Abstract
The dual role of CD8+ T cells in influenza control and lung pathology is increasingly appreciated. To explore whether protective and pathological functions can be linked to specific subsets, we dissected CD8+ T responses in influenza-infected murine lungs. Our single-cell RNA-sequencing (scRNA-seq) analysis revealed notable diversity in CD8+ T subpopulations during peak viral load and infection-resolved state. While enrichment of a Cxcr3hi CD8+ T effector subset was associated with a more robust cytotoxic response, both CD8+ T effector and central memory exhibited equally potent effector potential. The scRNA-seq analysis identified unique regulons regulating the cytotoxic response in CD8+ T cells. The late-stage CD8+ T blockade in influenza-cleared lungs or continuous CXCR3 blockade mitigated lung injury without affecting viral clearance. Furthermore, adoptive transfer of wild-type CD8+ T cells exacerbated influenza lung pathology in Cxcr3−/− mice. Collectively, our data imply that CXCR3 interception could have a therapeutic effect in preventing influenza-linked lung injury.
The CXCR3-expressing CD8+ T subset promotes severe lung pathology and exacerbates influenza disease severity.
INTRODUCTION
Influenza exhibits a complex disease phenotype, ranging from a self-limiting mild infection to severe life-threatening pneumonia. Seasonal influenza vaccines offer limited efficacy (1), and influenza remains a major public health problem with >30,000 annual deaths and more than $10.4 billion health care costs (2). Severe influenza pneumonia manifestations are profound airway lung and vascular injuries, affecting gas exchange and requiring hospitalization (3–5). The recovery is often slow and may leave patients with permanent lung damage (6–8). Most of these complications are attributed to the host’s own defense mechanisms because host responses while executing control of viral load drive collateral lung damage (9, 10). Viral load in the lung is one of the factors that affect the balance between infection resolution and immune-mediated lung pathology, but paradoxically, the acute injury continues and persists in the lungs after viral clearance (11, 12). Despite numerous studies exploring the pathogenesis of influenza, there remains a knowledge gap in how the host response to influenza turns pathologic and which subsets of immune cells are the major drivers of lung pathology.
Although CD8+ T cells are indispensable for influenza control, the role of CD8+ T cells in lung pathology is increasingly reported (9, 13–15). Data from the 2009 H1N1 pandemic show a strong correlation of the increased numbers/responses of CD8+ T cells with influenza disease severity (16). However, mice with impaired CD8+ T cell responses eventually succumb to infection due to their inability to control viral load (17). Thus, paradoxically, CD8+ T cells while indispensable to influenza control, also contribute to lung pathology that exacerbates the disease. These responses rely on the development of cytotoxic CD8+ T cells armed in cytotoxic molecules such as perforin and granzyme B (GzmB), which exert a cytolytic function to kill virus-infected cells (18–20). However, the magnitude of CD8+ T cell cytotoxic function needs to be appropriately gauged for effective virus removal without causing excessive collateral damage (13). A recent report of influenza-specific cytotoxic CD8+ T cells, causing bystander damage to noninfected alveolar epithelial cells (14), provides evidence that the balance between protective and pathological cytotoxic functions can be easily disturbed. Besides their direct interaction with epithelial cells, CD8+ T cells produce a range of inflammatory cytokines that can promote inflammation and damage structural cells, compromising the lung barrier (14, 21, 22).
Interferons (IFNs) are crucial to regulating CD8+ T cell responses via IFN-inducible chemokines, C-X-C motif chemokine 9 (CXCL9) and 10 (CXCL10) (23), and, consequently, regulate the CXCR3-dependent effector function (24, 25). While the role of CXCR3 has been described in the context of influenza CD8+ T cell effector/memory generation, there is a knowledge gap if CXCR3-CD8+ T cell axis regulates the development of pathologic host responses during primary influenza infection. In addition, it remains to be determined whether IFN response diversity differentially regulates the development of CD8+ T cytotoxic responses during primary influenza infection. Therefore, the pathologic function of CD8+ T cells in the context of influenza disease warrants further investigation.
In this study, we sought to elucidate the role of CD8+ T cells during influenza A virus (IAV) infection, using a mouse model of severe but nonlethal IAV that displays a robust lung injury during the peak viral load at 7 days postinfection (dpi) as well as in postviral resolution period (14 dpi), mimicking severe human influenza with persisting lung injury. We characterized the CD8+ T cell transcriptional diversity and studied the regulation of CD8+ T cell pathways in the lung during the peak viral load and viral resolved phases. We found that CD8+ T cells expressing IFN-inducible chemokine receptor, CXCR3 (Cxcr3hi CD8+ T cells), exhibited a dominant enrichment of IFN-I–induced molecular pathways and produced higher levels of cytolytic molecules in the lung even after viral clearance at 14 dpi. The antibody-mediated blockade of CXCR3 (during active infection) resulted in reduced lung injury, disease severity, and faster resolution of lung injury. Furthermore, the CD8+ T blockade in virus-cleared lungs led to an expedited resolution of lung injury, supporting the findings from the CXCR3 blockade experiments. The adoptive transfer of Cxcr3+/+ [wild-type (WT)] CD8+ T cells enhanced IAV lung vascular pathology in Cxcr3−/− mice, mimicking the lung pathologic phenotype of WT mice. These findings provide strong support that the CXCR3-CD8 axis is dispensable for IAV clearance but mediates IAV-associated lung pathology and suggest that interference with the CXCR3 pathway could be explored therapeutically.
RESULTS
Activated CD8+ T cells persist in the lung after the viral clearance and correlate with lung injury
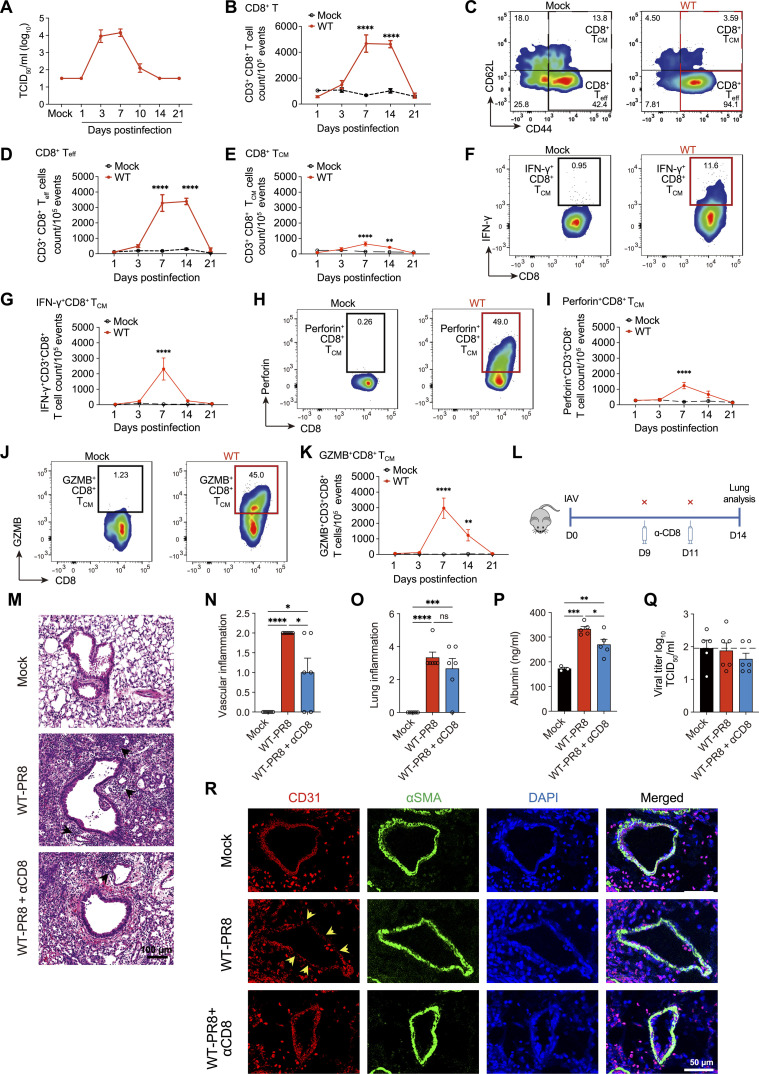
To establish spatiotemporal relation between lung pathology and the presence of CD8+ T cells, we conducted studies in the severe IAV model, characterized by substantial lung inflammation, airway, and vascular damage with a substantial loss of body weight (~25%). The inflammation and acute lung damage were determined by a combined metric of hematoxylin and eosin (H&E) staining of lung sections and the levels of lactate dehydrogenase (LDH) and albumin in the bronchoalveolar lavage fluid (BALF). While the measure of acute lung injury, LDH, and albumin levels peaked at 7 dpi and normalized by 14 dpi, histological analysis showed sustained lung inflammation and consolidation throughout 14 dpi before substantial resolution by 21 dpi (fig. S1, A to G). IAV load in the lung was mostly cleared by day 10 (Fig. 1A). We next characterized the CD8+ T cells in mock- and IAV-infected lungs during different stages of lung injury, at 3 to 21 dpi. The CD8+ T cells demonstrated a peak quantitative increase by 7 dpi, with their level remaining high until 14 dpi (Fig. 1B). Further analysis revealed that CD8+ T cells remained activated until 14 dpi (CD44hi CD62Llow) (Fig. 1, C to E). Next, we characterized the effector and cytolytic function of CD8+ T cells by examining the intracellular expression of IFN-γ, perforin, and GzmB during different stages of IAV infection. The lung single cells from mock- and IAV-infected mice were stimulated in vitro with IAV peptide NP366–374 (10 μg/ml) for 6 hours before intracellular staining (26). We observed a peak of IFN-γ response on 7 dpi and its complete contraction by 14 dpi (Fig. 1, F and G). The expression of cytolytic molecules, perforin, and GzmB also peaked at 7 dpi. However, contrary to the IFN-γ–expressing CD8+ T cells, perforin- and GzmB-expressing CD8+ T cells were detected in the lung well beyond the viral clearance time point (>day 14) (Fig. 1, H to K). These data show that while CD8+ T cells rapidly down-regulated their effector function (IFN-γ) after viral clearance, cytotoxic CD8+ T cells persist in virus-cleared lungs correlating with lung injury. Because IFN-γ is typically used to identify antigen-specific CD8+ T cells from the effector phase into memory (27), further studies are required to identify the pathophysiological mechanisms that could explain the uncoupling of effector and cytotoxic responses in CD8+ T subsets during different stages of IAV infection.
Fig. 1. Cellular diversity and relative distribution of lung CD8+ T cells in an influenza model.
Wild-type (WT) (C57BL/6) mice were mock-infected with phosphate-buffered saline (PBS) or infected with 1000 plaque-forming units (PFUs) of influenza A virus (IAV) (PR8) intranasally. At the indicated time points, mice were euthanized, and the bronchoalveolar lavage fluid (BALF) and lungs were aseptically collected. (A) TCID50 kinetics of influenza load in lung homogenates of PR8-infected mice. Data are shown as means ± SEM, representative of two different experiments with n = 5 per group and time point. (B) Kinetics of CD8+ T cells. (C) Representative graphs showing the gating for effector CD8+ Teff cells (CD44+CD62L−) and central memory CD8+ TCM (CD44+CD62L+) in mock-infected (black boxes) and PR8-infected WT (red boxes) mice. (D) Count of CD8+ Teff cells (CD44+CD62L). (E) CD8+ TCM (CD44+CD62L+) cells from mock- and PR8-infected mice, with gating shown in (C). (F to K) FACS representative graphs and kinetics of IFN-γ+–expressing [(F) and (G)], perforin+-expressing [(H) and (I)], and granzyme B+ (GzmB)–expressing [(J) and (K)] CD8+ T cells. Data pooled from two independent experiments in (B) to (K): with n = 5 to 10 per group per time point. (L) Timeline of late-stage CD8 antibody depletion in an IAV model. (M to O) H&E staining of lungs (M) with a score of vascular damage (N) and inflammation (O). (P) Albumin levels in BALF. (Q) Viral titer in lung homogenate measured by TCID50. (R) Microscroscopic images of immunofluorescent staining of lung endothelial cells. Data are representative of one independent experiment with n = 5 to 6 per group. The data are shown as means ± SEM. Two-way ANOVA with Sidak multiple comparisons test [(B) to (K)] or one-way ANOVA with Tukey post hoc test [(N) to (Q)] of the means was used for statistical significance; significant differences at indicated time points are shown, *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0 0.001. ns, not significant.
Late-stage blockade of CD8+ T cells promotes faster resolution of lung injury in virus-cleared lungs
Because CD8+ T cells can play a double-edged role in facilitating viral clearance and contributing to lung injury, we hypothesized that the persistence of cytotoxic CD8+ T cells causes bystander lung pathology in IAV-cleared lungs. To address it, we performed CD8+ T blockade using anti-CD8a antibody on 9 and 11 dpi. Most IAV clearance in our model was achieved by 10 dpi (Fig. 1L), justifying the CD8+ T blockade timeline.
We compared various parameters of lung injury, i.e., inflammation (cellular infiltration) and lung vascular damage, in IAV-infected mice treated with or without anti-CD8a antibody on 14 dpi. The CD8a antibody blockade resulted in a >95% reduction of total CD8+ T cell frequency in the lung (fig. S1, H and I). Compared to IAV-infected control mice (WT-PR8), mice treated with CD8a antibody (WT-PR8-αCD8) did not exhibit differences in lung inflammation as both groups had similar levels of cellular infiltration in lung parenchyma based on H&E inflammation scores (Fig. 1, M and O). However, WT-PR8-αCD8 mice (compared to WT-PR8) had significantly reduced vascular inflammation (Fig. 1N). The H&E vascular pathology data in WT-PR8-αCD8 mice were supported by reduced leakage of serum albumin in BALF and higher expression of platelet endothelial cell adhesion molecule (CD31) based on immunofluorescence (IF) staining of lung sections (Fig. 1, P and R). Furthermore, the CD8 blockade ameliorated bronchial epithelial injury, as evidenced by improved integrity of airway epithelial cellular adhesion molecule (EpCAM) by IF staining of lung sections (fig. S1J). Both IAV-infected groups (WT-PR8 versus WT-PR8-αCD8) did not show detectable viral load at 14 dpi (Fig. 1Q). These data suggest that cytotoxic CD8+ T cells persist in IAV-cleared lungs and cause bystander lung injury. Late-stage blockade of CD8+ T cells in virus-cleared lungs can lead to expedited resolution of lung vascular injury, which is crucial to improving lung function.
scRNA-seq reveals cellular and functional heterogeneity among CD8+ T cell subsets during peak IAV and post-IAV clearance phases
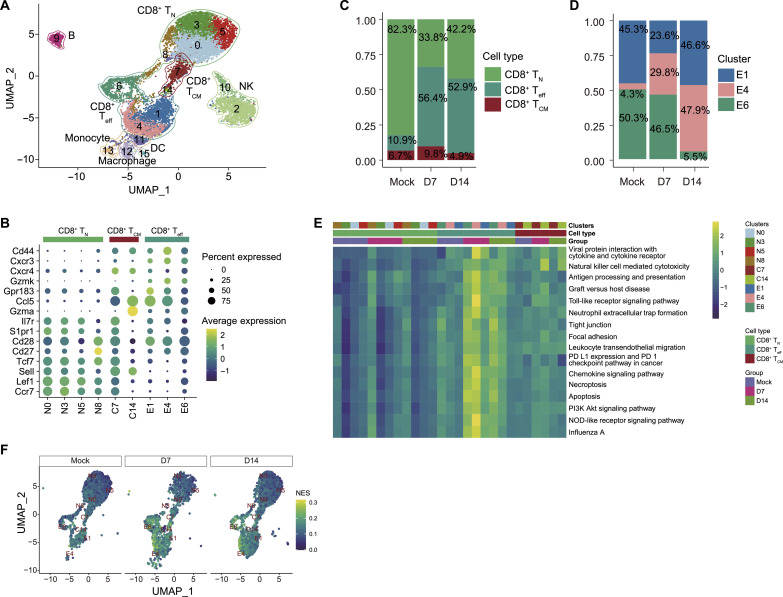
Because CD8+ T cells maintained a persistent cytotoxic phenotype even in virus-cleared lungs, we performed scRNA-seq to study the cellular heterogeneity and regulation of cellular response in CD8+ T cells during peak viral (day 7) and postviral resolution phase (day 14). After stringent filtration, the transcriptome data of a total of 12,113 high-quality cells (mock: 4,92, day 7: 3367, and day 14: 4354) were retained for further analysis. Through unsupervised clustering, a total of 16 cell clusters were identified and visualized as uniform manifold approximation and projection (UMAP) embeddings (Fig. 2A). Seven clusters (clusters 2, 9, 10, 11, 12, 13, and 15) were removed because of the low expression of Cd8a and Cd8b1 (Fig. 2A and fig. S2A). Further cell annotation (based on canonical lineage-defining markers) identified 7 clusters that were removed from the study as natural killer (NK) cells (clusters 2 and 10), B cells (cluster 9), monocytes (clusters 11 and 13), macrophage (cluster 12), and dendritic cells (cluster 15) (fig. S2D). The total number of cells belonging to these seven clusters is 2739, which is approximately 22.6% of total cells sequenced (fig. S2E). On the basis of the canonical lineage-defining markers, 10 cellular clusters expressing Cd8a and Cd8b1 were further grouped into three cell subsets as CD8+ T naïve (CD8+ TN: Ccr7, Lef1, Sell, Tcf7, Cd27, Cd28, and S1pr1), CD8+ T central memory (CD8+ TCM: Il7r, Gmza, and Ccl5), and CD8+ T effector/effector memory (CD8+ Teff: Gzmk, Cxcr4, Cxcr3, and Cd44) (Fig. 2, A and B, and fig. S2, B and C) (28).
Fig. 2. Cellular and functional heterogeneity among CD8+ T cell subsets during peak influenza A virus (IAV) and post-IAV clearance phases.
Single-cell RNA-sequencing (scRNA-seq) analysis of FACS-sorted CD8+ T cells from mock- and PR8-infected mice lungs at 7 and 14 dpi. (A) Unbiased UMAP of total CD8+ T cells from mock- and PR8-infected mice lungs at 7 and 14 dpi, revealing 15 clusters of CD8+ T cells. (B) Dot plot representing expression levels of canonical marker genes for the selected 10 clusters generated by unbiased analysis and their corresponding subsets clustering within CD8+ TN, TCM, or Teff. Clusters were renamed as N for naïve, C for central, and E for effectors corresponding with their CD8+ T cell subsets. (C and D) Distribution of the proportions of CD8+ TN, TCM, and Teff clusters (C) and clusters E1, E4, and E6 (D), corresponding to the CD8+ Teff subset; and their kinetics at day 0 (mock), 7 (D7) and 14 (D14) dpi. (E) Heatmap of GSVA enrichment scores of selected significant pathways between CD8+ Teff against TN and TCM. Color stands for the level of up-regulation (yellow) or down-regulation (dark green) in cells. (F) GSVA enrichment scores of CD8+ T cells with UMAP embedding NK cell–mediated cytotoxicity pathway. Color stands for the normalized enrichment score (NES).
We then compared the proportion of each cell subset during different stages of infection (7 dpi versus 14 dpi) (Fig. 2C). In both mock and infection groups, the Teff subset fell into three cellular clusters, designated as E1, E4, and E6. Compared to the mock group, IAV infection led to a notable enrichment of Teff cell clusters (E1, E4, and E6) at 7 and 14 dpi, whereas a simultaneous contraction of the naïve subset (TN) was observed (Fig. 2C and fig S2F). To determine the relative ratio of Teff cell clusters during different infection stages, we compared the cell proportion and cell numbers of the clusters E1, E4, and E6 in IAV-infected lungs at 7 and 14 dpi (Fig. 2D and fig S2G). Of these three clusters, cluster E6 had a similar abundance between mock (50.3%) and 7 dpi (46.5%) and a marked reduction at 14 dpi (5.5%) (Fig. 2D). A substantial reduction of cluster E1 was observed in IAV-infected lungs at 7 dpi (23.6%), and its restoration to almost the same level as mock by 14 dpi (46.6%) (Fig. 2D). The cluster E4 was enriched at 7 dpi (29.8%, compared to mock infection with 4.3%) and further expanded to become a dominant cluster at 14 dpi (47.9%) (Fig. 2D). These findings reveal that Teff exhibits robust cellular diversity with dynamic changes in cellular clusters over time during IAV infection.
Next, we performed gene set variation analysis (GSVA) analyses to identify functionally enriched pathways in CD8+ T cells between mock- and IAV-infected groups. Compared to CD8+ TN and CD8+ TCM, pathways related to inflammation, cytotoxicity, and cell death were up-regulated in CD8+ Teff (Fig. 2E and data S1), at both at 7 and 14 dpi. Among those pathways, the “NK cell–mediated cytotoxicity” pathway was highly enriched in CD8+ Teff (E1, E4, and E6) at 7 and 14 dpi (Fig. 2F). The scRNA-seq findings, while concurring with flow cytometry data that cytotoxic CD8+ T cells persist in viral resolved lungs, provide another layer of evidence vis-a-vis the distribution of cellular heterogeneity and molecular response of CD8+ Teff subsets. Because of our interest in understanding the CD8+ T cell pathologic response, we focused on Teff cells as a subject of the in-depth scRNA-seq analysis.
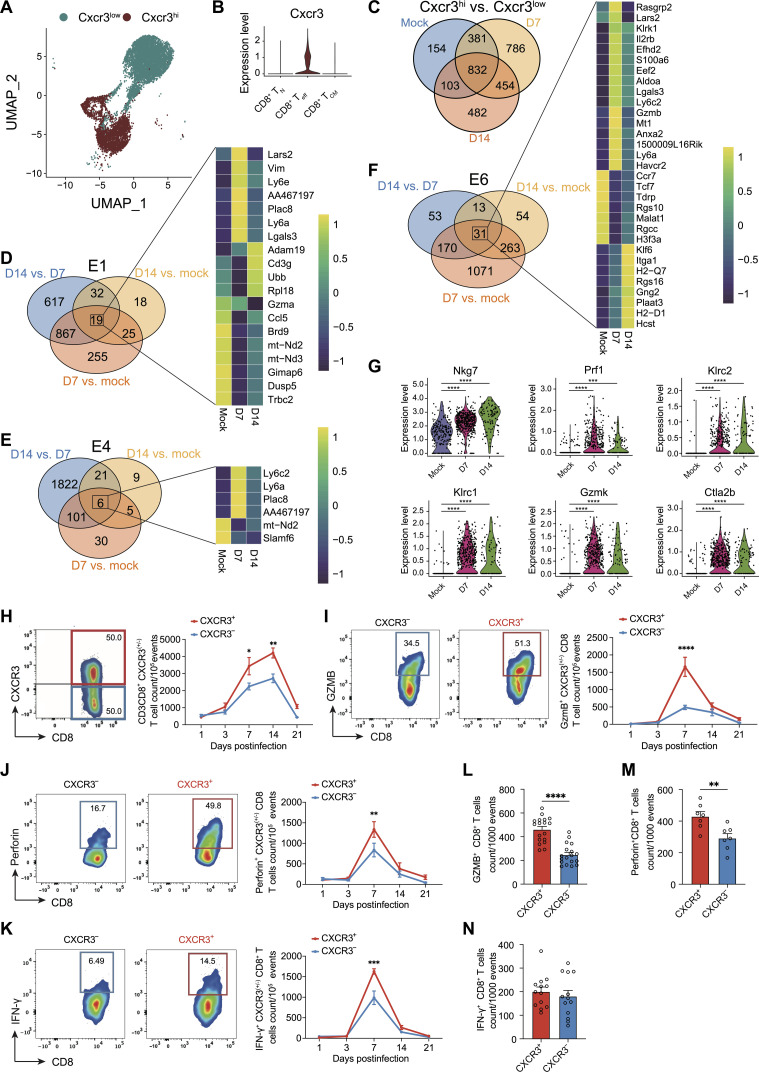
Cxcr3 expression is linked to enhanced CD8+ T cell–specific cytolytic gene expression in IAV-infected lungs
The scRNA-seq data show that the high expression of chemokine receptor Cxcr3 (Cxcr3hi) was predominantly associated with Teff subsets, in contrast with its low-level expression by TCM and TN CD8+ T cell subsets, designated as Cxcr3low (Fig. 3, A and B). To examine the shared and unique differentially expressed genes (DEGs) between Cxcr3hi and Cxcr3low CD8+ T cells, we performed differential gene expression analyses from the mock- and IAV-infected lungs at 7 and 14 dpi. A total of 832 DEGs between Cxcr3hi and Cxcr3low CD8+ T cells were common in these three sets (Fig. 3C and data S2). Notably, there were 154, 786, and 482 DEGs that were identified as unique DEGs (Cxcr3hi versus Cxcr3low) at mock, 7, and 14 dpi, respectively. We also found 57 shared pathways that were significantly enriched among the DEGs, all of which were highly up-regulated in Cxcr3hi clusters in mock at 7 and 14 dpi (fig. S3A and data S3). Several pathways were linked to cytotoxicity and to infections, which are known to induce profound inflammatory damage.
Fig. 3. CXCR3 expression and CD8+ T cell response in influenza-infected lungs.
(A) Cxcr3-annotated UMAP incorporating CD8+ single-cell transcriptomes from mock- and PR8-infected mice. (B) Violin plots of Cxcr3 expression within CD8+ TN, CD8+ Teff, and TCM cell clusters. (C) Venn diagram of shared and exclusively DEGs between Cxcr3hi with Cxcr3low CD8+ T cells from mock- and PR8-infected mice at 7 and 14 dpi. (D to F) Venn diagram illustrating the quantity of common or distinct DEGs found in the E1 (D), E4 (E), and E6 (F) clusters, respectively, between mock versus D7 PR8, mock versus D14 PR8, and D7 PR8 versus D14 PR8. Heatmaps showing the genes expressed in common across the three comparisons in the E1 (D), E4 (E), and E6 (F) clusters, individually. Color bar indicates the expression level for each gene. (G) Violin plots of Nkg7, Prf1, Klrc2, Klrc1, Gzmk, and Ctla2b expression in E6 cluster between mock and PR8 at 7 and 14 dpi. Statistical significance was determined by MAST. (H to K) FACS representative graphs (left) and kinetics (right) of CXCR3+ versus CXCR3− CD8+ T cells (H) and GzmB-expressing (I), perforin-expressing (J), and IFN-γ–expressing (K) cells in CXCR3+ versus CXCR3− CD8+ T cell populations in PR8-infected wild-type (WT) mice at indicated time points. (L to N) Number of CXCR3+ and CXCR3− CD8+ T cells from PR8-infected mice was normalized at 1000 events at 7 dpi. GzmB-expressing (L), perforin-expressing (M), and IFN-γ–expressing (N) cells within CXCR3+ versus CXCR3− CD8+ T cells were quantified. Two-way ANOVA with Sidak multiple comparison tests (I) to (K) and two-tailed t test (L) to (N) were used to determine the statistical significance. The data are representative of two independent experiments and expressed as means ± SEM with n = 5 to 10. For statistical significance: *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
Because Cxcr3hi CD8+ T cells were associated with a dominant effector and cytotoxic response, we next investigated the cluster-specific host response in Cxcr3hi CD8+ T cells. Venn diagrams represented the common and unique DEGs in Cxcr3hi cell clusters (clusters E1, E4, and E6) in mock- and IAV-infected lungs (Fig. 3, D to F). Nineteen shared DEGs were identified in cluster E1 among all three comparisons (Fig. 3D and data S4). Gzma and Ccl5 were highly expressed in mock Cxcr3hi cluster E1 and down-regulated in influenza-specific Cxcr3hi cluster E1 at 7 and 14 dpi (Fig. 3D), whereas the expression of Ly6a, Vim, and Ly6e was highly expressed at 7 dpi, compared to mock or 14 dpi Cxcr3hi cluster E1 (Fig. 3D). Gene set enrichment analysis (GSEA) identified 10 enriched pathways in all three comparisons. These include interleukin-17 (IL-17) signaling pathway, hematopoietic cell lineage, B cell receptor signaling pathway, and antigen processing and presentation (fig. S3B and data S5). The cluster E4 had six common DEGs between mock and 7 and 14 dpi time points (Fig. 3E and data S6). Among six DEGs, Ly6c2, Ly6a, and Plac8 were induced after influenza infection at 7 dpi, and a significant down-regulation was observed at 14 dpi (Fig. 3E). A total of 32 shared pathways were enriched in cluster E4 among all three comparisons (fig. S3C and data S7). Compared to clusters E1 and E4, cluster E6 had the highest number of DEGs, with 31 significant DEGs between mock- and IAV-specific Cxcr3hi cluster E6 at 7 and 14 dpi (Fig. 3F and data S8). Compared to mock and 14 dpi, cluster E6 exhibited higher expression of Il2rb, Lgals3, S100a6, Ly6c2, Gzmb, Ly6a, and Plac8. A remarkable down-regulation of Ccr7, Tcf7, Malat1, Rgs10, Rgcc, and H3f3a was observed in IAV-infected lungs compared to mock at 7 and 14 dpi. We also found that cytotoxicity-associated molecules, such as Nkg7, Prf1, Klrc1, Klrc2, Gzmk, and Ctla2b, were up-regulated at 7 and 14 dpi when compared to mock in cluster E6 (Fig. 3G). Furthermore, a total of 24 enriched pathways were highly enriched in cluster E6 (fig. S3D and data S9), including those related to cytokine-cytokine receptor interaction, chemokine signaling pathway, and apoptosis. Overall, our data show the cellular heterogeneity in Cxcr3 expressing CD8+ T cells and cluster E6 being more profoundly associated with the expression of cytotoxic genes and pathways. We further validated the scRNA-seq data to determine the functional differences between CXCR3+ and CXCR3− CD8+ T cells by flow cytometry (Fig. 3, H to N). Consistent with these findings, we found significantly higher levels of GzmB- and perforin-expressing CXCR3+ cells, based on both total (Fig. 3, I and J) and normalized counts (per 1000 cells of CXCR3+ and CXCR3−) (Fig. 3, L and M).
The CXCR3+ CD8+ T cells exhibited a peak cytotoxic response at 7 dpi and down-regulation of response by 14 dpi (Fig. 3, I and J). Both CXCR3+ and CXCR3− CD8+ T populations had comparable levels of IFN-γ–expressing cells (Fig. 3, K and N) (based on the normalized counts), suggesting that the central differences between CXCR3+ and CXCR3− CD8+ T cells were related to their cytotoxic rather than the effector functions.
Unique molecular pathways and regulons associated with cytotoxic response in CXCR3hi CD8+ T cells are revealed by scRNA-seq analysis
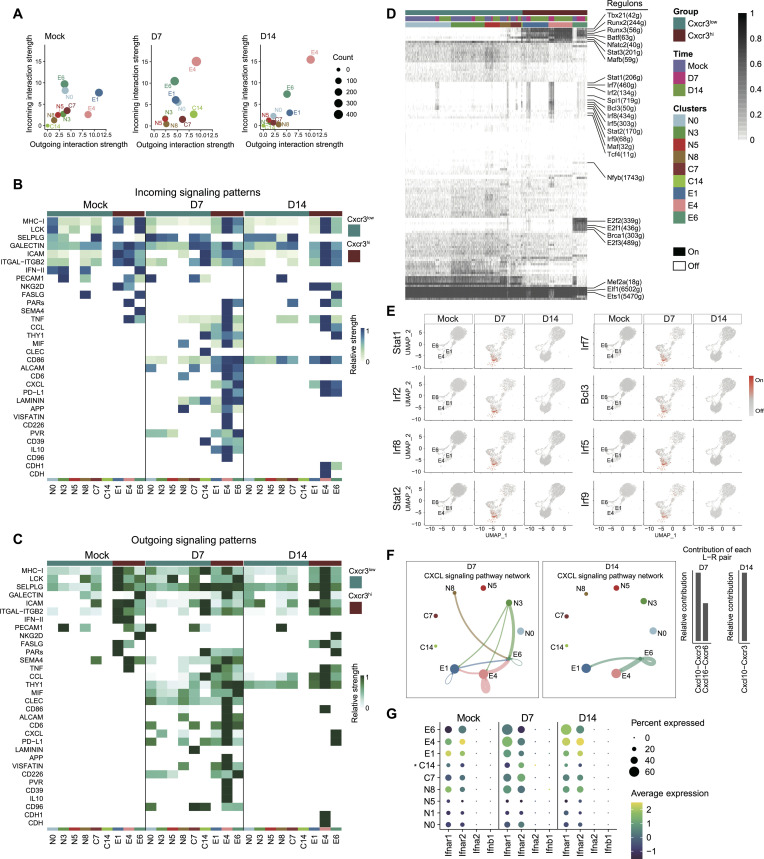
To further unravel the differences between Cxcr3hi and Cxcr3low CD8+ T cell functional properties, we analyzed ligand-receptor interactions in the scRNA-seq dataset. The ligand (outgoing signal) and receptor (incoming signal) in Cxcr3hi and Cxcr3low cellular clusters were compared among three groups, mock and IAV infection at 7 and 14 dpi. Compared to mock, CXCR3hi cellular clusters (E1, E4, and E6) exhibited notable differences in incoming and outgoing signals at 7 and 14 dpi. Among the three CXCR3hi clusters, the E4 cluster was the major source of incoming and outgoing signals at 7 and 14 dpi (Fig. 4A). Consistent with their enhanced cytotoxic properties, the CXCR3hi clusters also exhibited higher ligand-receptor interactions at 7 and 14 dpi leading to the regulation of crucial signaling pathways in these clusters (Fig. 4, B and C). We identified 38 signaling pathways with dominant ligand-receptor pairs in Cxcr3hi and Cxcr3low clusters, including CCL, THY1, MIF, CLEC, CD86, ALCAM, CD6, CXCL, PD-L1, LAMININ, APP, VISFATIN, CD226, PVR, CD39, IL-10, CD96, CDH1, and CDH (Fig. 4, B and C). To compare the patterns of outgoing (ligands) and incoming (receptors) signaling between Cxcr3hi and Cxcr3low clusters (mock and 7 and 14 dpi), we combined all identified signaling pathways from different datasets. We subsequently compared them in parallel, which allowed us to identify ligand-receptor pairing that exhibited different signaling patterns (Fig. 4, B and C). Compared with mock, most of the pathways such as CCL, CXCL, MIF, PVR, ALCAM, CD39, CD6, CD86, PD-L1, and THY1 were found to be active in Cxcr3hi cellular clusters at 7 dpi. Among the highly active pathways at 7 dpi, MIF, ALCAM, CD39, CD6, CLEC, and IL-10 pathways were down-regulated by 14 dpi, except for cytotoxicity-triggering receptor NKG2D, which was up-regulated in cluster E6 at 14 dpi (Fig. 4, B and C). Thus, Cxcr3hi CD8+ T cells display unique molecular pathways associated with cytotoxic response in the lungs during IAV infection.
Fig. 4. Single-cell RNA-sequencing (scRNA-seq) analysis of host response regulation in CXCR3+ CD8+ T cells.
Wild-type (WT) (C57BL/6) mice were mock-infected with phosphate-buffered saline (PBS) or infected with 1000 plaque-forming units (PFUs) of influenza A virus (IAV) (PR8) intranasally. At the indicated time points, mice were euthanized, and the bronchoalveolar lavage (BALF) and lungs were aseptically collected. The FACS-sorted CD8+ T cells were subjected to scRNA-seq analysis. (A) Scatterplot showing the major source and targets in mock- and PR8-infected mice at 7 and 14 dpi. The color indicates the cell types. Dot size is proportional to the number of inferred links (both outgoing and incoming) associated with each cell group. (B and C) Compare incoming (B) and outgoing (C) signaling associated with each CD8+ T cell cluster at 7 and 14 dpi. The colored bars show the relative importance of each cell group based on outgoing and incoming signaling patterns for pathways. (D) A heatmap of CD8+ cells showing high-confidence regulons at 7 and 14 dpi. “On” indicates active regulons; “Off” indicates inactive regulons. Active regulons per cell appear in black; the horizontal color bar indicates the subset associated with each cell and group. Numbers in parentheses represent the number of genes comprising the regulon. (E) UMAP plots showing the activity of the indicated regulons. Cells in which the indicated regulon is active (regulon activity exceeds a regulon-specific AUC threshold) are shown in red. (F) The inferred CXCL signaling networks and relative contribution of each ligand-receptor pair to the overall CXCL signaling network, respectively. Circle sizes are proportional to the number of cells in each cell group and edge width represents the communication probability. (G) Dot plot representing ligand and receptors expression levels of IFN-I signaling pathway for 10 clusters generated. The asterisk (*) indicates that C14 had less than 10 cells at one of the time points.
Because cytotoxic CD8+ T cell response is regulated by a coordinated function of several transcriptional factors (TFs) (29, 30), we examined the differences in TFs and their gene modules, also known as regulons, of Cxcr3hi versus Cxcr3low CD8+ T cells using the single-cell regulatory network inference and clustering (SCENIC) software. A total of 232 regulons with 9374 genes were identified across Cxcr3 clusters, which were further binarized and matched with cell types (Fig. 4D). Several important regulons, including IFN-regulated Tbx21, Nfatc2, and Batf, were uniquely activated in Cxcr3hi clusters. In addition, Runx3, Runx2, and Stat3 regulons were highly activated in Cxcr3hi clusters (Fig. 4E), and down-regulation of these regulons was observed in Cxcr3low cells (Fig. 4E). Several regulons such as Irf7, Irf2, Irf5, Irf9, Irf8, Stat1, Stat2, Bcl3, and Spi1 were commonly activated at 7 dpi and were turned off at 14 dpi in both Cxcr3hi and Cxcr3low cells (Fig. 4, D and E). Notably, consistent with the above findings of chemokine and IFN signaling, cluster E6 represented a majority of activated regulons in Cxcr3hi clusters, suggesting that cluster E6 is a major contributor of inflammatory response in Cxcr3hi CD8+ T cells (Fig. 4, D and E). Overall, these data identified several key regulons implicated in differential regulation of host response in Cxcr3hi and Cxcr3low clusters, further supporting that these cells are the major drivers of inflammation and cytotoxicity during IAV infection.
CXCR3 pathway is a major element of the late-phase CD8+ T cell communication and a source of the IFN-I–responsive CD8+ T cell subset recruited to IAV-infected lungs
Because chemokine signaling plays an instrumental role in shaping the CD8+ T cell responses (31), we determined the cellular communications for CXCL signaling in Cxcr3hi and Cxcr3low CD8+ T cells. Our data show that compared to mock, clusters N3 and N8 from Cxcr3low and clusters E1 and E4 from Cxcr3hi cells were more profoundly associated with the expression of CXCL chemokine ligands at 7 dpi (Fig. 4F). The cluster E6 was the only expressor of CXCL chemokines at day 14 after infection, and the clusters E1 and E4 acted as the recipient cells of Cxcr3hi clusters at 14 dpi (Fig. 4F). We did not detect the chemokine signaling in Cxcr3low cells at 14 dpi. The Cxcr3-Cxcl10 and Cxcr6-Cxcl16 ligand-receptor pairs were predominantly expressed at 7 dpi, suggesting that both pathways were involved in the early recruitment of CD8 T cells in the lungs. In contrast, the Cxcl10-Cxcr3 ligand-receptor pair was the only contributor to the CXCL communication pathway at 14 dpi (Fig. 4F, bottom and right), suggesting that the late-phase CD8+ T cell recruitment was relying solely on the CXCR3 pathway. We further examined the expression of CXCR3 ligand, Cxcl9, and Cxcl10 in CD8+ T cell clusters. While Cxcl10 expression was detected in each cluster (Cxcr3hi and Cxcr3low), the expression of Cxcl9 remained undetectable (fig. S4A), suggesting the CD8+ T cell–independent association of Cxcl9 expression in our model.
Next, we analyzed the ligand-receptor interactions of IFN-I and IFN-II. While CD8+ T subsets did not express IFN-I, Cxcr3hi CD8+ T cells were the main recipient of IFN-I signaling based on the expression of IFN-I–receptor and ligand-receptor interactions (Fig. 4G). In particular, clusters E4 and E6 were the major Cxcr3hi clusters associated with dominant IFN-I signaling correlating with enhanced cytotoxic response at 7 dpi. However, cluster E6 was uniquely associated with the persistence of cytotoxic response in Cxcr3hi CD8+ T cells even at 14 dpi. We did not detect IFN-II receptor signaling among Cxcr3hi or Cxcr3low cell clusters in mock- or IAV-infected mice at 7 and 14 dpi time points (fig. S4B). To validate scRNA-seq findings, we performed intracellular cytokine staining to investigate cytolytic (GzmB and perforin) response in CD8+ T cells from IAV-infected WT and IFN-γ−/− mice. The CD8+ T cells from WT and IFN-γ−/− mice did not differ in intracellular expression of GzmB and perforin (fig. S5, A and B). These data support the notion that IFN-γ is dispensable in regulating the cytotoxic phenotype of CD8+ T cells during IAV pathogenesis. Thus, the CXCR3/CXCL10 axis is the main pathway for both early and late CD8+ “T cell” recruitment in the IAV-infected mice, and among Cxcr3hi cells, cluster E6 appears to be the main driver of IFN-I–dependent inflammatory response in Cxcr3hi CD8+ T cells.
The antibody blockade of CXCR3 mitigates influenza lung injury and disease severity without affecting viral clearance
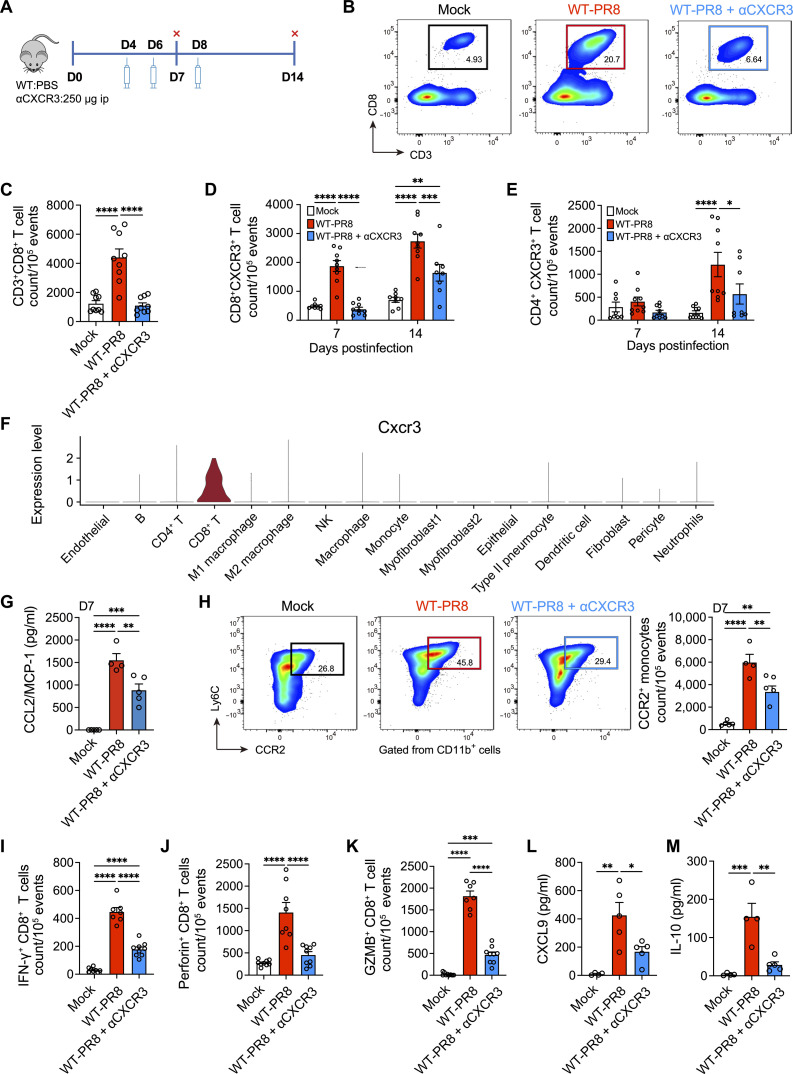
Because CXCR3+ CD8+ T cells showed all the attributes of cells specialized in the most robust cytotoxic response, we postulated that CXCR3+ CD8+ T cells contributed to IAV lung injury. We performed antibody-mediated CXCR3 blockade in IAV-infected mice (Fig. 5A) and examined multiple parameters associated with lung inflammation, injury, and disease severity. The CXCR3 blockade led to an approximately 70% reduction of CD8+ T cells in IAV-infected lungs at 7 dpi (Fig. 5, B and C). In contrast, we did not observe a quantitative increase in the frequency of CXCR3+ CD4+ T cells at 7 dpi and CXCR3 blockade did not reduce CD4+ T cell frequency at 7 dpi (Fig. 5E and fig. S7, E and F). Furthermore, even at the 14-dpi time point, CXCR3 blockade resulted in a much greater loss of CD8+ T cells than CD4+ T cells. (Fig. 5E), suggesting that CD8+ T cells acquired effector and cytolytic phenotype (Fig. 5, D and E) as early as 7 dpi. Furthermore, scRNA-seq analysis of total lung cells revealed that while cells such as neutrophils, CD4+ T cells, and monocytes expressed Cxcr3, CD8+ T cells had the highest degree of Cxcr3 expression at 7 dpi (Fig. 5F).
Fig. 5. The effect of CXCR3 neutralization on CD8+ T cell cytotoxic responses and inflammation in influenza A virus (IAV)-infected lungs.
(A) Wild-type (WT) (C57BL/6) mice were mock-infected with phosphate-buffered saline (PBS) or infected with 1000 plaque-forming units (PFUs) of IAV (WT-PR8) intranasally. PR8-infected mice received CXCR3 neutralizing antibody (WT-PR8 + αCXCR3) every other day starting day 4 after infection, and mice were euthanized at 7 and 14 dpi. The levels of cytokines/chemokines were measured in homogenized lungs. The frequencies of immune cells were determined in lung single cells using flow cytometry. ip, intraperitoneally. (B and C) Flow cytometry representative graphs (B) and number/quantification (C) of CD8+ T cells at 7 dpi. (D and E) Count of CXCR3+ CD8+ T cells (D) and CXCR3+ CD4+ T cells (E) at 7 and 14 dpi. (F) The expression of Cxcr3 was determined from a dataset of single-cell RNA-sequencing (scRNA-seq) from total lung cells of mock- and PR8-infected mice at 7 dpi. (G) Levels of CCL2/MCP-1 at 7 dpi, measured using the BiolegendPlex kit. Data are shown as means ± SEM, representative of two experiments with n = 4 to 5 per group. (H) Flow cytometry representative graphs (left) and count (right) of CCR2+ monocytes at 7 dpi. CCR2+ monocytes were gated from CD45+ CD11b+ Ly6C+ cells. (I to K) Count of IFN-γ–expressing (I), perforin-expressing (J), and GzmB–expressing (K) CD8+ T cells at 7 dpi. Data are shown as means ± SEM, pooled data of two experiments with n = 7 to 9 per group. (L and M) Levels of CXCL9 (L) and IL-10 (M) at 7 dpi, measured using the BiolegendPlex kit. Data are shown as means ± SEM with n = 4 to 5. One-way ANOVA with Tukey’s post hoc test [(C) and (G) to (M)] or two-way ANOVA with Sidak multiple comparisons test (means) was used for statistical significance in (C) to (L). *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
The CXCR3 blockade resulted in the reduced level of CCL2 chemokine and, consequently, reduced CCR2+ monocytes (Fig. 5, G and H). The CXCR3 blockade also resulted in significantly reduced levels of CD8+ T cell–specific effector (i.e., IFN-γ) and cytolytic molecules (i.e., perforin and GzmB) (Fig. 5, I to K). Notably, while CXCR3 antibody blockade resulted in a significant loss of CD8+ T cell cytotoxic response, it did not abolish the CD8+ T effector and cytotoxic response altogether. We further measured the levels of CXCR3 cognate–binding chemokines CXCL9 and CXL10, as well as cytokines in the lung homogenates of mock- and influenza-infected mice at 7 dpi. We found that CXCR3 antibody blockade resulted in reduced levels of CXCL9 and IL-10 (Fig. 5, L and M). The lower IL-10 level is indicative of reduced lung inflammation in mice with CXCR3 antibody blockade. Overall, these data demonstrated that CXCR3 blockade dampened the expression of inflammatory and cytolytic molecules in IAV-infected lungs. To corroborate our CXCR3 blockade approach, we compared CD8+ T cell cytotoxic response in IAV-infected WT with those of CXCR3-deficient (CXCR3−/−) mice. The results demonstrated a substantial loss of CD8+ T cells (approximately 70%) in the lung (fig. S6A) and a marked reduction in overall CD8+ T cell cytotoxic response (fig. S6, B to D) as well as decreased CCR2+ monocyte recruitment (fig. S6E) in CXCR3−/− mice. Together, these data show that CXCR3 amplifies the magnitude of inflammatory response during IAV pathogenesis likely via augmenting the recruitment and cytotoxic function of CD8+ T cells.
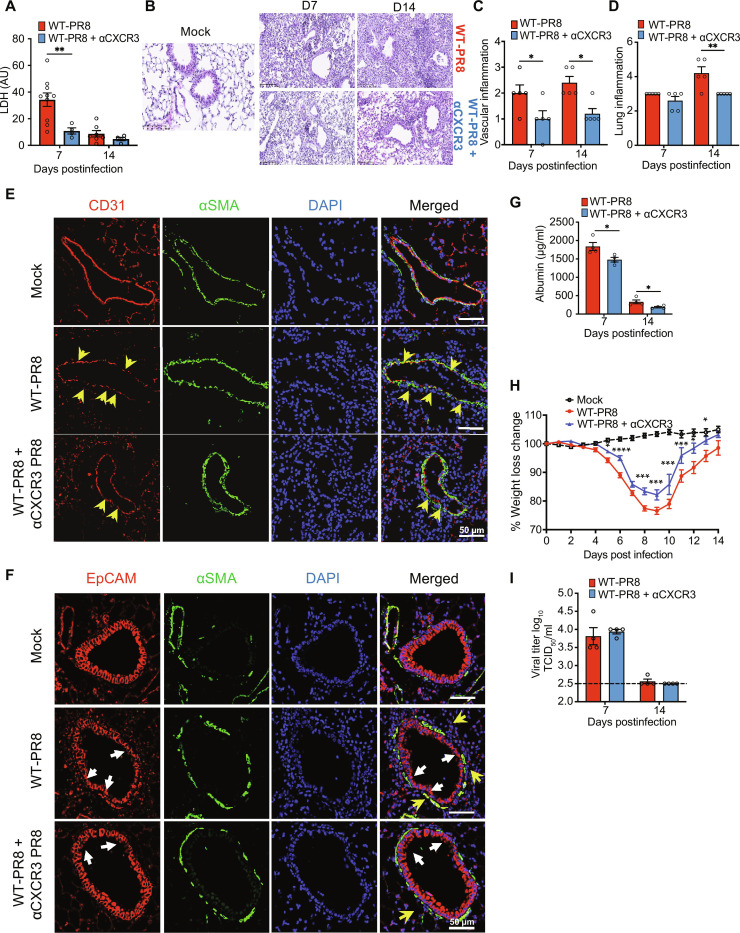
We further hypothesized that CXCR3 blockade would reduce lung inflammation and pathology while sustaining antiviral responses through the function of CXCR3− CD8+ T cells. We compared multiple parameters of IAV disease severity, i.e., weight loss, inflammation, and acute lung damage, in IAV-infected mice treated with or without anti-CXCR3 antibody. Compared to IAV-infected control mice (WT-PR8), mice treated with CXCR3 antibody (WT-PR8-αCXCR3) exhibited significantly reduced lung injury, evidenced by a reduced level of LDH in BALF (Fig. 6A) and reduced inflammation shown in and H&E tissue pathology at 7 and 14 dpi (Fig. 6, B to D). CXCR3-depleted infected mice also exhibited reduced vascular damage and lung permeability based on higher expression of platelet endothelial cell adhesion molecule (CD31) (IF lung sections) (Fig. 6E) and reduced leakage of serum albumin level in the BALF (Fig. 6G). Furthermore, CXCR3 blockade ameliorated bronchial epithelial injury, evidenced by improved anti-EpCAM (epithelial) IF staining of lung sections (Fig. 6F). While LDH and albumin levels peaked at 7 dpi and significantly reduced by 14 dpi, the H&E tissue-pathology assessments showed a nonresolving lung injury and vascular damage in IAV-infected mice even at 14 dpi.
Fig. 6. The effect of CXCR3 depletion on lung inflammation and pathology in an influenza model.
PR8-infected wild-type (WT) mice received CXCR3 neutralizing antibody (WT-PR8 + αCXCR3) every other day starting day 4 after infection and euthanized at 7 and 14 dpi. (A) LDH level in bronchoalveolar lavage fluid (BALF). BALF Data are representative of two experiments, n = 4 to 9 mice per group and shown as means ± SEM. Statistics were calculated using one-way ANOVA with Tukey’s post hoc analysis. (B) H&E staining of lungs. Endothelial (C) and lung (D) inflammatory scores. Data are representative of two independent experiments, n = 5 per group. Two-way ANOVA with Sidak multiple comparisons was used to compare the groups. (E and F) Immunofluorescent staining of lung sections for endothelial (E) and epithelial (F) damage. Mock-infected (top) and PR8-infected mice (WT-PR8) (middle) and PR8-infected mice recipient of αCXCR3 (WT-PR8 + αCXCR3) (bottom). (E) CD31 (red) for endothelial cells, αSMA (green) for α-smooth muscle cells; CD31 (red) to highlight the integrity of endothelial layer, where yellow arrowheads show the loss of endothelial cells marked by the discontinuity of the red line. (F) EpCAM (red) was used to probe airway bronchoepithelial cells. White arrows highlight the loss of epithelial integrity, and yellow arrows show the infiltrating inflammatory cells around airways. (G) BALF albumin. t test was used at each time point. Data are representative of two independent experiments, n = 4 per group. (H) Kinetics of body weight change. Data are shown as pooled data of two independent experiments, n = 5 to 6 per group. Statistics were calculated using two-way ANOVA with Tukey’s post hoc test. (I) TCID50 viral titer in lung homogenates. Data are representative of two independent experiments, n = 4 per group. Black dashed line indicates the baseline value (mock) negative control. Statistics were calculated using one-way ANOVA with Tukey’s post hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
Consistent with reduced lung inflammation and injury, IAV-infected mice with CXCR3 blockade also exhibited a significantly reduced weight loss (Fig. 6H). These findings were subsequently validated in CXCR3−/− mice showing similarly reduced lung inflammation and pathology evident on histological sections and through the reduced levels of LDH in the BALF compared to the WT mice (fig. S7, A to C). These data demonstrated that the CXCR3 pathway is an important driver of lung injury during the peak viral load (7 dpi), and its disruption facilitates the expedited resolution of lung injury following the viral clearance. Because abolished CXCR3 signaling had such a profound impact on limiting lung pathology, we assessed its role in viral clearance to explore its usefulness as a therapeutic target. Both CXCR3 antibody blockade (Fig. 6I) and CXCR3 gene deletion (fig. S7C) did not hinder viral clearance, as a similar viral load was detected among all IAV-infected groups at 7 dpi (peak viral load), and a lack of detectable viral load at 14 dpi. Although CXCR3 blockade did not affect viral clearance at 7 dpi, we further examined whether CXCR3 blockade interfered with viral clearance at earlier time points (before day 7) or between 7 and 14 dpi by comparing viral load in WT-PR8 and WT-PR8-αCXCR3 on 5 and 10 dpi (fig. S7, D to F). Our data show that CXCR3 blockade did not affect viral clearance as both WT-PR8 and WT-PR8-αCXCR3 mice had similar viral load by 5 dpi and cleared most of the viral load by 10 dpi (fig. S7D). While CXCR3 signaling is an important driver of immune-mediated lung pathology, the residual inflammation in WT-PR8-αCXCR3 mice is sufficient to clear IAV and does not adversely affect viral clearance throughout the course of infection.
CXCR3+ CD8+ T cells are sufficient to exacerbate IAV lung pathology in CXCR3−/− mice
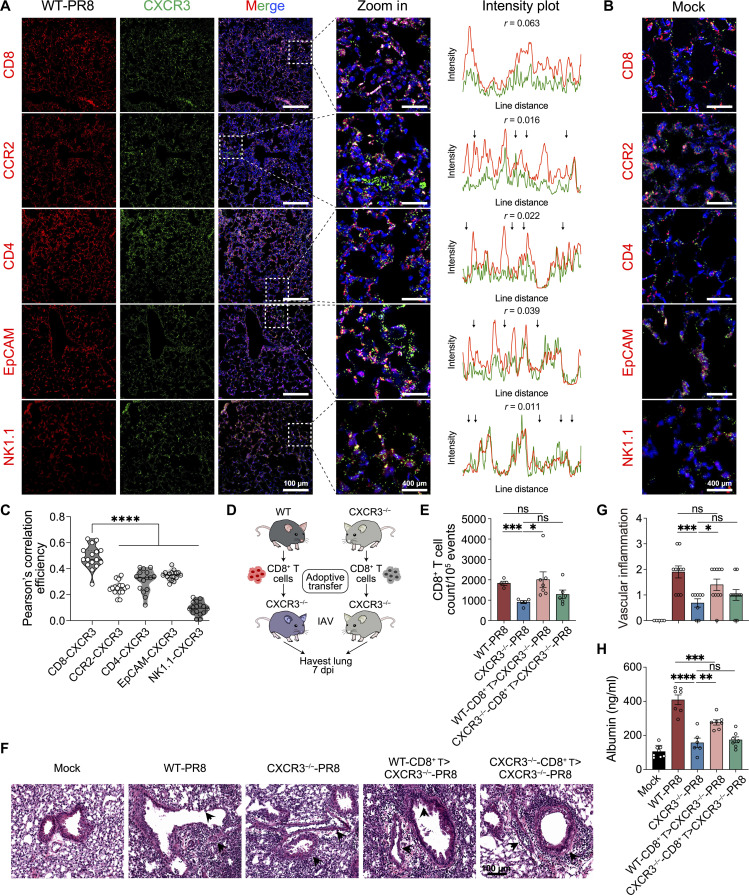
scRNA-seq of total lung cells reveals that CXCR3 is primarily expressed by CD8+ T cells in our model at peak IAV lung inflammation and pathology at 7 dpi (Fig. 5F), suggesting that CXCR3+ CD8+ T cells are the major driver of pathology in our model. However, several immune cells and epithelial cells can express CXCR3 in certain biological circumstances (32–37). To explore these cell subsets, we performed IF staining and colocalized CXCR3 with a number of immune cells (CD8+, CD4+, monocytes, and NK1.1) and epithelial cells during peak IAV lung injury at 7 dpi. Our IF colocalization data show that CD8+ T cells represented the primary cell type with most significant distribution of CXCR3 expression, although CD4+ T cells, monocytes, and epithelial cells expressed CXCR3 to some lesser degree (Fig. 7, A to C, and fig. S8A). This validated our initial conclusion from scRNA-seq analysis that CD8+ T cells are the major subset expressing CXCR3 during peak IAV lung injury.
Fig. 7. CXCR3 cellular colocalization and adoptive transfer of Cxcr3+/+ and Cxcr3−/− CD8+ T cells in CXCR3−/− recipient mice.
(A) Immunofluorescent staining (left) of lung sections of influenza A virus (IAV)-infected mice at 7 dpi. Box indicated location of zoom. Scale bars, 100 μm and 400 μm (zoom in). Intensity plots (right) indicated fluorescence intensity from a rectangle drawn within the dashed box in each image for CXCR3 (green) and CD8, CCR2, CD4, EpCAM, or NK1.1 (red) obtained using plot profile tool in Fiji/ImageJ. Arrow: No colocalization. (B) Immunofluorescent staining of lung sections of mock mice. Scale bars, 400 μm. (C) Pearson’s correlation coefficients between CXCR3 and CD8, CCR2, CD4, EpCAM, or NK1.1 from randomly selected areas (n = 16) were measured using Coloc2 plug-in in Fiji/ImageJ. (D to H) CD8+ T cells were sorted from the spleen of WT (C57BL/6) and CXCR3−/− mice and retroorbitally injected into IAV-infected CXCR3−/− mice. Mice were euthanized at 7 dpi, and bronchoalveolar lavage fluid (BALF) and lung sections were preserved. (D) Timeline of adoptive transfer experiment. (E) Counts of CD8+ T cells in lungs determined by flow cytometry. (F) and (G) H&E staining of lung sections (F) with inflammatory score (G). (H) Albumin levels present in BALF. Data are shown as means ± SEM, data are representative of two independent experiments with n = 6 to 8 mice per group per experiment. Statistics were calculated using one-way ANOVA with Tukey’s post hoc analysis [(C) and (H)] or t test [(E) and (G)]. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
To establish the direct mechanistic link between CXCR3-expressing CD8+ T cells and lung pathology in our model, we performed adoptive transfer of CXCR3+/+ (WT) and CXCR3−/− CD8+ T cells into CXCR3−/− recipient mice. The CD8+ T cells were magnetically sorted from spleens of naïve WT and CXCR3−/− mice, and 1 × 106 cells were retro-orbitally transferred into CXCR3−/− mice 24 hours after IAV infection. The adoptively transferred mice were euthanized at 7 dpi to study the parameters of lung inflammation and injury (Fig. 7D). The adoptive transfer of CXCR3+/+ CD8+ T cells resulted in significantly increased CD8+ T (lung) frequency in CXCR3−/− mice (WT CD8+ T > CXCR3−/−). In contrast, the adoptive transfer of CXCR3−/− CD8+ T cells did not change lung CD8+ T cell frequency in CXCR3−/− mice (Fig. 7E and fig. S8B) (CXCR3−/− CD8+ T > CXCR3−/−). Furthermore, only the adoptive transfer of the WT but not the CXCR3−/− CD8+ T cells enhanced the lung vascular inflammation/injury in CXCR3−/− mice detected by histopathological analysis (Fig. 7, F and G). This increased vascular injury correlated with higher serum albumin leakage in BALF of CXCR3−/− mice recipient of WT CD8+ T cells (Fig. 7H). No difference in CD8+ T cell frequency or vascular inflammation was detected in IAV-infected WT or Cxcr3−/− mice recipient of WT CD8+ T cells, demonstrating that transferred CXCR3+/+ CD8+ T cells could drive acute lung pathology in CXCR3/− mice.
DISCUSSION
In this study, we dissected the CD8+ T cell responses in influenza-infected lungs during the peak viral (acute) and virus-cleared states. The major finding of this study is that the CD8+ T cell population recruited to IAV-infected lungs represents notable transcriptional and functional diversity, with a subset not required for viral clearance but instead driving the severe lung pathology. The pathological CD8+ T cell subset is characterized by high CXCR3 expression, the enhanced cytotoxic pathway signature, and their persistence in the lungs, resulting in increased lung epithelium and vascular damage and the extended time of inflammatory infiltrate lung consolidation. Intercepting CXCR3 with either antibody or genetic deletion prevented the development of severe influenza lung pathology, without affecting viral clearance. Thus, the temporal blocking of the CXCR3 pathway could be a viable candidate for therapeutic intervention that may prevent the development of severe lung injury during influenza-induced pneumonia.
A growing body of literature suggests that exuberant CD8+ T cells could contribute to lung injury and exacerbate influenza disease severity (9, 38, 39). Furthermore, influenza-specific CD8+ T cells have been shown to cause bystander damage to noninfected alveolar epithelial cells in vitro (14), supporting the in vivo data on the contribution of CD8+ T cells in influenza lung pathology. Paradoxically, mice with impaired CD8+ T cell responses fail to clear influenza and succumb to high viral load–driven lung pathology (17). These contrasting findings suggest that while CD8+ T cells are indispensable to controlling influenza (9, 16), the exuberant CD8+ T cell responses can enhance lung pathology and exacerbate the disease. Therefore, the goal of this study was to conduct a detailed analysis of CD8+ T cells to gain insight into their role in both viral clearance and the development of influenza-associated lung pathology. To this end, we used a combined approach involving scRNA-seq and flow cytometry to unravel the cellular diversity and regulation of inflammatory response in CD8+ T cells and their role in maintaining a balance between viral clearance and tissue pathology.
Our data show that CD8+ T cell functional heterogeneity can be distinguished on the basis of the expression of the chemokine receptor, CXCR3. While Cxcr3hi CD8+ Teff cells are associated with enhanced cytotoxic response, CD8+ T with low Cxcr3 expression (TCM) show less profound expression of cytotoxic molecules and cytotoxic pathways. However, on the basis of IFN-γ+ effector response, both Cxcr3hi and Cxcr3low CD8+ T cells displayed equally potent effector phenotype, suggesting that Cxcr3 expression did not dictate the development of effector response in CD8+ T cells. Instead, Cxcr3 expression was a crucial determinant of cytotoxic response in CD8+ T cells. There is an abundance of literature on CXCR3 in maintaining the balance between T effector and memory generation following influenza (31, 40), but studies of the CXCR3 axis with pathological outcomes and immune cell types implicated in CXCR3-mediated lung pathology during primary influenza infection are limited and not always consistent. Fadel et al. (41) showed that while CXCR3 deficiency protected the CCR5-deficient mice from influenza mortality, CXCR3 deficiency on its own did not affect the survival of influenza-infected mice in a lethal challenge model. In contrast, another study showed that CXCR3-deficient mice had improved survival following lethal influenza challenge, and neutrophils were reported to be the primary CXCR3-expressing cells (42). These contrasting data necessitate further evaluating the role of CXCR3 as a pathologic framework in influenza lung injury. Our model is different from those prior studies because we used a severe but nonlethal model that allowed us to study the lung injury during both the peak viral load and resolution phase. It is crucial because postinfluenza complications involve persistent lung injury and impaired repair with compromised lung functions after the virus is already cleared (43, 44). In this context, antibody blockade of CXCR3 ameliorated lung injury during the peak viral load and led to a faster resolution of postinfection lung injury without interfering with viral clearance. Furthermore, late-stage CD8+ T blockade also resulted in a faster resolution of injury in virus-cleared lungs, consistent with the CXCR3 blockade data. These findings suggest that residual inflammation and cytotoxic responses in CXCR3−/− mice are sufficient to clear viral load. Furthermore, interception of either CXCR3 (during peak viral load) or total CD8+ T cells (late stage) can lead to faster resolution of lung injury following influenza pneumonia.
CXCR3 can be expressed by immune cells and epithelial cells, which can participate in viral clearance or lung pathology depending on the stage and severity of infection (32, 33, 40). Our scRNA-seq and IF colocalization data show that CD8+ T cells constitute the primary cell type expressing CXCR3 during peak lung injury at 7 dpi. CD8+ T cells can exert pathologic effects primarily in two ways. While CD8+ T cytolytic response directly kills virus-infected epithelial cells in the lung parenchyma, the CD8+ T effector response (IFN-γ) can modulate the inflammatory function of myeloid cells, contributing to viral clearance or lung pathology (18, 45). We have previously shown that CD8+ T cell–derived IFN-γ is a main driver of pathologic monocyte responses during influenza (45). Both CXCR3−/− and a-CXCR3-WT PR8 (IAV-infected) mice had reduced CD8+ T cytolytic (Gzm+) and effector response (IFN-γ). It is likely that CD8+ T–mediated lung injury at the peak IAV lung inflammation (7 dpi) stems from a combined effect of CD8+ cytotoxic and effector function, while cytotoxic CD8+ T cells continue to cause bystander lung pathology in virus-cleared lungs (14 dpi). However, the immune modulatory effects of CD8+ T cell subsets, including CXCR3, on regulating antiviral or pathological myeloid cell responses deserve further investigation. Similarly, NK cells are a major source of IFN-γ and a crucial antiviral framework during the early stage of influenza infection (46). Some studies have shown the association of CXCR3 with NK cell responses in influenza models (35). The limitation of our study is that we did not perform scRNA-seq in earlier time points of IAV infection (<7 dpi) to investigate the role of early lung immune environment in shaping the CD8+ T cell responses. Furthermore, we did not investigate whether CXCR3 blockade had any effects on NK or myeloid cells during the early phase of CXCR3 treatment. Nonetheless, the competent viral clearance in CXCR3 blockade mice suggests that modulation of CXCR3-mediated host response does not impede viral clearance or has any adverse consequences on lung pathology throughout the course of infection. The adoptive transfer of CXCR3+ CD8+ T cells exacerbated lung pathology and restored WT-like lung vascular pathology in Cxcr3−/− mice, linking CXCR3-pathway–mediated lung injury with CD8+ T cells in our model. These data, along with published literature (31, 32), suggest that influenza disease severity likely regulates the contribution of specific immune cells in CXCR3-induced lung pathology, and CXCR3 appears to be a main driver of acute lung damage, implicating CD8+ T cells during influenza sublethal infection.
IFNs are crucial in regulating the antiviral function of CD8+ T cells (47–49). Our data agree with prior reports that IFN-γ deficiency did not impact the recruitment of CD8+ T cells in influenza models (50), suggesting the IFN-γ–independent chemokine signaling driving the CD8+ T cell recruitment in influenza-infected lungs. Furthermore, similar expression levels of perforin and GzmB in CD8+ T cells from WT and IFN-γ−/− mice suggest the dispensability of IFN-γ in regulating the expression of cytolytic molecules in CXCR3+ CD8+ T cells. These data are further substantiated by the scRNA-seq analysis of cell-cell communication, demonstrating the weak ligand-receptor interactions for IFN-II in CXCR3hi CD8+ T cell clusters. Instead, scRNA-seq revealed enrichment of IFN-I receptor signaling and activation of multiple IFN-I–dependent regulons in Cxcr3hi CD8+ T cells. Our scRNA-seq analysis identified the functional diversity in CXCR3+ CD8+ T cells with a specific subset E6 exhibiting a major contributor to host responses, including the enrichment of IFN-I signaling in CXCR3+ CD8+ T cells. These findings suggest the key role of IFN-I–receptor signaling as a potential determinant of the inflammatory response of Cxcr3hi CD8+ T cells that warrant further investigation.
In summary, this study unveiled the CD8+ T cellular heterogeneity associated with pathologic host response in influenza model during active infection with peak viral and during virus-cleared infection recovery. Our findings have shown that the CXCR3 blockade approach benefited the host, improving inflammatory balance in the lung, preventing excessive CD8+ T cell–driven lung injury during the peak viral load, and a faster resolution of lung injury without inhibiting viral clearance. These data suggest that CXCR3 and CD8+ interceptions could be explored clinically in patients with severe influenza as a treatment option for reducing lung damage and accelerating postinfluenza recovery.
MATERIALS AND METHODS
Mice and influenza infection model
WT C57BL/6J, CXCR3−/−, and IFN-γ−/− mice were originally bought from the Jackson Laboratory and bred in-house. An equal proportion of 6- to 8-week-old sex-matched mice were included in this study. All experiments were performed according to the approved protocol by the University of North Dakota Animal Care and Use Committee (IACUC) (protocol no. 1808-8) and University of Florida Institutional Animal Care and Use Committee (protocol no. IACUC202100000057). Mice received food and water ad libitum at all times. IAV (H1N1 A/Puerto Rico/8/1934 or PR8) was purchased from Charles River (Norwich, CT), and a plaque assay (51) was performed to determine the plaque-forming units (PFUs) for IAV. For IAV infection, mice were lightly anesthetized with 4% (v/v) isoflurane/oxygen mixture and intranasally inoculated with 1000 PFUs of IAV in a 50-μl volume. At the indicated time points (days 3 to 21), mice were euthanized by CO2 exposure followed by cervical dislocation, and then lungs and BALF were aseptically isolated and processed for downstream applications.
Histopathology
Mice were euthanized, and after perfusion, the left lobe was fixed in 10% neutral buffered (pH 7.4) formalin for 24 hours at room temperature before transferring into 70% ethanol. The lung tissues were embedded in paraffin, sliced into 5-mm sections to reveal the maximum longitudinal view of the main intrapulmonary bronchus of the left lobe, and stained with H&E. H&E staining was performed by the histology core, University of North Dakota. Lung inflammation was evaluated and quantitatively by two blinded pathologists, on a scale of 0 to 5, with increments of 0.5 (15), with 0 as no inflammation and 4 as the highest degree of tissue infiltration of immune cells. Three parameters were focused on H&E analysis, Inflammation index (alveolar inflammation and inflammatory cell infiltration), bronchial epithelial damages (thickness, thinning, necrosis of bronchial epithelium, mucosal, and inflammatory plugs), and vascular endothelium change (52).
BALF albumin and LDH
BALF was collected at each end point by instillation and aspiration of lung with 1 ml of sterile ice-cold phosphate-buffered saline (PBS) through a 20-gauge tracheal catheter (BD Biosciences). BALF supernatants were collected and stored at −80°C until use. Albumin concentration in BALF samples was determined using an albumin enzyme-linked immunosorbent assay kit (ALPCO, Salem, NH) following the manufacturer’s instructions. The level of LDH was measured in the BALF of mock- and IAV-infected mice using a colorimetric LDH Assay kit following the manufacturer’s recommendations (ab102526, Abcam).
Measurement of cytokines and chemokines
At the indicated time points, lungs were homogenized in tissue protein extraction reagent using the tissue homogenizer 850 (Thermo Fisher Scientific). The levels of cytokines and chemokines (CXCL9, CXCL10, and CCL2/MCP-1) were determined in the lung lysates or BALF samples using the LegendPlex mouse proinflammatory chemokine panel (13-plex, catalog no. 740451) and the mouse inflammatory panel (13-plex, catalog no. 740446), respectively, following the manufacturer’s instructions. Samples were acquired on a BD FACSymphony A3 flow cytometer, and data were analyzed using LEGENDplex V8.0 Data Analysis Software (BioLegend).
Flow cytometry
The lungs were aseptically collected from mock- and IAV-infected mice, and single cells were prepared after collagenase digestion of lungs, as previously described (52). One million cells per sample were stained with Ghost Dye BV510 (Tonbo Biosciences, San Diego, CA) and anti-mouse CD16/CD32 (Fc-blocked) (BD Biosciences, San Jose, CA) at 4°C for 30 min. For cell surface staining, cells were incubated for 30 min at room temperature with anti-mouse CD3–allophycocyanin (APC)–Cyanine 7 (Cy7), CD3–Alexa Fluor 488, CD4–Brilliant Violet 786, CD8–phycoerythrin (PE)–Cy7 (Tonbo Biosciences), CD8-PE-Cy5, CD44-PerP-Cy5.5, CD44-PE-Cy7, CD62L–fluorescein isothiocyanate (FITC), CXCR3-BV650, and CXCR3-PE-Dazzle. For monocytes, cells were stained with Cd11b APC-Cy7, Ly6C-BV711, Ly6G-FITC, and CCR2-PE-Cy7. All the antibodies were purchased from BioLegend unless specified.
Intracellular staining
For intracellular cytokine staining, 1 million lung single cells were Fc-blocked with anti-CD16/32 and LIVE/DEAD-stained with Ghost Dye BV510 (Tonbo Biosciences) and surface-stained as described above. The cells were fixed, permeabilized with CytoFix/CytoPerm, and stained with anti-mouse GzmB-conjugated BV421 and APC-conjugated perforin. The perforin and GzmB expression represent the response from bulk CD8+ T cells. For IFN-γ detection (IFN-γ), lung single cells prepared from mock- and IAV-infected mice were stimulated with 10 μM IAV peptide NP366–374 for 5 hours with brefeldin A, in RPMI medium containing 10% fetal bovine serum (FBS) and supplemented with antibiotics, before intracellular staining with anti-mouse IFN-γ–PE–Dazzle 594 (26). A BD FACSymphony or SONY MA900 flow cytometer was used to acquire 100,000 events, and data were analyzed using FlowJo (Tree Star Inc.). The list of the antibodies used in this study including their clone and catalog number is provided in the Supplementary Materials (table S1).
IF staining and quantification
Formalin-fixed and paraffin-embedded lung sections from mock- and IAV-infected mice were prepared as previously described (52) and probed for epithelial or endothelial cell detection using monoclonal mouse anti–α-smooth muscle actin (αSMA) (1:10,000; A2547, Sigma-Aldrich, Darmstadt, Germany), rabbit anti-mouse CD31 (1:50; ab28364, Abcam, Cambridge, UK), or rabbit anti-mouse EpCAM (1:50; ab71916, Abcam, Cambridge, UK) antibodies. Tissues were incubated with corresponding secondary antibodies goat anti-mouse immunoglobulin G2a (IgG2a)–AF488 (1:200; A-21131, Thermo Fisher Scientific) and anti-rabbit IgG-AF546 (1:200; Invitrogen, Carlsbad, CA) in 7% goat serum/PBS for 1 hour at room temperature, and nuclei were counterstained with 4′,6-diamidino-2-phenylindole. All the images were acquired using a Leica DMi8 Thunder Imager fluorescent microscope and analyzed using the Fiji/ImageJ software. For colocalization analysis, a plot profile tool was used to measure and generate the intensity plots of the boxed regions, and Coloc2 plug-in was used to quantify colocalization parameters using Pearson’s correlation efficiency recorded in Fig. 7 and fig. S8.
In vivo CXCR3 and CD8+ T blockade
IAV-infected WT mice were intraperitoneally injected with 250 μg (200 μl) of anti-mouse CXCR3 monoclonal antibody (WT-PR8 + αCXCR3) (Bio X Cell, clone CXCR3-173) (50) every other day starting day 2 after IAV infection. Mice were euthanized on days 7 and 14 after infection for inflammatory characterization and viral load determination. Mice were also euthanized on days 5 and 10 for measuring viral load in IAV-infected WT or WT-PR8 + αCXCR3 mice. For CD8+ T blockade, IAV-infected WT mice were intraperitoneally injected with 250 μg of anti-mouse CD8a monoclonal antibody (WT-PR8 + αCD8a) (Bio X Cell, clone 53-6.7) (50) on days 9 and 11 after infection. Mice were euthanized on day 14 after infection for further investigations.
Adoptive transfer
Spleens were isolated from naïve WT and Cxcr3−/− mice. A single-cell suspension was prepared, and CD8+ T cells were magnetically sorted using CD8 isolation kit from Miltenyi Biotec. The purity of magnetically sorted CD8+ T cells was determined with flow cytometry. CD8+ T cells (1 × 106 cells per mouse) from naïve WT or Cxcr3−/− mice were retro-orbitally injected into IAV-infected Cxcr3−/− mice at 1 dpi. Mice were euthanized at day 7 after infection.
IAV titers
The lung IAV load was determined via end-point dilution assay and expressed as 50% tissue culture-infective dose (TCID50). The partial lobe of the right lung from mock- or IAV-infected mice was homogenized in PBS with volume normalized to the lung weight and stored at −80°C until use. The 10-fold dilutions of lung lysate supernatants were mixed with 3 × 103 Madin-Darby canine kidney cells (MDCK), in four replicates, in Dulbecco’s modified Eagle’s medium containing 0.0002% l-1-(tosylamido-2-phenyl) ethyl chloromethyl ketone–treated trypsin (Worthington Biochemical) with antibiotics. After 6 days of incubation (37°C with 5% CO2), the cells were fixed with formalin and stained with 0.3% crystal violet solution. For each animal, viral titers were obtained using serial dilutions on MDCK monolayers and normalized to the total volume of lung homogenate supernatant (53).
scRNA-seq library preparation and sequencing
From single-cell suspensions of mock- and IAV-infected lungs at 7 and 14 dpi, CD8+ T cells were fluorescence-activated cell sorting (FACS)–sorted by positive staining with anti-mouse CD8-APC and CD3-FITC using BD Aria II flow cytometer sorter. Cells (2 × 106) were resuspended in medium containing 10% dimethyl sulfoxide and 20% FBS and allowed to slow freeze in Mr. Frosty. 3′ single-cell gene expression libraries (v3.0) were constructed using the 10x Genomics Chromium system. Single-cell library preparation was done by Singulomics Corporation (https://singulomics.com/). A 150–base pair (bp) paired-end sequencing was performed to produce high-quantity data on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA).
Data alignment and sample aggregating
Sample demultiplexing, barcode processing, and unique molecular identifier counting were performed by the Cell Ranger v5.0 (54). Briefly, FASTQ files were first extracted from the raw bcl files with the cellranger mkfastq demultiplexing pipeline. Then, the FASTQ files were mapped to the mouse reference genome (mm10) to generate the count matrices by using the “cellranger count” pipeline with the default filtering parameters. Last, the count matrices of all three time points were loaded into R 4.0.2 with the Seurat v3 package (55). To obtain high-quality data for the downstream analysis, we discard cells with less than 200 and more than 6000 expressed genes or the fraction of transcripts mapped to mitochondrial genes larger than 25%. The expression level of each gene was normalized by using NormalizeData function of Seurat. Last, the datasets were integrated using the Seurat integration workflow.
Dimensionality reduction and clustering
Principal components analysis (PCA) was performed to identify the cell clusters based on the highly variable genes. The principal component (PC) number where the percent change in variation between the consecutive PCs with less than 0.1% was selected as the optimal PC number. To create the UMAP, the graph-based clustering was performed on the PCA-reduced data for clustering analysis (resolution = 0.8) with the FindClusters function.
Cell-type identification and differential expression analysis
To determine the cell type of each cluster, the gene markers for each cluster were identified by using the FindAllMarkers function in Seurat. Then, the cell type for each cluster was assigned by using the CellMarker and PanglaoDB database as the reference based on marker genes, with manual correction. DEGs (adjusted P value <0.05 and the average |log2 fold change| > 0.2) were identified by MAST (56) across experimental groups. GSEA of Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed by using the richR package (https://github.com/hurlab/richR), and significant pathways were identified with the P value <0.05.
Gene set variation analysis
Pathway activities in individual cells were assessed using the GSVA package (v1.40.1) (57) with standard settings based on the KEGG dataset. To assess differential activities of pathways between different types of cells, we contrasted the activity scores for CD8+ Teff against the CD8+ TN and CD8+ TCM cells by using the Wilcoxon test, and an adjusted P value <0.05 was used as the cutoff value for significant pathways identification.
Cell-cell communication
To determine global communications among cells, CellChat (58) was used for the cell-cell communication analysis. The gene expression data of cells were used as the input, and then the CellChat object was created with the metadata, including the cell type and group information. Then, the significant ligand-receptor pairs among cell groups were identified by performing a permutation test and categorized into signaling pathways. Pathways were filtered out if there were only 10 cells in certain cell groups. Next, the key incoming and outgoing signals were predicted for each cell group and global communication patterns by leveraging pattern recognition approaches. To cluster the signaling pathways, the similarity of pathways was measured, and manifold learning was performed from both functional and topological perspectives. The overall communication probability analysis was performed across all the datasets.
TF regulon prediction
Gene regulatory network was generated using the SCENIC package (59). Briefly, the raw expression matrix for the cells of all samples was filtered by keeping genes with default parameters, and then the coexpressed gene modules and the potential TF targets (regulons) for each module were identified with the GENIE3 based on the expression matrix. Cis-regulatory motif analysis was performed by scanning two TF databases (https://resources.aertslab.org/cistarget/) (mm10__refseqr80__10kb_up_and_down_tss.mc9nr.feather,mm10__refseqr80__500bp_up_and_100bp_down_tss.mc9nr.feather) with the RcisTarget implemented in SCENIC. The modules with significant motif enrichment were kept and termed as regulons. To visualize the regulon activity for each cell, the area under the curve (AUC) scores (regulon activities) in each cell were computed and binarized the regulon network activity based on the AUCell algorithm, and we used binary regulon activity matrix to visualize regulon activity.
Statistical analysis
Statistical analysis was performed using Prism 9 (GraphPad Software), and significance was determined by unpaired two-tailed t test for the comparison of means between two groups, by one-way analysis of variance (ANOVA) with Tukey’s post hoc test for multiple comparisons between groups, and by two-way ANOVA test for flow cytometric kinetic data; body weight change data with only the significance of comparisons by time point are shown in the figures for clarity. P values less than 0.05 were considered significant. All the experiments were repeated at least twice.
Acknowledgments
We thank J. Kolls (Center for Translational Research in Infection and Inflammation Tulane School of Medicine) for valuable inputs in scRNA-seq and overall interpretation of data. We thank R. Varma and T. Arekar for performing TCID50 assay. We thank S. Solaymani-Mohammadi (University of North Dakota School of Medicine and Health Sciences) for critical reading of the manuscript. We also thank the support of the scRNA-seq data analysis from the Computational Data Analysis Core at the University of North Dakota (supported by NIH COBRE grant P20GM113123).
Funding: This work was supported by NIH grants R01 AI143741 and R21 AI151522 to N.K. M.A.O. was supported by Biomedical Laboratory Research and Development (BLRD) Research Career Scientist Award IK6BX005966. J.X. was supported by the American Lung Association Grant CA-827199 and the Pandemic Research Recovery Program from the University of Michigan Medical School. J.H. was supported by the National Institute of General Medical Sciences awards P20GM113123.
Author contributions: N.K. conceived the research and designed the experiments. D.J.K.Y., K.G., T.S., J.T., Z.W., N.A., and Z.N. performed and analyzed the experiments. K.G. and N.K. analyzed and interpreted the scRNA-seq data. N.K., K.G., D.J.K.Y., M.A.O., J.X., J.H., J.S., B.D.K., N.A., R.M., and Z.W. wrote and revised the manuscript. All authors provided comments on the manuscript. All authors have seen and approved the manuscript, which has not been accepted or published elsewhere.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The scRNA-seq data reported in this study have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) under the accession number GSE186839. The scRNA-seq datasets for the whole lung were collected from NCBI’s BioProject database (PRJNA733762). The script for the preprocessing of the data is publicly available on GitHub (https://github.com/guokai8/Cd8_T_cell) and deposited in Zenodo (https://zenodo.org/records/10139369).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Table S1
Legends for data S1 to S9
Other Supplementary Material for this manuscript includes the following:
Data S1 to S9
REFERENCES AND NOTES
- 1.Houser K., Subbarao K., Influenza vaccines: Challenges and solutions. Cell Host Microbe 17, 295–300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Putri W., Muscatello D. J., Stockwell M. S., Newall A. T., Economic burden of seasonal influenza in the United States. Vaccine 36, 3960–3966 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Kalil A. C., Thomas P. G., Influenza virus-related critical illness: Pathophysiology and epidemiology. Crit. Care 23, 258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taubenberger J. K., Morens D. M., The pathology of influenza virus infections. Annu. Rev. Pathol. 3, 499–522 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clementi N., Ghosh S., de Santis M., Castelli M., Criscuolo E., Zanoni I., Clementi M., Mancini N., Viral respiratory pathogens and lung injury. Clin. Microbiol. Rev. 34, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeler S. P., Agapov E. V., Hinojosa M. E., Letvin A. N., Wu K., Holtzman M. J., Influenza a virus infection causes chronic lung disease linked to sites of active viral RNA remnants. J. Immunol. 201, 2354–2368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothberg M. B., Haessler S. D., Brown R. B., Complications of viral influenza. Am. J. Med. 121, 258–264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Wu J., Hao S., Yang M., Lu X., Chen X., Li L., Long term outcomes in survivors of epidemic influenza A (H7N9) virus infection. Sci. Rep. 7, 17275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers M. A., Smith A. P., Lane L. C., Moquin D. J., Aogo R., Woolard S., Thomas P., Vogel P., Smith A. M., Dynamically linking influenza virus infection kinetics, lung injury, inflammation, and disease severity. eLife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klomp M., Ghosh S., Mohammed S., Nadeem Khan M., From virus to inflammation, how influenza promotes lung damage. J. Leukoc. Biol. 110, 115–122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brauer R., Chen P., Influenza leaves a TRAIL to pulmonary edema. J. Clin. Invest. 126, 1245–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Luna X., Hartshorn K. L., Influenza casts a lung shadow. Am. J. Pathol. 187, 697–699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan S., Thomas P. G., Balancing immune protection and immune pathology by CD8+ T-cell responses to influenza infection. Front. Immunol. 7, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Sandt C. E., Bárcena M., Koster A. J., Kasper J., Kirkpatrick C. J., Scott D. P., de Vries R. D., Herold S., Rimmelzwaan G. F., Kuiken T., Short K. R., Human CD8+ T cells damage noninfected epithelial cells during influenza virus infection in vitro. Am. J. Respir. Cell Mol. Biol. 57, 536–546 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Zhao M. Q., Stoler M. H., Liu A. N., Wei B., Soguero C., Hahn Y. S., Enelow R. I., Alveolar epithelial cell chemokine expression triggered by antigen-specific cytolytic CD8+ T cell recognition. J. Clin. Invest. 106, R49–R58 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mauad T., Hajjar L. A., Callegari G. D., da Silva L. F. F., Schout D., Galas F. R. B. G., Alves V. A. F., Malheiros D. M. A. C., Auler J. O. C. Jr., Ferreira A. F., Borsato M. R. L., Bezerra S. M., Gutierrez P. S., Caldini E. T. E. G., Pasqualucci C. A., Dolhnikoff M., Saldiva P. H. N., Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Respir. Crit. Care Med. 181, 72–79 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Bender B. S., Croghan T., Zhang L., Small P. A. Jr., Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J. Exp. Med. 175, 1143–1145 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moskophidis D., Kioussis D., Contribution of virus-specific CD8+ cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. J. Exp. Med. 188, 223–232 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullen S. P., Martin S. J., Mechanisms of granule-dependent killing. Cell Death Differ. 15, 251–262 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Topham D. J., Tripp R. A., Doherty P. C., CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J. Immunol. 159, 5197–5200 (1997). [PubMed] [Google Scholar]
- 21.Major J., Crotta S., Llorian M., McCabe T. M., Gad H. H., Priestnall S. L., Hartmann R., Wack A., Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369, 712–717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson S., Crotta S., McCabe T. M., Wack A., Pathogenic potential of interferon αβ in acute influenza infection. Nat. Commun. 5, 3864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzemaekers M., Vanheule V., Janssens R., Struyf S., Proost P., Overview of the mechanisms that may contribute to the non-redundant activities of interferon-inducible CXC chemokine receptor 3 ligands. Front. Immunol. 8, 1970 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groom J. R., Luster A. D., CXCR3 in T cell function. Exp. Cell Res. 317, 620–631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurachi M., Kurachi J., Suenaga F., Tsukui T., Abe J., Ueha S., Tomura M., Sugihara K., Takamura S., Kakimi K., Matsushima K., Chemokine receptor CXCR3 facilitates CD8+ T cell differentiation into short-lived effector cells leading to memory degeneration. J. Exp. Med. 208, 1605–1620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brincks E. L., Katewa A., Kucaba T. A., Griffith T. S., Legge K. L., CD8 T cells utilize TRAIL to control influenza virus infection. J. Immunol. 181, 4918–4925 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui W., Kaech S. M., Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol. Rev. 236, 151–166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J. Y., Konno H., Guo X., Ye Y., Gao S., Wang S., Hu X., Ren X., Shen Z., Ouyang W., Zhang Z., Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin A., Masson F., Liao Y., Preston S., Guan T., Gloury R., Olshansky M., Lin J. X., Li P., Speed T. P., Smyth G. K., Ernst M., Leonard W. J., Pellegrini M., Kaech S. M., Nutt S. L., Shi W., Belz G. T., Kallies A., A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17, 422–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohlmeier J. E., Reiley W. W., Perona-Wright G., Freeman M. L., Yager E. J., Connor L. M., Brincks E. L., Cookenham T., Roberts A. D., Burkum C. E., Sell S., Winslow G. M., Blackman M. A., Mohrs M., Woodland D. L., Inflammatory chemokine receptors regulate CD8+ T cell contraction and memory generation following infection. J. Exp. Med. 208, 1621–1634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aksoy M. O., Yang Y., Ji R., Reddy P. J., Shahabuddin S., Litvin J., Rogers T. J., Kelsen S. G., CXCR3 surface expression in human airway epithelial cells: Cell cycle dependence and effect on cell proliferation. Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L909–L918 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Bangs D. J., Tsitsiklis A., Steier Z., Chan S. W., Kaminski J., Streets A., Yosef N., Robey E. A., CXCR3 regulates stem and proliferative CD8+ T cells during chronic infection by promoting interactions with DCs in splenic bridging channels. Cell Rep. 38, 110266 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler K. L., Clancy-Thompson E., Mullins D. W., CXCR3+ monocytes/macrophages are required for establishment of pulmonary metastases. Sci. Rep. 7, 45593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlin L. E., Hemann E. A., Zacharias Z. R., Heusel J. W., Legge K. L., Natural killer cell recruitment to the lung during influenza A virus infection is dependent on CXCR3, CCR5, and virus exposure dose. Front. Immunol. 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelsen S. G., Aksoy M. O., Yang Y., Shahabuddin S., Litvin J., Safadi F., Rogers T. J., The chemokine receptor CXCR3 and its splice variant are expressed in human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 287, L584–L591 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Kohlmeier J. E., Cookenham T., Miller S. C., Roberts A. D., Christensen J. P., Thomsen A. R., Woodland D. L., CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J. Immunol. 183, 4378–4384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connors T. J., Ravindranath T. M., Bickham K. L., Gordon C. L., Zhang F., Levin B., Baird J. S., Farber D. L., Airway CD8+ T Cells are associated with lung injury during infant viral respiratory tract infection. Am. J. Respir. Cell Mol. Biol. 54, 822–830 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goplen N. P., Wu Y., Son Y. M., Li C., Wang Z., Cheon I. S., Jiang L., Zhu B., Ayasoufi K., Chini E. N., Johnson A. J., Vassallo R., Limper A. H., Zhang N., Sun J., Tissue-resident CD8+ T cells drive age-associated chronic lung sequelae after viral pneumonia. Sci. Immunol. 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slütter B., Pewe L. L., Kaech S. M., Harty J. T., Lung airway-surveilling CXCR3hi memory CD8+ T cells are critical for protection against influenza A virus. Immunity 39, 939–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fadel S. A., Bromley S. K., Medoff B. D., Luster A. D., CXCR3-deficiency protects influenza-infected CCR5-deficient mice from mortality. Eur. J. Immunol. 38, 3376–3387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichikawa A., Kuba K., Morita M., Chida S., Tezuka H., Hara H., Sasaki T., Ohteki T., Ranieri V. M., dos Santos C. C., Kawaoka Y., Akira S., Luster A. D., Lu B., Penninger J. M., Uhlig S., Slutsky A. S., Imai Y., CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am. J. Respir. Crit. Care Med. 187, 65–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia G. L., Valenzuela A., Manzoni T., Vaughan A. E., Lopez C. B., Distinct chronic post-viral lung diseases upon infection with influenza or parainfluenza viruses differentially impact superinfection outcome. Am. J. Pathol. 190, 543–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Goplen N. P., Sun J., Aging and respiratory viral infection: From acute morbidity to chronic sequelae. Cell Biosci. 11, 112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmit T., Guo K., Tripathi J. K., Wang Z., McGregor B., Klomp M., Ambigapathy G., Mathur R., Hur J., Pichichero M., Kolls J., Khan M. N., Interferon-γ promotes monocyte-mediated lung injury during influenza infection. Cell Rep. 38, 110456 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Björkström N. K., Strunz B., Ljunggren H. G., Natural killer cells in antiviral immunity. Nat. Rev. Immunol. 22, 112–123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolumam G. A., Thomas S., Thompson L. J., Sprent J., Murali-Krishna K., Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202, 637–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welsh R. M., Bahl K., Marshall H. D., Urban S. L., Type 1 interferons and antiviral CD8 T-cell responses. PLOS Pathog. 8, e1002352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prabhu N., Ho A. W., Wong K. H. S., Hutchinson P. E., Chua Y. L., Kandasamy M., Lee D. C. P., Sivasankar B., Kemeny D. M., Gamma interferon regulates contraction of the influenza virus-specific CD8 T cell response and limits the size of the memory population. J. Virol. 87, 12510–12522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graham M. B., Dalton D. K., Giltinan D., Braciale V. L., Stewart T. A., Braciale T. J., Response to influenza infection in mice with a targeted disruption in the interferon-γ gene. J. Exp. Med. 178, 1725–1732 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaush C. R., Smith T. F., Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl. Microbiol. 16, 588–594 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmit T., Ghosh S., Mathur R. K., Barnhardt T., Ambigapathy G., Wu M., Combs C., Khan M. N., IL-6 deficiency exacerbates allergic asthma and abrogates the protective effect of allergic inflammation against Streptococcus pneumoniae pathogenesis. J. Immunol. 205, 469–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benton K. A., Misplon J. A., Lo C. Y., Brutkiewicz R. R., Prasad S. A., Epstein S. L., Heterosubtypic immunity to influenza A virus in mice lacking IgA, all Ig, NKT cells, or γδ T cells. J. Immunol. 166, 7437–7445 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Zheng G. X., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., Mc Dermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., Farland C. M., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A. K., Slichter C. K., Miller H. W., McElrath M. J., Prlic M., Linsley P. S., Gottardo R., MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin S., Guerrero-Juarez C. F., Zhang L., Chang I., Ramos R., Kuan C. H., Myung P., Plikus M. V., Nie Q., Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van de Sande B., Flerin C., Davie K., De Waegeneer M., Hulselmans G., Aibar S., Seurinck R., Saelens W., Cannoodt R., Rouchon Q., Verbeiren T., De Maeyer D., Reumers J., Saeys Y., Aerts S., A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1
Legends for data S1 to S9
Data S1 to S9