Abstract
This work determined and analyzed the complete chloroplast genome sequence of Amorphophallus konjac K. Koch ex N.E.Br 1858 from Yunnan, China. The genome size was 167,470 bp, of which contains a large single-copy region (LSC 93,443 bp), a small single-copy region (SSC 21,575 bp), and a pair of inverted repeat regions (IR 26,226 bp). The chloroplast genome has 131 genes, including 86 protein-coding genes, 37 tRNAs, and eight rRNAs. A previous study reported deletion of accD, psbE, and trnG-GCC genes in the A. konjac chloroplast genome. Our study supports the conservative structure of A. konjac and does not support the gene deletion mentioned above. Phylogenetic analysis indicated that A. konjac shares a close relationship with another A. konjac (collected from Guizhou) and A. titanium by forming a clade in the genus Amorphophallus. Our results provide some useful information to the evolution of the family Araceae.
Keywords: Araceae, amorphophallus konjac K. Koch ex N.E.Br 1858, chloroplast genome, phylogenetic analysis
Introduction
Amorphophallus konjac K. Koch ex N.E.Br 1858 is a perennial, herbaceous monocot that is mainly distributed in Southeast Asia and Africa and belongs to the genus Amorphophallus (Chua et al. 2010; Hu et al. 2019). A. konjac also is an important economic crop widely used in health products and biomaterials because of its tuber contains a large amount of konjac glucomannan (KGM) (Gao et al. 2022). The high quality and purity of KGM obtained from A. konjac makes this species the first most cultivated Amophophallus species in China, especially in Yunnan, Guizhou, and Hubei (Sun et al. 2023). Previously, two chloroplast genomes of A. konjac from different regions have been reported (Hu et al. 2019; Liu et al. 2019). Moreover, a chloroplast genome of A. konjac reported by Liu et al. (2019) found that some genes deletion, including deletion of accD, psbE, and trnG-GCC. This phenomenon may be attributed to errors in either the assembly, or the annotations, or both (Henriquez et al. 2021). Yunnan province is one of the largest plantation areas of A. konjac in China and has many endemic local A. konjac resources. The local A. konjac resources of Fuyuan are the most representative local varieties in Yunnan, and also an economically important crop for rural revitalization in Yunnan Province. To date, the chloroplast genome of local A. konjac resources of Fuyuan prefecture is still lacking. To develop and utilize local A. konjac resources of Fuyuan prefecture in Yunnan, and to compare the chloroplast of A. konjac from different distributed regions, we sequenced the complete chloroplast genome of A. konjac from Yunan, China.
Materials and methods
The corms of Fuyuan local A. konjac resources were cultivated in Kunming University. Then, the fresh leaves of A. konjac were collected from the Konjac Genetic Resources Garden of Kunming University, Yunnan Province (Figure 1, 24.97406°N, 102.79605°E), and a specimen was deposited at the Herbarium of Yunnan Urban Agricultural Engineering and Technological Research Center (Kunming, China, Website: https://www.kmu.edu.cn/zzjg/kyjg.htm, Li-Fang Li, lilf0215@163.com) under the voucher number HMY20230616. Total genomic DNA was extracted and sequenced on the Illumina HiSeq 2500 platform (Illumina, CA, USA). The quality of the short reads was checked with FastQC. De novo assembly was conducted using metaSPAdes (Sun et al. 2022). The complete chloroplast genome of A. konjac (MK611803) from Guizhou was used as a reference. The assembled chloroplast genome was annotated by CPGAVAS2 (Shi et al. 2019), whereas the tRNA genes were further confirmed by tRNAscan-SE v.2.0 (Chan et al. 2021). MAFFT v7.419 was employed to align the chloroplast genome sequence of three Amorphophallus species and adjusted manually with BioEdit (Katoh et al. 2013). Then, DnaSP v6 software (Rozas et al. 2017) was used to identify high variation regions with a sliding window analysis with a window length of 600 bp and a step size of 200 bp. Furthermore, the maps of the annotated chloroplast genome and cis-splicing/trans-splicing genes of A. konjacwere processed by CPGview (Liu et al. 2023).
Figure 1.
Species reference image of Amorphophallus konjac. (A) Plant of A. konjac. (B) flower of A. konjac. (C) corm of A. konjac. These photos were taken by Penghua Gao and Yu Lei at Kunming city, Yunnan Province, China. It is a perennial herb with dark purple flowers and green palmate compound leaves. The petiole has the background color dirty cream-colored, often nearly entirely covered by blackish green spots.
The complete chloroplast genomes of 26 Araceae species were downloaded from GenBank and aligned using MAFFT v7. Then, the maximum likelihood (ML) method was used to construct the phylogenetic tree by using IQ-TREE v1.6.12 (Nguyen et al. 2015) with 1000 bootstrap replicates. The Atractylodes chinensis (MT834519) and Iris missouriensis as outgroups.
Results
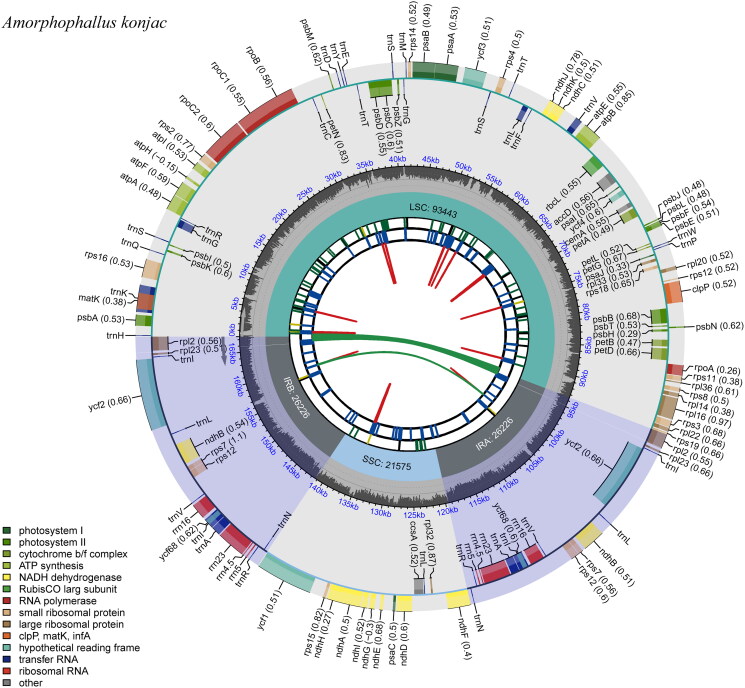
The chloroplast genome of A. konjac was 167,470 bp in length, like other reported plant chloroplast genomes, the genome has a conserved quadripartite structure, including a large single copy (LSC, 93,443 bp), a small single copy (SSC, 21,575 bp) and a pair of inverted repeats (IRa and IRb, 26,226 bp) (Table S1, Figure 2). Finally, the obtained chloroplast genome sequence was constructed with a 4500 coverage depth (Figure S1) and submitted to the NCBI database (Accession number: OR438675). Additionally, the overall GC content in complete chloroplast genomes was 35.4%, and the GC contents of the LSC, SSC, IRa, and IRb regions were 35.4%, 29.7%, 41.5%, and 41.5%, respectively (Table S1). The genome encodes a total of 131 genes, including 86 protein-coding genes, 37 transfer-RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA) (Table S1). Among them, six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) and 13 protein-coding genes (petB, petD, atpF, ndhA, ndhB, rps12, rps16, rpl16, atpF, rpoC1, rpl2, accD, ycf68) containing one intron and two genes (ycf 3, clpP) having two introns, whereas all other genes contained one intron (Table S2). Moreover, most of the gene species occurred in a single copy, while 19 gene species occurred in double copies, including all rRNA species, eight tRNA species, and 7 protein-coding species (Table S2). Meanwhile, the chloroplast genome contained 16 cis-splicing genes (Figure S2) and one trans-splicing gene (Figure S3). In addition, the nucleotide variability (Pi) values of A. konjac and the closely related species A. konjac from Guizhou and A.titanum were analyzed. The pi value of nucleotide diversity ranged from 0 to 0.13, and a total of seven highly variable sites were found, including rps16 (0.13), ycf1 (0.068), trnS-GCU (0.055), ycf4-cemA (0.053), trnG-UCC (0.043), trnC-GCA-petN (0.422) and psbD (0.036) (Figure S4).
Figure 2.
The chloroplast genome map of Amorphophallus konjac. From the center, the first track shows the dispersed repeats, including direct repeats (red) and palindromic repeats (green). The second and third tracks show the long and short tandem repeats, respectively. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted in the fifth track. The genes are shown on the sixth track.
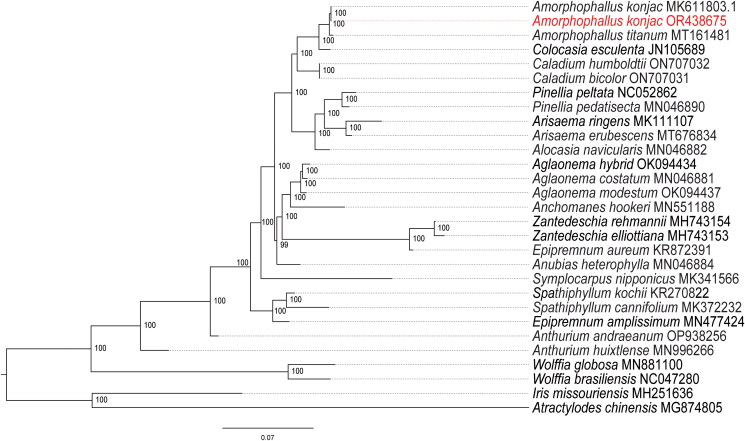
To analyze the phylogenetic relationship of Araceae species, we constructed a phylogenetic tree using whole chloroplast genome sequences. The maximum likelihood (ML) tree showed that species in the genus Amorphophallus were grouped into a monophyletic group with high support value, and A. konjac is a sister to A. konjac (MK611803) which was collected in Guizhou, China. In addition, A. konjac was clustered with A. titanum into a subclade with 100% supported value and the genus Amorphophallus was sister to the genus Colocasia, Caladium and Pinellia (Figure 3).
Figure 3.
Phylogenomic tree of Amorphophallus konjac (in red) and 28 species constructed using maximum-likelihood method based on complete chloroplast genome sequences. Iris missouriensis and Atractylodes chinensis are used as an outgroups. The best-fit model according to the bayesian information criterion (BIC) was TVM + F+R6. Numbers at each node represent the bootstrap values for 1000 replicates. The following sequences were used: Amorphophallus konjac MK611803.1 (Hu et al. 2019), Amorphophallus titanum MT161481 (Henriquez et al. 2021), Colocasia esculenta JN105689 (Ahmed et al. 2012), Caladium humboldtii ON707032 (Ye et al. 2022), Caladium bicolor ON707031 (Ye et al. 2022), Pinellia peltata NC052862 (unpublished), Pinellia pedatisecta MN046890 (Henriquez, Ahmed, et al. 2020), Arisaema ringens MK111107 (unpublished), Arisaema erubescens MT676834 (Zhang et al. 2020), alocasia navicularis MN046882 (Henriquez, Ahmed, et al. 2020), Aglaonema hybrid OK094434 (Li et al. 2022), Aglaonema costatum MN046881 (Henriquez, Ahmed, et al. 2020), Aglaonema modestum OK094437 (Li et al. 2022), anchomanes hookeri MN551188 (Henriquez, Ahmed, et al. 2020), Zantedeschia rehmannii MH743154 (He et al. 2020), Zantedeschia elliottiana MH743153 (He et al. 2020), Epipremnum aureum KR872391 (Tian et al. 2018), anubias heterophylla MN046884 (Henriquez, Ahmed, et al. 2020), symplocarpus nipponicus MK341566 (Kim et al. 2019), Spathiphyllum kochii KR270822 (Han et al. 2016), Spathiphyllum cannifolium MK372232 (Liu et al. 2019), Epipremnum amplissimum MN477424(unpublished), Anthurium andraeanum OP938256 (Wan et al. 2023), Anthurium huixtlense MN996266 (Henriquez, Mehmood, et al. 2020),Wolffia globosa MN881100 (unpublished), Wolffia brasiliensis NC047280 (unpublished), Iris missouriensis MH251636 (unpublished), Atractylodes chinensis MG874805 (Wang et al. 2020).
Discussion and conclusion
In the current study, the chloroplast genome of A. konjac was 167,470 bp in length which was similar to the published one (A. konjac from Wuhan, SRR7938681, 167, 424 bp) (Table S1). On the contrary, the chloroplast genome assembled in this study was 1548 bp longer than that of another published one (A. konjac MK611803, 161,647 bp) mainly due to the expansion of the LSC and IR regions (Table S1). Furthermore, the overall GC content (35.63%) and genome quadripartite structure were highly similar to those in other Araceae chloroplast genomes, including A. konjac and A. titanium (Hu et al. 2019; Henriquez et al. 2021). Significantly, a previous study reported deletion of accD, psbE and trnG-GCC genes in A. konjac from Wuhan (Liu et al. 2019). Henriquez et al. (2021) speculate that this phenomenon may be caused by either assembly errors, or annotation errors, or both. The Organellar Genome Annotator (DOGMA) annotation software they use is prone to errors (Henriquez et al. 2021). Our study shows similar gene content to previous reports in aroids as well as to A. konjac (MK611803.1) and A. titanium (MT161481), which supports the conservative structure of chloroplast genomes in the Amorphophallus species and does not support gene deletion mentioned above. Our phylogeny showed that A. konjac was closely related to A. konjac (MK611803.1) and A. titanium (MT161481), which is congruent with the previous study (Henriquez et al. 2021). Finally, this study improve our understanding of the characteristics of the chloroplast genome of A. konjac and are valuable for future breeding and research efforts.
Supplementary Material
Funding Statement
This study was funded by Yunnan Province Youth Talent Support Program [YNWR-QNBJ-2018-324], Yunnan Provincial Science and Technology Department [202101BA070001-163], Yunnan Education Department Research Project [2022J0644, 2023J0827].
Authors contributions
Lifang Li conceived the project and wrote the manuscript. Ying Qi and Penghua Gao conceived the project and collected the samples. Shaowu Yang, Yongteng Zhao, Jianwei Guo, and Jiani Liu performed the data analysis. Feiyan Huang conceived, designed the study, and edited the paper. Lei Yu conceived the study, interpreted the data, and revised the manuscript critically. All authors read and approved the final manuscript.
Ethical approval
Amorphophallus konjac is not an endangered or protected species; therefore, permission is not required to collect this species. Research on this species, including the collection of plant materials has been carried out in accordance with guidelines provided by Kunming University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The assembled chloroplast genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number of OR438675. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1008633, SRR25741018, and SAMN37123742, respectively.
References
- Ahmed I, Biggs PJ, Matthews PJ, Collins LJ, Hendy MD, Lockhart PJ.. 2012. Mutational dynamics of aroid chloroplast genomes. Genome Biol Evol. 4(12):1316–1323. doi: 10.1093/gbe/evs110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lin BY, Mak AJ, Lowe TM.. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49(16):9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua M, Baldwin TC, Hocking TJ, Chan K.. 2010. Traditional uses and potential health benefits of Amorphophallus konjac K. Koch ex N.E.Br. J Ethnopharmacol. 128(2):268–278. doi: 10.1016/j.jep.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Feng C, Chu H, Feng C, Wang H, Wu L, Yin S, Liu C, Chen H, et al. 2022. A chromosome-level genome assembly of Amorphophallus konjac provides insights into konjac glucomannan biosynthesis. Comput Struct Biotechnol J. 20:1002–1011. doi: 10.1016/j.csbj.2022.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Wang B, Wang ZZ.. 2016. The complete chloroplast genome sequence of Spathiphyllum kochii. Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2973–2974. doi: 10.3109/19401736.2015.1060466. [DOI] [PubMed] [Google Scholar]
- He S, Yang Y, Li Z, Wang X, Guo Y, Wu H.. 2020. Comparative analysis of four Zantedeschia chloroplast genomes: expansion and contraction of the IR region, phylogenetic analyses and SSR genetic diversity assessment. PeerJ. 8:e9132. doi: 10.7717/peerj.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez CL, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR.. 2020. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics. 112(3):2349–2360. doi: 10.1016/j.ygeno.2020.01.006. [DOI] [PubMed] [Google Scholar]
- Henriquez CL, Mehmood F, Carlsen MM, Islam M, Waheed MT, Poczai P, Croat TB, Ahmed I.. 2020. Complete chloroplast genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): unique inverted repeat expansion and contraction affect rate of evolution. J Mol Evol. 88(7):562–574. doi: 10.1007/s00239-020-09958-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez CL, Mehmood F, Hayat A, Sammad A, Waseem S, Waheed MT, Matthews PJ, Croat TB, Poczai P, Ahmed I.. 2021. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics. 113(1 Pt):183–192. doi: 10.1016/j.ygeno.2020.12.016. [DOI] [PubMed] [Google Scholar]
- Hu H, Liu J, Wang B, An J, Wang Q.. 2019. Characterization of the complete chloroplast genome of Amorphophallus konjac (Araceae) and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 4(1):1658–1659. doi: 10.1080/23802359.2019.1606683. [DOI] [Google Scholar]
- Katoh K, Standley D, Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Yang J, Park J, Yamada T, Maki M, Kim SC.. 2019. Comparison of whole plastome sequences between thermogenic skunk cabbage Symplocarpus renifolius and Nonthermogenic S. nipponicus (Orontioideae; Araceae) in East Asia. Int J Mol Sci. 20(19):4678. doi: 10.3390/ijms20194678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DM, Zhu GF, Yu B, Huang D.. 2022. Comparative chloroplast genomes and phylogenetic relationships of Aglaonema modestum and five variegated cultivars of Aglaonema. PLoS One. 17(9):e0274067. doi: 10.1371/journal.pone.0274067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E, Yang C, Liu J, Jin S, Harijati N, Hu Z, Diao Y, Zhao L.. 2019. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci Rep. 9(1):809. doi: 10.1038/s41598-018-37456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C.. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen W, Li F, Li C, Xie X, Chao Z, Tian E.. 2019. The complete chloroplast genome sequence of Spathiphyllum Cannifolium. Mitochondrial DNA Part B. 4(2):4041–4042. doi: 10.1080/23802359.2019.1613191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A.. 2017. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol Biol Evol. 34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C.. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qiu Z, Egan R, Ho H, Li Y, Wang Z.. 2022. Persistent memory as an effective alternative to random access memory in metagenome assembly. BMC Bioinformatics. 23(1):513. doi: 10.1186/s12859-022-05052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu S, Lu Y, Wu F, Chen X.. 2023. First Report of Anthracnose Caused by Colletotrichum camelliae on Amorphophallus konjac in Hunan Province of China. Plant Dis. doi: 10.1094/PDIS-01-23-0150-PDN. [DOI] [PubMed]
- Tian N, Han L, Chen C, Wang Z.. 2018. The complete chloroplast genome sequence of Epipremnum aureum and its comparative analysis among eight Araceae species. PLoS One. 13(3):e0192956. doi: 10.1371/journal.pone.0192956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Ge Y, Pan G, Tian D.. 2023. The complete chloroplast genome sequence of Anthurium andraeanum Linden (Araceae; Pothoideae). Mitochondrial DNA B Resour. 8(3):379–382. doi: 10.1080/23802359.2023.2185081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang H, Wu X, Wang Z, Fang W, Jiang M, Chen H, Huang L, Liu C.. 2020. Phylogenetic relationships of Atractylodes lancea, A. chinensis and A. macrocephala, revealed by complete plastome and nuclear gene sequences. PLOS One. 15(1):e0227610. doi: 10.1371/journal.pone.0227610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Liu J, Zhou Y, Zhu G, Tan J, Xu Y.. 2022. Complete chloroplast genome sequences of four species in the Caladium genus: comparative and phylogenetic analyses. Genes. 13(12):2180. doi: 10.3390/genes13122180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo X, Yan B.. 2020. Characterization of the complete chloroplast genome of Arisaema erubescens (Wall.) Schott, a traditional Chinese medicinal herb. Mitochondrial DNA B Resour. 5(3):3149–3150. doi: 10.1080/23802359.2020.1797577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
Supplementary Materials
Data Availability Statement
The assembled chloroplast genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number of OR438675. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1008633, SRR25741018, and SAMN37123742, respectively.