing (ATACseq) in siZNF189 and siC adipocytes. From 198,395 ATAC windows identified across four replicates, we found 22,048 up-regulated and 40,583 down-regulated windows upon ZNF189 depletion (Fig. 5D). However, while changes in chromatin accessibility and gene expression correlated well (Fig. 5E), only seven genes displayed (i) proximal ZNF189 binding (within 3 kb of the transcriptional start sites), (ii) altered gene accessibility, and (iii) perturbed expression in siC versus siZNF189 adipocytes. This suggests that much of the altered transcription from ZNF189 silencing reflects secondary consequences and/or binding to more distal gene regulatory elements, which is in concordance with previous studies of transcriptional repressors (20, 21).
The ZNF189-interacting protein TRIM28 regulates chromatin architecture by recruiting corepressor machinery, including SETDB1, which trimethylates H3K9 (H3K9me3) in euchromatin regions (22), and the nucleosome remodeling and deacetylase (NuRD) complex, which deposits nucleosomes and deacetylates histones at H3K27 (H3K27ac) (23, 24). To test whether the ZNF189-TRIM28 complex affects these chromatin modifications in adipocytes, we measured H3K9me3 and H3K27ac profiles using Cleavage Under Targets and Tagmentation (CUT&Tag) in siZNF189 versus siC adipocytes. We found 30,183 H3K9me3 peaks and 38,332 H3K27ac peaks across six replicates, with no overlap between these markers of gene repression and activation, respectively. Of these, the 1058 differentially regulated H3K9me3 peaks and the 545 H3K27ac peaks are displayed in Fig. 5F and fig. S4A. Our integrated genomics data at the PDE1B locus in ZNF189-depleted adipocytes revealed decreased H3K9me3, increased H3K27ac, and increased accessibility in siZNF189 versus control cells (Fig. 5G). Together, this suggests that ZNF189 regulates the accessibility and thereby gene transcription at this locus.
ZNF189 represses the expression of an adipocyte-specific isoform of PDE1B
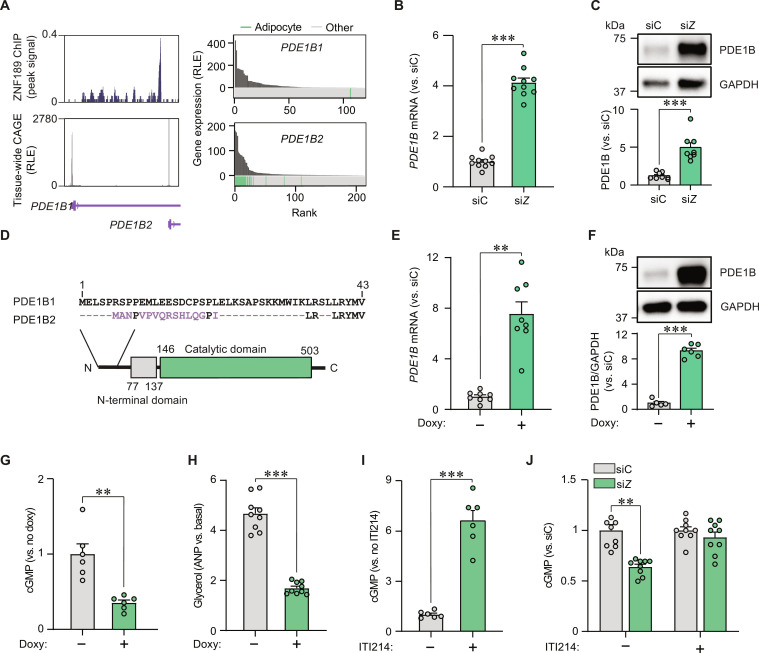
To better understand the regulation of chromatin architecture at the PDE1B locus, we examined our ChIPseq results more closely. Our analyses revealed that ZNF189 binds in the first intron of the canonical gene (which encodes PDE1B1 with 536 amino acids), specifically ~1 kB upstream of an alternative promoter (encoding PDE1B2 with 516 amino acids) (Fig. 6A). To validate these results, we performed ChIP quantitative polymerase chain reaction (qPCR) and found an enrichment in the corresponding peak region (fig. S4B). As phosphodiesterases display tissue- and cell type–specific expression patterns (25), we established which PDE1B transcripts are expressed in adipocytes using the FANTOM5 database. Promoter expression across tissues and cell types demonstrated that while the isoform encoding PDE1B1 is primarily expressed in neuronal tissues, the PDE1B2-encoding transcript is specifically expressed in fat cells (Fig. 6A). To confirm these findings, we performed qPCR analyses in adipocytes and found that the canonical transcript was barely detectable (Ct values 32 to 35). In contrast, the expression of the alternative isoform was detectable and up-regulated upon ZNF189 knockdown (Fig. 6B). These findings were confirmed at the protein level (Fig. 6C). Furthermore, overexpression of ZNF189 resulted in a reduction in PDE1B mRNA levels with a concomitant increase in basal lipolysis (fig. S5A). To predict the putative role of the adipocyte-enriched PDE1B2 protein, we aligned amino acid sequences of the two PDE1B isoforms and mapped out protein domains (Fig. 6D). These in silico analyses showed that PDE1B2 differs from PDE1B1 in short parts of the N-terminal region. However, the catalytic domain is intact in both variants, suggesting that both isoforms retain the capacity to hydrolyze cAMP and cGMP.
Fig. 6. The ZNF189-PDE1B2 axis regulates adipocyte lipolysis.
(A) Left: ZNF189 ChIP signal shown above tissue-wide cap analysis of gene expression signals [expressed as relative log expression (RLE)] at the two PDE1B promoters (encoding PDE1B1 and PDE1B2). Right: Gene expression of PDE1B1 and PDE1B2 across tissue/cell samples from the FANTOM5 database. Adipose samples are highlighted in green. (B and C) PDE1B mRNA (B) and protein (C) levels for siZNF189 (siZ) versus control cells (siC). (D) Sequence alignments for PDE1B1 and PDE1B2 shown above protein domains from InterPro. Purple characters represent deviations from the canonical protein isoform. Conserved protein domains are indicated with colored boxes. (E to H) PDE1B mRNA (E) and protein (F) as well as cGMP (G) and glycerol (H) in cells engineered to overexpress PDE1B2 under a doxycycline (doxy)-inducible promoter. In (G) and (H), cells were incubated with ANP. (I and J) Effects of the PDE1 inhibitor ITI214 on ANP-stimulated cGMP levels in untransfected (I) or siC versus siZ adipocytes (J). Bar charts are expressed relative to the indicated controls and are presented as means ± SEM. In these panels, replicates are highlighted by dots and are based on at least three independent experiments. Statistical analyses were performed using Welch’s two-tailed t test. **P < 0.01 and ***P < 0.001.
To test if increased levels of the PDE1B2 protein phenocopied the effects of ZNF189 depletion, we generated adipocytes with doxycycline-inducible expression of this isoform. Incubations with doxycycline up-regulated PDE1B2 mRNA and protein levels ~8- to 10-fold (Fig. 6, E and F). Compared to control cells cultured in the absence of doxycycline, PDE1B2 overexpression resulted in decreased cyclic nucleotide levels and glycerol release after lipolytic stimulation (Fig. 6, G and H, and fig. S5B). Our results therefore suggest that increased PDE1B2 levels may explain the reduction in lipid mobilization observed in ZNF189-depleted adipocytes. We confirmed this by incubating control and ZNF189-depleted cells with ITI214, a selective PDE1 inhibitor. As expected, ITI214 treatment in control cells increased cGMP levels (Fig. 6I), indicating that PDE1 is active in adipocytes. Furthermore, cyclic nucleotide levels were lower in siZNF189 versus siC cells upon lipolytic stimulation (Fig. 6J and fig. S5C)—an effect that was abrogated in the presence of ITI214 (Fig. 6J). Thus, PDE1B2 induction alone is sufficient to reduce lipolysis and is required for the effects of ZNF189 depletion on lipid mobilization in adipocytes.
DISCUSSION
In this work, we demonstrate through a targeted perturbation screen that ZNF189 stimulates lipolytic signaling in primary human fat cells. By combining genomic and proteomic mapping approaches, we find that ZNF189 represses chromatin accessibility and expression of PDE1B2, an adipocyte-specific phosphodiesterase isoform that hydrolyzes cAMP and cGMP. Thus, our work uncovers the ZNF189-PDE1B2 axis as a regulator of lipolytic signaling in human white adipocytes.
Transcription factors and the basal transcriptional machinery interface with chromatin to regulate gene expression and consequently cellular function (26). In mice, remodeling of chromatin architecture has been shown to regulate thermogenic gene programs in adipocytes, at least partly through altered chromatin accessibility by the switch/sucrose‐non‐fermentable (SWI/SNF) chromatin remodeling complex (27, 28). Here, we use MS, ChIPseq, ATACseq, and CUT&Tag to link the ZNF189-TRIM28 repressor complex to the regulation of adipocyte lipolysis through PDE1B2. More specifically, we find increased chromatin accessibility, increased H3K27 acetylation, and decreased H3K9 trimethylation with the loss of ZNF189 binding at the locus encoding PDE1B2. As ZNF189 contains an N-terminal KRAB domain and interacts with TRIM28, it is likely that these two proteins together promote the formation of heterochromatin-like regions to regulate target gene expression. We base this hypothesis on the well-established fact that TRIM28 in other cell types recruits repressive methyltransferases and deacetylases, including the NuRD complex, HP1, and SETDB1 (29). In support of our model, the KRAB domain of ZNF189 has been shown through systematic screening to be sufficient to repress gene transcription when fused to an endonuclease-deficient Cas9 protein (19). As TRIM28 haploinsufficiency has been linked to stochastic development of obesity through a complex and as-yet undefined gene network in adipose tissue, this work highlights one potential transcriptional axis linking TRIM28 to obesity (30).
The second messengers, cAMP and cGMP, relay circulating signals to subcellular compartments to regulate physiological processes, including adipocyte lipolysis (31). After production at the cell membrane, cyclic nucleotide concentration is fine-tuned primarily by phosphodiesterases, which constitute a large family of proteins that bind and degrade cAMP and/or cGMP (25). Phosphodiesterase family members control cyclic nucleotide hydrolysis through distinct substrate binding affinities and tissue-specific expression patterns (25). While canonical suppression of adipocyte lipolysis occurs through insulin stimulation of PDE3B (31), other PDE family members have recently been implicated in different aspects of adipocyte biology (32, 33). Here, we identify a PDE1B isoform that regulates cGMP levels and hormone-stimulated lipolysis and is highly expressed under an adipocyte-specific alternative promoter in human fat cells. Alternative promoters are gene features that often evolve to offer increased transcriptional and translational control (34) and have been shown in other instances to serve important metabolic functions [e.g., PPARγ2 (35), ChREBP-β (36), PGC-1α4 (37), and CLSTN3β (38)]. Similarly, we show that the adipocyte-specific PDE1B2 variant is under independent transcriptional control from the canonical PDE1B1 variant in adipocytes.
We and others have linked impairments in hormone-stimulated lipolysis to metabolic derangements in humans (39). It is therefore possible that treatments restoring hormone-stimulated lipolysis could mitigate adipose tissue-related metabolic disease. In addition to physical activity, which is known to improve WAT lipolysis and has beneficial effects on cardiovascular risk markers (40), our data suggest that PDE1B2 inhibition could be explored. In line with this, a recent study in mice showed that the nonspecific PDE1 inhibitor, vinpocetine, had positive effects on adiposity and in vivo measures of lipolysis, including circulating fatty acids and triacylglycerols (41). The development of selective PDE inhibitors is an ongoing challenge due to high structural similarities in PDE substrate-binding pockets between family members. Nevertheless, rapid advancements in phosphodiesterase pharmacology make the development of a selective PDE1B2 inhibitor feasible (25).
Our screen was designed to broadly capture regulators of lipid mobilization by measuring glycerol levels in adipocyte culture media after gene depletion. This allowed us to identify the ZNF189-PDE1B2 axis as a regulator of both isoprenaline- and ANP-stimulated lipolysis. Our findings underscore the complexity of lipolytic regulation, and the importance of studying screen hits using an extensive and appropriate panel of functional assays that probes elements of lipolytic control beyond basal glycerol release. Among the lipolysis regulators uncovered in our screen, we found an enrichment for JUN/SMAD family members, which are growth-related proteins that have been linked to lipolytic suppression in adipocytes through lipase gene networks (8, 9, 42). Together with identifying several transcription factors that have been linked to adiposity or metabolic disease in other tissues [KLF2 (43), KLF7 (44), REXO4 (45), and TSC22D1 (46)], these findings validate the utility of our human adipocyte screening platform as a resource of transcriptional regulators of adipocyte lipid mobilization. In this vein, we use protein network mapping to uncover a previously unknown role for histone chaperone proteins and mRNA elongation/splicing factors in regulating adipocyte lipolysis. Therefore, in addition to linking chromatin regulation and transcriptional activation to lipolysis, our systematic approach identifies several other pathways and targets that likely regulate adipocyte lipolysis through modifying chromatin architecture and gene transcription.
In conclusion, our targeted perturbation screen in human adipocytes systematically uncovers transcriptional regulators of adipocyte lipid mobilization including the lipolytic stimulator ZNF189. Through mechanistic studies, we show that a ZNF189 protein complex represses the expression of PDE1B2, an adipocyte-specific phosphodiesterase isoform that regulates cyclic nucleotide signaling. Thus, ZNF189 and PDE1B2 constitute a regulatory axis, whose pathophysiological relevance warrants further study.
MATERIALS AND METHODS
Experimental design
The objective of the study was to perform a targeted perturbation screen to identify regulators of lipolysis in human adipocytes. For this, glycerol levels in cell culture media after RNAi (details below) was used as a proxy to identify hits regulating lipid mobilization. Follow-up metabolic and molecular characterizations of the candidate gene ZNF189 were performed in adipocyte cultures.
Biological materials
Human subjects
Stromal vascular fractions for the perturbation screen were collected from three non-obese women undergoing cosmetic liposuction (age 41 ± 9) and one 38-year-old woman for independent validation experiments. Adipose tissue biopsies for lipolysis and qPCR analyses were collected from 12 individuals who participated in the Swedish cardiopulmonary bioimage study (eight males and four females; SCAPIS DOI: 10.1111/joim.12384, www.scapis.org/) with an average body mass index of 27.2 kg/m2 (subject characteristics are presented in fig. S2A). WAT-derived CD55+/DPP4+ progenitor cells were isolated from a lean 17-year-old male donor. The experimental protocols received ethical approval by the regional board of ethics and conformed to the Declaration of Helsinki, and all subjects provided written informed consent to participate.
Isolation and culture of mature adipocytes
Adipose tissue biopsies were collected by needle aspiration with local anesthesia at the paraumbilical region. A portion of the biopsies was saved for RNA analysis when applicable. Adipocytes were isolated from the remainder of the tissue by digesting tissues with type II collagenase (Sigma-Aldrich, C6885), filtering the digest through a 200 μM nylon strainer, and washing the floating fraction from this filtered digest. Adipocytes were then loaded by volume (2% v/v) into Krebs ringer phosphate buffer with 2% bovine serum albumin (BSA) (Sigma-Aldrich, A4503-500G), d-glucose (1 mg/ml; Merck, 1.08337.0250), and ascorbic acid (0.5 mg/ml; VWR Chemicals, 20150.184) supplemented with isoprenaline (Sigma-Aldrich, I5627-5G) or ANP (Sigma-Aldrich, A1663-1MG).
Analyses of intact adipose tissue samples
Total RNA was extracted from adipose tissue samples using the QIAGEN RNeasy kit (catalog no. 74104). Total RNA yield and purity was measured by spectrometry with the NanoDrop 2000 (Thermo Fisher Scientific, Lafayette, CO), and cDNA synthesis was performed with Bio-Rad iScript cDNA synthesis kits (1708891BUN). qPCR analysis was performed with Bio-Rad iQ SYBR Green Supermix (1708882) and analyzed by the comparative Ct method (47). Gene levels were expressed as the ratio versus the average Ct values of PPIA and B2M. Predesigned primers were purchased from Sigma-Aldrich for genes hits from the lipolysis screen (MilliporeSigma KiCqStart SYBR Green Primers, KSPQ12012). Primers for eight screen hits did not meet criteria for specific PCR products based on melt curve assessment, so these genes were excluded from correlational analyses. Primer sequences are listed in table S4 for detectable hits and house-keeping genes: ANKRD54, ARID1B, ASB8, EDF1, EPC1, HIRA, JUN, JUNB, KLF2, KLF7, LMCD1, LRCH4, NAA15, PLRG1, PTRF, REXO4, SMAD1, SMAD5, SRCAP, STK16, SUPT6H, TCERG1, TDG, TGS1, UBB, URI1, ZMIZ2, ZMYND8, ZNF189, ZNF326, PPIA, and B2M.
Primary cell cultures
Differentiation of human adipocytes from the stromal vascular fraction
The stromal vascular fraction was isolated by digestion of subcutaneous abdominal adipose tissue samples with type II collagenase (Sigma-Aldrich, C6885), erythrocyte lysis, and filtration before plating in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, 31885049) with 5% fetal calf serum for 24 hours. Differentiation was induced the next day with DMEM/Ham’s F12 media (mixed 1:1; Thermo Fisher Scientific, 31885049 and 21765037) supplemented with transferrin (10 μg/ml; Sigma-Aldrich, T8158), 0.86 μM insulin (Sigma-Aldrich, catalog no. I9278), 0.2 nM triiodothyronine (Sigma-Aldrich, T6397), 1 μM dexamethasone (Sigma-Aldrich, D1756), 100 μM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich, I5879), and 1 μM rosiglitazone (Cayman Chemicals, 71740). Rosiglitazone was removed from the differentiation cocktail after 6 days.
Differentiation of human adipocytes from CD55+/DPP4+ progenitor cells
CD55+/DPP4+ progenitor cultures were maintained and passaged in DMEM with 10 mM Hepes (Gibco, 15630-056), 10% fetal bovine serum (HyClone, SV30160.03), penicillin-streptomycin (50 μg/ml; Thermo Fisher Scientific, 15-140-122), and fibroblast growth factor-2 (2.5 ng/ml; Sigma-Aldrich, F0291-25UG) or differentiated with insulin (5 mg/ml), 0.25 mM dexamethasone, 0.5 mM IBMX, transferrin (10 μg/ml; T8158, Sigma-Aldrich), 0.2 nM triiodothyronine (T6397, Sigma-Aldrich), and 10 mM rosiglitazone. Dexamethasone and IBMX were removed after 2 days, and rosiglitazone was removed after 9 days.
Inducible PDE1B2-overexpressing cells
Inducible PDE1B2-overexpressing cells were generated by lentiviral transduction of proliferating CD55+/DPP4+ progenitor cells. A codon-optimized PDE1B2 sequence with a Kozak sequence (GCCACC) before the start codon was subcloned into the pCW-Cas9 backbone under a tight tetracycline-responsive element promoter (Eric Lander & David Sabatini -Addgene plasmid #50661; http://n2t.net/addgene:50661; RRID:Addgene_50661) (48). For this, the plasmid backbone and insert were digested with Bam HI and Nhe I restriction enzymes (New England Biolabs, catalog nos. R3136S and BR3131S, respectively) and ligated with a T4 DNA ligase (New England Biolabs, catalog no. M0202S) to yield ligated products for bacterial transformation. Sanger sequencing was performed to ensure correct insertion of the gene fragment. Lentiviruses were generated in HEK293T cells by transfecting the purified plasmid together with pMD2.G and psPAX2 (Addgene #12259 and #12260, respectively) with Lipofectamine 3000 (Invitrogen, L3000001). Harvested viruses (700 ng of p24 per 500,000 cells) and polybrene (8 μg/ml; Sigma-Aldrich, catalog no. TR-1003-G) were added onto the progenitors for 24 hours. Three days after virus removal, transformed cells were selected with puromycin (1 to 2 μg/μl for 4 to 7 days; Sigma-Aldrich, P8833). Following adipogenesis, the expression of PDE1B2 was induced by incubation with doxycycline (0.5 to 2 μg/ml; Sigma-Aldrich, D3447-500MG) for 2 days.
Methods
Confocal microscopy
Adipocytes from the stromal vascular fraction were differentiated in 24-well plates from Zell Kontakt (5232-20) with 100,000 cells per well. Following adipogenesis, cells were washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde (Santa Cruz Biotechnology, sc-281692) for 15 min, and washed again 3× in PBS. For PLIN1 immunofluorescence, cells were permeabilized with 0.1% Triton X-100 in PBS with 3% BSA for 20 min at room temperature before incubating with an anti-PLIN1 antibody (1:20,000 in PBS with 3% BSA; Cell Signaling Technology, D1D8) overnight at 4°C. Cells were washed and incubated with Alexa Fluor 568–conjugated anti-rabbit secondary antibody (1:500 in PBS with 3% BSA; Invitrogen, A11011) for 60 min at room temperature. Subsequently, lipid droplets and nuclei were stained by incubating the cells for 5 min with BODIPY 493/503 (1:2500 in PBS; Invitrogen, D3922) and Hoechst (1:10,000 in PBS; Thermo Fisher Scientific, 33342). Cells were thereafter washed and imaged at 20× magnification with a Nikon Eclipse Ti2 single point scanning confocal microscope. For PLIN1 knockdown cells, Z stacks were taken spanning 18 × 1 μM steps and representative images were selected from one plane representing the middle of the lipid-containing cell volume. PLIN1 intensity per cell and lipid droplets per cell (>11μm2) were calculated using ImageJ 1.53 (National Institutes of Health, Bethesda, MD).
Triglyceride quantitation
Triglycerides were quantified in adipocytes from the stromal vascular fraction after 1- and 15-day induction with differentiation media in 24-well plates using the Triglyceride Quantification Kit from Sigma-Aldrich (MAK266). After washing with PBS, cells were lysed in 150 μl of 5% NP-40 (in H2O), heated to 85°C for 8 min, and then diluted with 600 μl of H2O for subsequent analysis (50 μl per sample) following the assay protocol.
Glycerol release
Lipolysis was assessed by fluorometric detection of glycerol in phenol red–free media (Thermo Fisher Scientific, 21041-033) collected from cells after siRNA transfection and after various lipolytic stimuli. Glycerol release was measured in basal conditions for the lipolysis screen (concentration in the media 72 hours after the last transfection). When stimulated lipolysis was assessed, drugs were applied in media supplemented by ascorbic acid (0.1 mg/ml; VWR Chemicals, 20150.184) and 2% BSA (w/v) (Sigma-Aldrich A4503-500G). Cells were stimulated for 3 hours with isoprenaline (1 μM; Sigma-Aldrich, I5627-5G) or ANP (10 μM; MedChemExpress, HY-P2281) and 20 μl of media was loaded into reactions with 50 μl of free glycerol reagent (Sigma-Aldrich, F6428-40ML) prepared with Invitrogen Amplex UltraRed Reagent (10737474) for fluorometric assessment of glycerol based on a standard solution (Sigma-Aldrich, G7793-5ML).
RNA interference
RNAi optimization in differentiated stromovascular fractions was performed with siRNA targeting PLIN1 (positive control) or a nontargeting control with siRNAs from three different vendors (Dharmacon, Ambion, and QIAGEN) using QIAGEN transfection reagent (HiPerFect, 301707). Transfections were performed on days 10 and 11 after adipogenic induction, and cells were assayed on day 14. The main screening experiment was performed with the siGENOME SMARTpool siRNA Human Transcription Factor Library (Dharmacon, Lafayette, CO, USA) using 2 μl of siRNA and 0.9 μl of HiPerFect complexed in 60 μl of differentiation media for 15 min, with 50 μl subsequently loaded to differentiated stromovascular fraction in 96-well plates (final siRNA concentration of 20 nM). Each plate had nontargeting siRNA transfected in six replicates, siPLIN1 (positive control) transfected in triplicate, and siRNA transfection of 16 screen targets in triplicate for a total of 44 plates.
Transfection of siRNA in adipocyte differentiated from CD55+/DPP4+ progenitor cells was performed by electroporation with the Neon Transfection System (Thermo Fisher Scientific, MPK5000) on day 8 of adipogenic induction. One million cells were transfected per reaction in 100 μl of R buffer with 2 μM siRNA [Dharmacon, M-021350-00-0005 (ZNF189) and D-001206-13-05 (nontargeting control)] and diluted 40× into differentiation media. Cells were plated at 65,790 cells/cm2, received fresh media the next day, and were collected for functional assays 4 days after transfection. Gene silencing effects on stimulated lipolysis were validated using siRNA from QIAGEN (GeneGlobe GS7743 and control SI03650318) and Thermo Fisher Ambion (s15256 and control 4390843).
Western blotting
Cells were collected from six-well plates for protein isolation in 100 μl of Pierce radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific, 89910) with protease inhibitor cocktail (Roche, 11873580001) by incubating for 10 min on ice, passing through a 23-gauge needle 10 times, and removing debris by centrifugation at 14,000g for 10 min (4°C). Protein concentrations were then measured by colorimetric reaction with bicinchoninic acid (Thermo Fisher Scientific, 23227) detected at 562 nm (Tecan Infinite M200) for loading equal total protein amounts per sample except for assessment of protein localization between fractions, in which case the equivalent total organellar protein (5% cytosol and nuclei) was loaded to each lane by volume. Proteins were denatured by Laemmli buffer (Bio-Rad, 1610747) for 8 min (95°C), separated in SDS–polyacrylamide gel electrophoresis gels (Bio-Rad, 4568084), and transferred to polyvinylidene difluoride membranes (Bio-Rad, 1704274) with the Bio-Rad Trans-Blot Turbo (1.3A, 25 V, 7 min) before blocking in Tris-Buffered Saline, 0.1% Tween 20 (TBS-T) with 5% Blotto, non-fat dry milk (Santa Cruz Biotechnology, sc-2325). Antibody binding was performed overnight (4°C) as follows: PLIN1 (1:1000 in TBS-T with 5% BSA; Cell Signaling Technology, D1D8), lamin A/C (1:1000 in TBS-T with 5% milk; Cell Signaling Technology, 4C11), ZNF189 [1:500 in TBS + 0.2% Tween 20 with 3% Amersham ECL prime blocking agent (Cytiva, RPN418); Sigma-Aldrich, HPA034814-100UL], glyceraldehyde-3-phosphate dehydrogenase (1:1000 in TBS-T with 5% BSA; Cell Signaling Technology, 14C10), Phospho-HSL (Ser660) (1:1000 in TBS-T with 5% milk; Cell Signaling Technology, 4126S), HSL (1:1000 in TBS-T with 5% milk; Cell Signaling Technology, 4107), and PDE1B (1:250 in TBS-T with 3% milk; Sigma-Aldrich, HPA018492-100UL). Following incubations with primary antibodies, membranes were washed 3× for 10 min in TBS-T, incubated with the horseradish peroxidase–linked secondary antibodies targeting the respective primary antibody host species (1:10,000 in TBS-T with 5% milk; Cell Signaling Technology, 7074S or Invitrogen, G21040) for 1 hour and washed again 3× for 10 min in TBS-T. Membranes were lastly incubated in pico or femto chemiluminescence substrate (Thermo Fisher Scientific, 34580 or GE Healthcare, 12644055) for 5 min and detected with the Bio-Rad ChemiDoc MP Imaging System. Images were analyzed in ImageJ using the Gel Analyzer tool to obtain relative densities from lane profile plots after subtracting local background.
Cytotoxicity
Lactate dehydrogenase (LDH) activity was measured with the Cytotoxicity Detection KitPLUS (Roche, 4744926001) according to the manufacturer’s instructions.
Cellular fractionation
Cellular fractionation for the assessment of ZNF189 localization was performed by differential centrifugation. Adipocytes were rinsed and collected in homogenization buffer [20 mM tris-HCl (pH 7.5), 6 mM EDTA (pH 8.0), 250 mM sucrose in H2O with cOmplete, and EDTA-free protease inhibitor cocktail]. Subsequently, cells were disrupted with 20 strokes through a 23-gauge needle. The crude nuclear fraction was separated from the post-nuclear supernatant by pelleting with centrifugation at 1000g for 15 min (4°C). This pellet was then cleaned from whole cells and debris by incubating for 15 min (4°C) in 200 μl of hypotonic buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 10 mM KCl2 supplemented with 0.05% NP-40, and protease inhibitor cocktail]. After hypotonic swelling, the crude nuclear fraction was passed through a 23-gauge needle 3×, shaken vigorously by hand, and diluted 1:1 in dilution buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 10 mM KCl2, 0.6 M sucrose with 0.05% NP-40, and protease inhibitor cocktail) for nuclear isolation by centrifugation at 500g for 10 min (4°C). The membrane/mitochondrial fraction was separated from the cytoplasm by centrifuging the post-nuclear supernatant at 14,000g for 15 min (4°C). Nuclear fractions were lysed by pipet mixing in RIPA buffer supplemented with 1% SDS and benzonase nuclease (Sigma-Aldrich, E1014-25KU) before proceeding to Western blot analysis.
qPCR on adipocyte cultures
Total RNA was extracted from cultured adipocytes using NucleoSpin RNA columns (Macherey-Nagel, 740955.250). Analyses were otherwise performed the same way as outlined for adipose tissue pieces, but gene expression was normalized to the housekeeping genes indicated in each figure. Primer sequences are listed in table S4 for target genes: ZNF189, PLIN1, PPARG, ADIPOQ, FABP4, PDE1B, LRP10, 18S, and PPIA.
Adiponectin ELISA
Secreted adiponectin was measured in conditioned media collected from siZNF189 versus siC adipocyte cultures collected after 2 days of incubation in 24Kell plates using the Human Total Adiponectin kit (R&D Systems, DRP300).
Lipidomics
Control and ZNF189-depleted adipocytes were stimulated for 3 hours in the presence and absence of ANP following the same steps as the stimulated lipolysis protocol. After stimulation, cells were washed with PBS and collected with methanol-rinsed materials into a final volume of 350 μl of cells + PBS. Cell homogenates were obtained by repeated freezing/thawing and extracted according to Folch’s method (49). The internal standard SPLASH LIPIDOMIX Mass Spec Standard mix was spiked in before extraction and was used for normalization. The organic phase of the lipid extract was transferred to a new glass tube and dried under N2 flow. Last, the lipids were dissolved in 75 μl of chloroform/methanol (1:2, v/v).
Lipid extracts were subjected to MS analysis using a Orbitrap ID-X Tribrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with a TriVersa NanoMate (Advion Biosciences, Ithaca, NY). In 96-well plates, 10 μl of lipid extracts was mixed with 90 μl of 7.5 mM ammonium acetate in chloroform/methanol/propanol (1:2:4, v/v/v) for positive and negative ionization mode analysis. Lipids were infused using a backpressure of 1.25 psi and ionization voltage of 0.95 kV.
Chromatin Immunoprecipitation followed by deep sequencing
ChIP was performed on nuclei isolated from 14 × 15–cm2 dishes by hypotonic swelling. In brief, cells were rinsed 1× each in PBS and hypotonic buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 10 mM KCl2 with 0.05% NP-40, and protease inhibitor cocktail] before swelling for 15 min in 2 ml of hypotonic buffer per dish (4°C). Samples were then pooled together and passed through a 23-gauge needle 3×, shaken vigorously by hand, and diluted 1:1 with dilution buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 10 mM KCl2, 0.6 M sucrose with 0.05% NP-40, and protease inhibitor cocktail] for nuclear purification by centrifugation at 500g for 10 min (4°C). The pelleted nuclei were immediately crosslinked in 1 ml of PBS with 1% formaldehyde (Pierce, 28908) on rotation for 10 min at room temperature and quenched with PBS supplemented with 137 mM glycine on rotation for 5 min at room temperature. Crosslinked nuclei were collected by centrifugation (2000g, 5 min, 4°C) and washed twice in PBS before lysis in 200 μl of denaturing buffer [50 mM tris (pH 8), 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 150 mM NaCl, and protease inhibitor cocktail] with 15 strokes through a 27-gauge needle. The crosslinked nuclear lysate had its volume adjusted to 2 ml before being transferred to 15 ml of Bioruptor Pico Tubes and sonication beads (Diagenode, C01020031). Sonication was performed rotating in 4°C water for 30 min (30-s on, 30-s off) in the Bioruptor Pico sonication device (Diagenode, B01060010). Lysates were cleared from sonication beads and insoluble material by centrifugation at 14,000g for 10 min (4°C), and this and all subsequent steps were performed in DNA low bind tubes (Eppendorf, 022431021). Protein and DNA were quantified at this time, and input samples were collected before proceeding to ChIP.
ChIP was performed with equal parts protein A/G–conjugated magnetic beads (Invitrogen, 10003D and 10001D). Beads were washed twice (by 5-min rotation at room temperature) in PBS with 0.02% Tween 20 and incubated overnight for immunoprecipitation or used to preclear crosslinked nuclear lysate by rotation for 1 hour (4°C). Precleared lysates were removed from beads by magnet separation and incubated rotating overnight (4°C) with 16 μg of ZNF189 antibody (Sigma-Aldrich, HPA034814-100UL) for 5.6 μg of chromatin with 100 μl of protein A/G beads in a total reaction volume of 500 μl or with 10 μg of Rabbit immunoglobulin G (IgG) (MilliporeSigma, 12-370) for 0.56 μg of chromatin with 100 μl of protein A/G beads in a total reaction volume of 1000 μl. The next morning, beads were isolated with a magnet and washed by rotating for 2 min at room temperature with (i) 20 mM tris-HCl (pH 8), 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, and 0.1% SDS; (ii) 20 mM tris-HCl (pH 8), 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, and 0.1% SDS; (iii) 20 mM tris-HCl (pH 8), 1 mM EDTA, 250 mM LiCl, 1% NP-40, and 1% Na-deoxycholate; and (iv) 10 mM tris-HCl (pH 7.4) and 1 mM EDTA. ChIP material was eluted twice from beads by 15-min incubations (rotating at room temperature) in 100 μl of 1% SDS with 0.1 M NaHCO3. DNA was reverse crosslinked from protein by incubating overnight at 65°C with 200 mM NaCl followed by protein degradation by adding 10 μl of concentrated TE buffer [0.1 M tris-HCl (pH 8) and 0.1 M EDTA] supplemented with proteinase K (Invitrogen, AM2546) for 1 hour (45°C). DNA was isolated using the QIAquick PCR Purification Kit (QIAGEN, 28106). Libraries were prepared by the NEBNext Ultra Library Prep Kit for Illumina with 12 cycles using dual index oligos for Illumina (New England Biolabs, E6440S). Library size and abundance were assessed by TapeStation (Agilent High Sensitivity D1000) and Qubit (Thermo Fisher Scientific, Q32851). Sequencing was performed on a NextSeq 2000, P2 flow cell, with paired end reads 2 × 61 cycles. qPCR assays for validation of the ChIPseq peak at the PDE1B locus was performed from a separate experiment using 4% of ChIPed DNA from both IgG and ZNF189 ChIP samples using primer sequences presented in table S4.
Microarray
Total RNA was isolated from adipocytes electroporated with siRNA targeting ZNF189 or nontargeting control using NucleoSpin RNA columns (Macherey-Nagel, 740955.250). RNA quality was assessed by TapeStation (Agilent, RNA ScreenTape), and all samples had RNA integrity numbers ≥ 9.3. Samples were hybridized to Clariom S Arrays (Applied Biosystems, 902916).
mRNA overexpression
ZNF189 mRNA was synthesized from purified PCR products from a codon-optimized ZNF189 gBlock (sequence and primers in table S4) using the HiScribe T7 ARCA mRNA Kit (New England Biolabs, E2060S). The ZNF189 mRNA was electroporated into adipocytes at a concentration of 70.16 nM on day 8 of differentiation. Cells were harvested 5 days later.
Cyclic GMP
Cyclic GMP levels were measured by Competitive enzyme-linked immunosorbent assay (ELISA) (Invitrogen, EMSCGMPL). Adipocytes were lysed in 120 μl in 0.1 M HCl with 1% Triton X-100 after stimulating for 20 min with ANP to induce cyclic GMP.
PDE1B inhibition
PDE1 inhibition was performed with 1 hour pre-incubation of adipocytes in media containing 1 μM ITI214 (MedChemExpress, HY-12501A), which was replaced with fresh assay media containing 1 μM ITI214 at the time of glycerol or cGMP induction.
Immunoprecipitation MS
Crosslinked, sonicated nuclei were isolated following the ChIPseq protocol outlined above. In total, 100 μg of protein per replicate was incubated overnight (rotating at 4°C) with 1 μg of ZNF189 antibody (Sigma-Aldrich, HPA034814-100UL) or Rabbit IgG (MilliporeSigma, 12-370) in total reaction volumes of 200 μl of denaturing buffer [50 mM tris (pH 8), 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 150 mM NaCl, and protease inhibitor cocktail]. The next morning, samples were incubated (1 hour rotating at 4°C) with 100 μl of protein A/G beads (Invitrogen, 10003D and 10001D) that were first washed twice with PBS with 0.02% Tween 20. Antibody-bead complexes were washed following the identical washing conditions to the ChIP protocol with two additional tris-EDTA washes [10 mM tris-HCl (pH 7.4) and 1 mM EDTA] to remove salts before proceeding to on-bead protein digestion for MS.
A 50 μl of trypsin-urea buffer [2 M urea, 50 mM tris-HCl (pH 7.5), 2 mM dithiothreitol, and trypsin (20 μg/ml)] was added to protein-bead complexes, and samples were mixed for 30 min at 1300 rpm in an orbital mixer (37°C). Liquid was removed from beads, which underwent a second incubation in 50 μl of chloroacetamide-urea buffer [2 M urea, 5 mM tris-HCl (pH 7.5), and 10 mM chloroacetamide] for 5 min at 1300 rpm (37°C) in the dark. Both liquid fractions collected from the beads were combined and further incubated by mixing overnight at 1300 rpm (room temperature).
Tryptic peptides were acidified to 1% trifluoro acetic acid and desalted using triple layers of C18 reverse phase membranes (Empore) in the STAGE-tipping format. On the membrane, peptides were washed twice with 0.1% formic acid and eluted with 2× 50 μl of 50% acetonitrile/0.1% formic acid followed by 50 μl of 80% acetonitrile/0.1% formic acid. After evaporation of the elution buffer, samples were resuspended in 2% acetonitrile/0.1% trifluoro acetic acid for measurement on the mass spectrometer. A total of four replicates were detected by liquid chromatography tandem MS across two separate instrument runs.
Assay for transposase-accessible chromatin using sequencing
ATACseq was performed on the basis of the Omni-ATAC protocol by Corces et al. (50). First, nuclei were isolated from ZNF189-depleted cells and control cells following the same steps outlined in the ChIPseq protocol above. Nuclei were resuspended in ATAC resuspension buffer with 0.1% Tween 20 for counting. Subsequently, 50,000 nuclei were washed for duplicate transposition reactions per siRNA. Transposition, DNA cleanup, PCR amplification, qPCR to determine additional cycles, and library purification were performed using reagents indicated in the Omni-ATAC protocol. Library size and abundance were assessed by TapeStation (Agilent High Sensitivity D1000) and Qubit (Thermo Fisher Scientific, Q32851). Libraries were sequenced on a NextSeq 2000, P2 flow cell, with paired end reads 2 × 64 cycles.
Cleavage under targets and Tagmentation
CUT&Tag was performed using a published protocol (51). In brief, nuclei were isolated from ZNF189-depleted and control cells following the same steps used for ChIPseq and ATACseq. CUT&Tag was performed on 500,000 cell nuclei per reaction. Primary antibodies were H3K27ac (1 μl; Abcam, ab4729) and H3K9me3 (1 μl; Abcam, ab8898); the secondary antibody was anti-rabbit (1 μl; EpiCypher, 13-0047). Samples were incubated with pAG-Tn5 (1 μl; EpiCypher, 15-1117) for 1 hour. After segmentation, the cleaved DNA was extracted using the DNA Clean & Concentrator-5 Kit (Zymo Research, D4013). Integrated DNA Technologies (IDT) primers (Illumina, 20027213) and PCR enzyme mix (New England Biolabs, M0541S) were used for library preparation, and AMPure beads (Beckman Coulter, A63881) were used for PCR cleanup. The prepared library samples were checked by the Qubit Fluorometric Quantification Kit (Thermo Fisher Scientific, Q33238) and TapeStation System 4200. Library samples were sequenced on NextSeq 2000 and P2 flow cell with paired end reads 2 × 59 cycles.
Quantifications
RNAi screen
For the RNAi screen, glycerol release and LDH data were analyzed using R v4.2.2. Outliers were identified using the interquartile range (IQR) method, additionally triplicates with coefficient of variation (CV) > 20% were excluded from further analysis. The reproducibility of the glycerol release assays was assessed by calculation of the Pearson correlation coefficient between the experimental replicates. Data were normalized and scaled using the robust z score method. Replicates with LDH release with robust z score > |5| were excluded from the statistical analysis. Targets that significantly affect glycerol release (“hits”) were identified using a one-way analysis of variance (ANOVA) with Dunnet’s post hoc test.
Screen hits were classified into functional clusters by entering all 37 proteins into a STRING network search (String Consortium, version 11.5) for proteins with medium confidence evidence for interactions. This identified clusters centered on JUNB, SRCAP, and TCERG1. These interactions were then supplemented by literature searches in Pubmed for all 37 hits to validate and identify additional interactions not included in the STRING database. In this way, final clusters were manually classified by experimental evidence for related functions. Spearman correlations between the expression of lipolysis hits and both isoprenaline- and ANP-stimulated lipolysis were computed by the Hmisc package in R.
Lipidomics
Samples were analyzed in both positive and negative ion modes with a resolution of mass-to-charge ratio (Rm/z) = 200 = 500,000 for MS and Rm/z = 200 = 30,000 for MS/MS experiments. MS/MS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1-Da increments (52). Both MS and MS/MS data were combined to monitor CE, DAG, and TAG ions as ammonium adducts; PC and PC O- as acetate adducts; and CL, PA, PE, PE O-, PG, PI, and PS as deprotonated anions. MS only was used to monitor LPA, LPE, LPE O-, LPI, and LPS as deprotonated anions and Cer, HexCer, SM, LPC, and LPC O- as acetate adducts. Data were analyzed with in-house developed lipid identification software based on LipidXplorer (53). Data postprocessing and normalization were performed using an in-house developed data management system. Only lipid identifications with a signal-to-noise ratio > 5 and a signal intensity fivefold higher than in corresponding blank samples were considered for further data analysis. Lipid species were included in statistical analysis if they had detectable values across all six replicates for both siRNA conditions. Principal components analysis was performed with the FactoMineR package in R. Confidence ellipses were calculated for a 0.95 confidence level. Species level differences in lipids were assessed by pairwise comparisons using eBayes with the limma package in R, with false discovery rate (FDR) correction.
Microarray
Microarray data were analyzed with packages available from R Bioconductor (www.bioconductor.org), R version 4.2.0. Normalization and calculation of gene expression were performed with the Robust Multichip Average expression measure using the oligo package (54). Before further analysis, a nonspecific filter was applied to include only coding genes with log2 expression signal > 5 in at least 50% of all samples. To identify differentially expressed genes between the two sample groups, the remaining 26,787 transcripts were analyzed using the Linear Models for Microarray Data (limma) package [version 3.52.2.; (55)]. Pathway analysis was performed for differentially expressed genes (adjusted P value < 0.05) with overrepresentation and pathway topology analysis in Reactome (56). Significantly enriched pathways by overrepresentation analysis were ranked by the number of differentially expressed entities found in the pathway, and the largest cluster of pathways (signal transduction) is presented.
Chromatin immunoprecipitation
The nf-core/chipseq pipeline [https://zenodo.org/record/7139814#.ZC3eMXtByUk (57)] was used to perform the quality control, mapping, and peak calling of the ChIPseq analysis in conjunction with Docker. All data were processed relative to the human genome hg38. For a condensed overview of the pipeline, the bioinformatics tools used in each step and an extensive list of citations, please see the pipeline homepage (https://github.com/nf-core/chipseq). The Homer (58) package was used to perform the motif enrichment analysis within the identified ChIPed regions using default settings. For genomic annotations, the bam files were converted to tag directories using Homer (58), which was also subsequently used for peak calling and annotation of the identified peaks. The genomic annotation produced by Homer was used to calculate the enrichment of the peaks in the various functional genomic regions using hypergeometric testing. Bedgraph files were produced using Homer and normalized to 10 million reads. The BETA package (15) was used to integrate ChIPseq data with differential gene expression data of siZNF189 versus siC adipocytes to predict whether ZNF189 has activating or repressive function and to infer direct target genes of ZNF189. The cutoff for significantly regulated genes in this analysis was an adjusted P value < 0.1 and absolute log2 fold change above 0.5. The heatmap of ChIP signals was plotted using computeMatrix and plotHeatmap packages in Deeptools (59).
Assay for transposase-accessible chromatin using sequencing
To process the raw Bcl files obtained from Illumina sequencing, we used the bcl2fastq program to perform conversion and demultiplexing, resulting in fastq files. We used STAR (60) to index to the human reference genome (hg38/GRCh38), which was obtained from the University of California, Santa Cruz and then aligned the fastq files. Next, we used a sliding window approach from the csaw (61) R package to count reads across the genome. Reads were counted in 400–base pair (bp) windows with a distance of 100 bp between consecutive windows. In addition, for normalization purposes, we counted reads in 10-kb bins across the genome. We then detected differential windows using the edgeR (62) library, with normalization factors calculated using the 10-kb bin counts. P values were adjusted for multiple hypothesis testing (FDR), and windows with P values < 0.05 were termed significant. The heatmap of ATACseq peak signals was plotted using computeMatrix and plotHeatmap packages in Deeptools (59). The heatmap of ATACseq and microarray signals was plotted with the ComplexHeatmap package in R.
Cleavage under targets and Tagmentation
Sequencing files (fastq) were aligned to the GRCh38/hg38 version of the human reference genome using Bowtie2 on the Galaxy platform. Peak calling was done using MACS2 (63). The statistical analysis for differential expression was further performed using edgeR. Peak changes with an adjusted P value < 0.05 were considered differential peaks. In the quantitative analysis (percentage) for CUT&Tag, total tags (107) were used as the normalization factor. The sequencing tags (SAM) were then imported to HOMER (58). Peak coverage was calculated with the HOMER tool Histograms Tag (annotatePeaks.pl) and visualized in R.
Immunoprecipitation MS
Raw MS data were analyzed with Spectronaut (v15.7.220308.50606) using the direct DIA analysis mode. All analysis parameters are documented as part of the PRIDE submission in the setup.txt file. In short, spectra were searched against UniProts human regular and “additional” FASTA databases (UP000005640_9606_2020-06; 20,609 and 77,157 entries, respectively) and the MaxQuant contaminants list (245 entries). Carbamidomethylation (C) was specified as a fixed- and acetylation (N-term) and oxidation (M) as variable modifications. For protein quantification, the MaxLFQ algorithm was used.
Transcript and protein comparisons
Cap analysis of gene expression of all 1530 genes from the siGENOME SMARTpool siRNA Library “Human Transcription Factor” library was assessed at days 4, 8, and 12 of adipogenesis. Measures were obtained from published data (64), and the cutoff for the selection of genes for screening was established by tags per million > 10.
Expression for the two promoters of the PDE1B gene (encoding PDE1B1 and PDE1B2) was downloaded from the Fantom5 CAGE tag start site (CTSS) dataset (7). The respective promoter expression was ranked for all samples in the dataset, and samples were identified as adipocyte versus other with a rug plot produced in R. Only samples with measurable gene expression are presented. To evaluate the conservation of the ZNF189 gene (ENSG00000136870), the Ensembl database containing 200 species was used to identify orthologs. The phylogenetic tree with significant gene gain/loss events was downloaded and mapped in R v4.2.2 using packages ggtree v3.6.2 (65) and treeio v1.22.0 (66). Next, to evaluate the protein conservation within the placentals taxon, 10 sequences representing the major clades within the taxon were obtained from Uniprot (A0A6P5PHN9, G1P627, A0A452EU80, A0A2Y9QEN8, A0A2I3T8W9, G1T8E4, G3TH83, A0A8I3NMD7, A0A287B140, and O75820) and multiple sequence alignment was performed in Jalview v2.11.2.6 (67) with MafftWS alignment tool. Conservation of the aligned amino acid sequences was calculated as the Valdar conservation score (68). The protein sequence corresponding to the adipocyte-specific PDE1B2 gene isoform identified in the Fantom dataset (NP_001159447.1) was obtained from National Center for Biotechnology Information (NCBI) Gene database and aligned with the canonical protein sequence (NP_000915.1) using Muscle (69) in Jalview (v2.11.2.6) (67).
Statistical analysis
Statistical testing was performed in GraphPad Prism version 9.5.0 (GraphPad Software, San Diego, CA, USA) and R v4.2.2. Data were tested for normality by D’Agostino and Pearson omnibus K2 normality testing or Shapiro-Wilk test in cases of ≤8 samples per group, and statistics were performed on log-transformed data when required to achieve normality. Group data are presented as means ± SEM. For the meta-analysis of the relationship between ZNF189 mRNA and clinical parameters, both random and common effects from Spearman correlations are presented.
Acknowledgments
We thank L. Massier for performing the meta-analysis of the clinical cohorts and our support staff, K. Hertel, N. Wang, and T. De Castro Barbosa (Department of Medicine H7, Karolinska Institutet), for the valuable help in collecting and processing clinical samples. We thank R. V. Farese Jr and T. C. Walther (Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center) for allowing us to use their lipidomic platform. Generation and processing of microarray and sequencing results were conducted together with the Bioinformatics and Expression Analysis core facility, and imaging was performed at the Live Cell Imaging core facility, both at Karolinska Institutet.
Funding: This work was supported by the CIMED (M.R. and N.M.), DFG Emmy Noether KR5166/2 (to N.K.), DFG BATenergy TRR 333/1-450149205 (to N.K.), EFSD/Lilly research fellowship (to Z.H.), ERC-SyG SPHERES 856404 (to M.R.), European Foundation for the Study of Diabetes Future Leaders award (to N.M., N.K., and R.F.), Knut & Alice Wallenberg’s foundation (to M.R.), Margareta af Uggla’s foundation (to M.R.), NovoNordisk Foundation MeRIAD consortium 0064142 (to M.R.), NovoNordisk Foundation NNF20OC0061149 (to N.M.), Novo Nordisk Foundation NNF21OC0070256 (to E.T.), Stockholm County Council (to M.R.), Strategic Research Program in Diabetes at Karolinska Institutet (to H.G., M.R., and N.M.), Swedish Diabetes Foundation (to M.R.), Swedish Research Council (to M.R. and N.M.), Swedish Research Council 2020-01150 (to E.T.), and Swedish Research Council 2019-01884 (to R.F.). A.C.L. is supported by a Novo Nordisk postdoctoral fellowship run in partnership with Karolinska Institutet. M.O.-H. is supported by postdoctoral fellowship from the strategic research program in diabetes at Karolinska Institutet.
Author contributions: Conceptualization: P.A., H.G., M.R. and N.M. Methodology: A.C.L., M.H., J.J., Z.H., M.O.-H., G.R., F.K., Y.A.A., S.B., and N.W. Investigation: A.C.L., M.H., D.Z., J.J., Z.H., M.O.-H., G.R., N.W., J.L., T.J., P.P., H.G., M.R., and N.M. Visualization: M.H., D.Z., J.J., Z.H., A.D., H.G., and N.M. Supervision: N.K., R.F., E.T., H.G., M.R. and N.M. Writing—original draft: A.C.L., M.R. and N.M. Writing—review and editing: All authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data for the lipidomics, the meta-analyses, and the perturbation screen as well as relevant codes are available at Dryad using this link: https://doi.org/10.5061/dryad.8pk0p2nvh. Genomics data are deposited in NCBI’s Gene Expression Omnibus (70) with the accession number GSE247818. Proteomic data are deposited in the PRIDE database (71) with the accession number PXD046964.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Tables S1 to S4
REFERENCES AND NOTES
- 1.Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., Zimmermann R., Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281, 40236–40241 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Grabner G. F., Xie H., Schweiger M., Zechner R., Lipolysis: Cellular mechanisms for lipid mobilization from fat stores. Nat. Metab. 3, 1445–1465 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Reilly S. M., Saltiel A. R., Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 13, 633–643 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Markussen L. K., Rondini E. A., Johansen O. S., Madsen J. G. S., Sustarsic E. G., Marcher A.-B., Hansen J. B., Gerhart-Hines Z., Granneman J. G., Mandrup S., Lipolysis regulates major transcriptional programs in brown adipocytes. Nat. Commun. 13, 3956 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rydén M., Jocken J., van Harmelen V., Dicker A., Hoffstedt J., Wirén M., Blomqvist L., Mairal A., Langin D., Blaak E., Arner P., Comparative studies of the role of hormone-sensitive lipase and adipose triglyceride lipase in human fat cell lipolysis. Am. J. Physiol. Endocrinol. Metab. 292, 1847–1855 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C., Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266, 11341–11346 (1991). [PubMed] [Google Scholar]
- 7.Forrest A. R. R., Kawaji H., Rehli M., Baillie J. K., de Hoon M. J. L., Haberle V., Lassmann T., Kulakovskiy I. V., Lizio M., Itoh M., Andersson R., Mungall C. J., Meehan T. F., Schmeier S., Bertin N., Jørgensen M., Dimont E., Arner E., Schmidl C., Schaefer U., Medvedeva Y. A., Plessy C., Vitezic M., Severin J., Semple C. A., Ishizu Y., Young R. S., Francescatto M., Alam I., Albanese D., Altschuler G. M., Arakawa T., Archer J. A. C., Arner P., Babina M., Rennie S., Balwierz P. J., Beckhouse A. G., Pradhan-Bhatt S., Blake J. A., Blumenthal A., Bodega B., Bonetti A., Briggs J., Brombacher F., Burroughs A. M., Califano A., Cannistraci C. V., Carbajo D., Chen Y., Chierici M., Ciani Y., Clevers H. C., Dalla E., Davis C. A., Detmar M., Diehl A. D., Dohi T., Drabløs F., Edge A. S. B., Edinger M., Ekwall K., Endoh M., Enomoto H., Fagiolini M., Fairbairn L., Fang H., Farach-Carson M. C., Faulkner G. J., Favorov A. V., Fisher M. E., Frith M. C., Fujita R., Fukuda S., Furlanello C., Furino M., Furusawa J.-i., Geijtenbeek T. B., Gibson A. P., Gingeras T., Goldowitz D., Gough J., Guhl S., Guler R., Gustincich S., Ha T. J., Hamaguchi M., Hara M., Harbers M., Harshbarger J., Hasegawa A., Hasegawa Y., Hashimoto T., Herlyn M., Hitchens K. J., Sui S. J. H., Hofmann O. M., Hoof I., Hori F., Huminiecki L., Iida K., Ikawa T., Jankovic B. R., Jia H., Joshi A., Jurman G., Kaczkowski B., Kai C., Kaida K., Kaiho A., Kajiyama K., Kanamori-Katayama M., Kasianov A. S., Kasukawa T., Katayama S., Kato S., Kawaguchi S., Kawamoto H., Kawamura Y. I., Kawashima T., Kempfle J. S., Kenna T. J., Kere J., Khachigian L. M., Kitamura T., Klinken S. P., Knox A. J., Kojima M., Kojima S., Kondo N., Koseki H., Koyasu S., Krampitz S., Kubosaki A., Kwon A. T., Laros J. F. J., Lee W., Lennartsson A., Li K., Lilje B., Lipovich L., Mackay-Sim A., Manabe R.-i., Mar J. C., Marchand B., Mathelier A., Mejhert N., Meynert A., Mizuno Y., de Lima Morais D. A., Morikawa H., Morimoto M., Moro K., Motakis E., Motohashi H., Mummery C. L., Murata M., Nagao-Sato S., Nakachi Y., Nakahara F., Nakamura T., Nakamura Y., Nakazato K., van Nimwegen E., Ninomiya N., Nishiyori H., Noma S., Noma S., Noazaki T., Ogishima S., Ohkura N., Ohimiya H., Ohno H., Ohshima M., Okada-Hatakeyama M., Okazaki Y., Orlando V., Ovchinnikov D. A., Pain A., Passier R., Patrikakis M., Persson H., Piazza S., Prendergast J. G. D., Rackham O. J. L., Ramilowski J. A., Rashid M., Ravasi T., Rizzu P., Roncador M., Roy S., Rye M. B., Saijyo E., Sajantila A., Saka A., Sakaguchi S., Sakai M., Sato H., Savvi S., Saxena A., Schneider C., Schultes E. A., Schulze-Tanzil G. G., Schwegmann A., Sengstag T., Sheng G., Shimoji H., Shimoni Y., Shin J. W., Simon C., Sugiyama D., Sugiyama T., Suzuki M., Suzuki N., Swoboda R. K., ‘t Hoen P. A. C., Tagami M., Takahashi N., Takai J., Tanaka H., Tatsukawa H., Tatum Z., Thompson M., Toyodo H., Toyoda T., Valen E., van de Wetering M., van den Berg L. M., Verado R., Vijayan D., Vorontsov I. E., Wasserman W. W., Watanabe S., Wells C. A., Winteringham L. N., Wolvetang E., Wood E. J., Yamaguchi Y., Yamamoto M., Yoneda M., Yonekura Y., Yoshida S., Zabierowski S. E., Zhang P. G., Zhao X., Zucchelli S., Summers K. M., Suzuki H., Daub C. O., Kawai J., Heutink P., Hide W., Freeman T. C., Lenhard B., Bajic V. B., Taylor M. S., Makeev V. J., Sandelin A., Hume D. A., Carninci P., Hayashizaki Y.; FANTOM Consortium and the RIKEN PMI and CLST (DGT) , A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modica S., Straub L. G., Balaz M., Sun W., Varga L., Stefanicka P., Profant M., Simon E., Neubauer H., Ukropcova B., Ukropec J., Wolfrum C., Bmp4 promotes a brown to white-like adipocyte shift. Cell Rep. 16, 2243–2258 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Yogosawa S., Mizutani S., Ogawa Y., Izumi T., Activin receptor-like kinase 7 suppresses lipolysis to accumulate fat in obesity through downregulation of peroxisome proliferator–activated receptor γ and C/EBPα. Diabetes 62, 115–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massier L., Jalkanen J., Elmastas M., Zhong J., Wang T., Nono Nankam P. A., Frendo-Cumbo S., Bäckdahl J., Subramanian N., Sekine T., Kerr A. G., Tseng B. T. P., Laurencikiene J., Buggert M., Lourda M., Kublickiene K., Bhalla N., Andersson A., Valsesia A., Astrup A., Blaak E. E., Ståhl P. L., Viguerie N., Langin D., Wolfrum C., Blüher M., Rydén M., Mejhert N., An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat. Commun. 14, 1438 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laakso M., Kuusisto J., Stančáková A., Kuulasmaa T., Pajukanta P., Lusis A. J., Collins F. S., Mohlke K. L., Boehnke M., The metabolic syndrome in men study: A resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 58, 481–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maqdasy S., Lecoutre S., Renzi G., Frendo-Cumbo S., Rizo-Roca D., Moritz T., Juvany M., Hodek O., Gao H., Couchet M., Witting M., Kerr A., Bergo M. O., Choudhury R. P., Aouadi M., Zierath J. R., Krook A., Mejhert N., Rydén M., Impaired phosphocreatine metabolism in white adipocytes promotes inflammation. Nat. Metab. 4, 190–202 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recazens E., Mouisel E., Langin D., Hormone-sensitive lipase: Sixty years later. Prog. Lipid Res. 82, 101084 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Najafabadi H. S., Mnaimneh S., Schmitges F. W., Garton M., Lam K. N., Yang A., Albu M., Weirauch M. T., Radovani E., Kim P. M., Greenblatt J., Frey B. J., Hughes T. R., C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C. A., Zhang Y., Liu X. S., Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 8, 2502–2515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke H., Wang H., Crystal structures of phosphodiesterases and implications on substrate specificity and inhibitor selectivity. Curr. Top. Med. Chem. 7, 391–403 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Urrutia R., KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ecco G., Imbeault M., Trono D., KRAB zinc finger proteins. Development 144, 2719–2729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alerasool N., Segal D., Lee H., Taipale M., An efficient KRAB domain for CRISPRi applications in human cells. Nat. Methods 17, 1093–1096 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Reynolds N., O’Shaughnessy A., Hendrich B., Transcriptional repressors: Multifaceted regulators of gene expression. Development 140, 505–512 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Huang Z., Efthymiadou A., Liang N., Fan R., Treuter E., Antagonistic action of GPS2 and KDM1A at enhancers governs alternative macrophage activation by interleukin 4. Nucleic Acids Res. 51, 1067–1086 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J., SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornelöv S., Reynolds N., Xenophontos M., Gharbi S., Johnstone E., Floyd R., Ralser M., Signolet J., Loos R., Dietmann S., Bertone P., Hendrich B., The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell 71, 56–72.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M.-M., Rauscher F. J., PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baillie G. S., Tejeda G. S., Kelly M. P., Therapeutic targeting of 3′,5′-cyclic nucleotide phosphodiesterases: Inhibition and beyond. Nat. Rev. Drug Discov. 18, 770–796 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narlikar G. J., Fan H.-Y., Kingston R. E., Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Liu T., Mi L., Xiong J., Orchard P., Yu Q., Yu L., Zhao X.-Y., Meng Z.-X., Parker S. C. J., Lin J. D., Li S., BAF60a deficiency uncouples chromatin accessibility and cold sensitivity from white fat browning. Nat. Commun. 11, 2379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajbhandari P., Thomas B. J., Feng A.-C., Hong C., Wang J., Vergnes L., Sallam T., Wang B., Sandhu J., Seldin M. M., Lusis A. J., Fong L. G., Katz M., Lee R., Young S. G., Reue K., Smale S. T., Tontonoz P., IL-10 signaling remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell 172, 218–233.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyengar S., Farnham P. J., KAP1 protein: An enigmatic master regulator of the genome. J. Biol. Chem. 286, 26267–26276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalgaard K., Landgraf K., Heyne S., Lempradl A., Longinotto J., Gossens K., Ruf M., Orthofer M., Strogantsev R., Selvaraj M., Lu T. T.-H., Casas E., Teperino R., Surani M. A., Zvetkova I., Rimmington D., Tung Y. C. L., Lam B., Larder R., Yeo G. S. H., O’Rahilly S., Vavouri T., Whitelaw E., Penninger J. M., Jenuwein T., Cheung C.-L., Ferguson-Smith A. C., Coll A. P., Körner A., Pospisilik J. A., Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell 164, 353–364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilherme A., Rowland L. A., Wang H., Czech M. P., The adipocyte supersystem of insulin and cAMP signaling. Trends Cell Biol. 33, 340–354 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra S., Sadagopan N., Dunkerly-Eyring B., Rodriguez S., Sarver D. C., Ceddia R. P., Murphy S. A., Knutsdottir H., Jani V. P., Ashok D., Oeing C. U., O’Rourke B., Gangoiti J. A., Sears D. D., Wong G. W., Collins S., Kass D. A., Inhibition of phosphodiesterase type 9 reduces obesity and cardiometabolic syndrome in mice. J. Clin. Investig. 131, e148798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sancar G., Liu S., Gasser E., Alvarez J. G., Moutos C., Kim K., van Zutphen T., Wang Y., Huddy T. F., Ross B., Dai Y., Zepeda D., Collins B., Tilley E., Kolar M. J., Yu R. T., Atkins A. R., van Dijk T. H., Saghatelian A., Jonker J. W., Downes M., Evans R. M., FGF1 and insulin control lipolysis by convergent pathways. Cell Metab. 34, 171–183.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry J.-R., Mager D. L., Wilhelm B. T., Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 19, 640–648 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Fajas L., Auboeuf D., Raspé E., Schoonjans K., Lefebvre A.-M., Saladin R., Najib J., Laville M., Fruchart J.-C., Deeb S., Vidal-Puig A., Flier J., Briggs M. R., Staels B., Vidal H., Auwerx J., The organization, promoter analysis, and expression of the human PPARγ gene. J. Biol. Chem. 272, 18779–18789 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Herman M. A., Peroni O. D., Villoria J., Schön M. R., Abumrad N. A., Blüher M., Klein S., Kahn B. B., A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484, 333–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruas J. L., White J. P., Rao R. R., Kleiner S., Brannan K. T., Harrison B. C., Greene N. P., Wu J., Estall J. L., Irving B. A., Lanza I. R., Rasbach K. A., Okutsu M., Nair K. S., Yan Z., Leinwand L. A., Spiegelman B. M., A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151, 1319–1331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian K., Tol M. J., Wu J., Uchiyama L. F., Xiao X., Cui L., Bedard A. H., Weston T. A., Rajendran P. S., Vergnes L., Shimanaka Y., Yin Y., Jami-Alahmadi Y., Cohn W., Bajar B. T., Lin C.-H., Jin B., DeNardo L. A., Black D. L., Whitelegge J. P., Wohlschlegel J. A., Reue K., Shivkumar K., Chen F.-J., Young S. G., Li P., Tontonoz P., CLSTN3β enforces adipocyte multilocularity to facilitate lipid utilization. Nature 613, 160–168 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentine J. M., Ahmadian M., Keinan O., Abu-Odeh M., Zhao P., Zhou X., Keller M. P., Gao H., Yu R. T., Liddle C., Downes M., Zhang J., Lusis A. J., Attie A. D., Evans R. M., Rydén M., Saltiel A. R., β3-Adrenergic receptor downregulation leads to adipocyte catecholamine resistance in obesity. J. Clin. Invest. 132, e153357 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You T., Berman D. M., Ryan A. S., Nicklas B. J., Effects of hypocaloric diet and exercise training on inflammation and adipocyte lipolysis in obese postmenopausal women. J. Clin. Endocrinol. Metab. 89, 1739–1746 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Kim N.-J., Baek J.-H., Lee J., Kim H., Song J.-K., Chun K.-H., The transcription factor NFIL3 controls regulatory T-cell function and stability. Exp. Mol. Med. 51, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinent M., Prokesch A., Hackl H., Voshol P. J., Klatzer A., Walenta E., Panzenboeck U., Kenner L., Trajanoski Z., Hoefler G., Bogner-Strauss J. G., Adipose triglyceride lipase and hormone-sensitive lipase are involved in fat loss in JunB-deficient mice. Endocrinology 152, 2678–2689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J.-L., Lu X.-J., Zou K.-L., Ye K., Krüppel-like factor 2 promotes liver steatosis through upregulation of CD36. J. Lipid Res. 55, 32–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanazawa A., Kawamura Y., Sekine A., Iida A., Tsunoda T., Kashiwagi A., Tanaka Y., Babazono T., Matsuda M., Kawai K., Iiizumi T., Fujioka T., Imanishi M., Kaku K., Iwamoto Y., Kawamori R., Kikkawa R., Nakamura Y., Maeda S., Single nucleotide polymorphisms in the gene encoding Krüppel-like factor 7 are associated with type 2 diabetes. Diabetologia 48, 1315–1322 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Chen W., Gao C., Shen J., Yao L., Liang X., Chen Z., The expression and prognostic value of REXO4 in hepatocellular carcinoma. J Gastrointest. Oncol. 12, 1704–1717 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jäger J., Greiner V., Strzoda D., Seibert O., Niopek K., Sijmonsma T. P., Schäfer M., Jones A., De Guia R., Martignoni M., Dallinga-Thie G. M., Diaz M. B., Hofmann T. G., Herzig S., Hepatic transforming growth factor-β 1 stimulated clone-22 D1 controls systemic cholesterol metabolism. Mol. Metab. 3, 155–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Wang T., Wei J. J., Sabatini D. M., Lander E. S., Genetic screens in human cells using the CRISPR-Cas9 system. Science. 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Folch J., Lees M., Stanley G. H. S., A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957). [PubMed] [Google Scholar]
- 50.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaya-Okur H. S., Janssens D. H., Henikoff J. G., Ahmad K., Henikoff S., Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Surma M. A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D., Simons K., Masoodi M., Sampaio J. L., An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 117, 1540–1549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herzog R., Schuhmann K., Schwudke D., Sampaio J. L., Bornstein S. R., Schroeder M., Shevchenko A., LipidXplorer: A software for consensual cross-platform lipidomics. PLOS One 7, e29851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvalho B. S., Irizarry R. A., A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., Deng C., Varusai T., Ragueneau E., Haider Y., May B., Shamovsky V., Weiser J., Brunson T., Sanati N., Beckman L., Shao X., Fabregat A., Sidiropoulos K., Murillo J., Viteri G., Cook J., Shorser S., Bader G., Demir E., Sander C., Haw R., Wu G., Stein L., Hermjakob H., D’Eustachio P., The reactome pathway knowledgebase 2022. Nucleic Acids Res. 50, D687–D692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ewels P. A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M. U., Di Tommaso P., Nahnsen S., The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lun A. T. L., Smyth G. K., csaw: A Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44, e45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ehrlund A., Mejhert N., Björk C., Andersson R., Kulyté A., Åström G., Itoh M., Kawaji H., Lassmann T., Daub C. O., Carninci P., Forrest A. R. R., Hayashizaki Y., Sandelin A., Ingelsson E.; FANTOM Consortium, Rydén M., Laurencikiene J., Arner P., Arner E., Transcriptional dynamics during human adipogenesis and its link to adipose morphology and distribution. Diabetes 66, 218–230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y., ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods. Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 66.Wang L.-G., Lam T. T.-Y., Xu S., Dai Z., Zhou L., Feng T., Guo P., Dunn C. W., Jones B. R., Bradley T., Zhu H., Guan Y., Jiang Y., Yu G., Treeio: An R package for phylogenetic tree input and output with richly annotated and associated data. Mol. Biol. Evol. 37, 599–603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valdar W. S. J., Thornton J. M., Protein–protein interfaces: Analysis of amino acid conservation in homodimers. Proteins 42, 108–124 (2001). [PubMed] [Google Scholar]
- 69.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yılmaz Ş., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., Vizcaíno J. A., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Tables S1 to S4