Abstract
Antiretroviral therapy has been effective in suppressing HIV viral load and enabling people living with HIV to experience longer, more conventional lives. However, as people living with HIV are living longer, they are developing aging-related diseases prematurely and are more susceptible to comorbidities that have been linked to chronic inflammation. Coincident with HIV infection and aging, drug abuse has also been independently associated with gut dysbiosis, microbial translocation, and inflammation. Here, we hypothesized that injection drug use would exacerbate HIV-induced immune activation and inflammation, thereby intensifying immune dysfunction. We recruited 50 individuals not using injection drugs (36/50 HIV+) and 47 people who inject drugs (PWID, 12/47 HIV+). All but 3 of the HIV+ subjects were on antiretroviral therapy. Plasma immune profiles were characterized by immunoproteomics, and cellular immunophenotypes were assessed using mass cytometry. The immune profiles of HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+ were each significantly different from controls; however, few differences between these groups were detected, and only 3 inflammatory mediators and 2 immune cell populations demonstrated a combinatorial effect of injection drug use and HIV infection. In conclusion, a comprehensive analysis of inflammatory mediators and cell immunophenotypes revealed remarkably similar patterns of immune dysfunction in HIV-infected individuals and in people who inject drugs with and without HIV-1 infection.
Keywords: inflammation, mass cytometry, Olink, PLWH, PWID, soluble mediators, speedball
1. Introduction
Injection drug use (IDU) of opioids and synthetic opioids (i.e. fentanyl) has become an ongoing epidemic in the United States and its territories. In the US territory of Puerto Rico, people who inject drugs (PWID) primarily use speedball, a mixture of cocaine and heroin or its derivatives.1,2 Moreover, speedball is often laced with fentanyl, which remains unknown to the majority of Puerto Rican PWID.3 This group also experiences high rates of infection with HIV and hepatitis C virus (HCV) that have been predicted to manifest in other regions of the United States with high incidences of IDU.4–6 The distinctive feature of untreated HIV infection is the depletion of CD4+ T cells, resulting in immune suppression. In addition, the depletion of CD4+ T cells in the gut lining reduces barrier integrity between the gut and the circulatory system, enabling microbes and microbial products to enter the bloodstream, where they may induce systemic inflammation.7–9 Further, HIV infection is known to induce dysbiosis of the gut microbiome, which could enhance microbial translocation and inflammation even in the virally suppressed.10,11
Antiretroviral therapy (ART) has effectively suppressed HIV viral load, allowing people living with HIV (PLWH), including those in Puerto Rico, to live longer, more typical lives.12,13 However, addiction and IDU among PLWH may counteract the effectiveness of ART. Moreover, chronic diseases such as diabetes, cardiovascular diseases, premature aging, and neurologic diseases seen in PLWH could further be enhanced due to IDU, coinfection, dysbiosis, and chronic inflammation.14,15 For example, methamphetamine injectors displayed moderate elevation in lipopolysaccharide (LPS)–binding protein, soluble CD163 (sCD163), IL-6, and soluble tumor necrosis factor–alpha receptor 1 (sTNF-αR1) compared to methamphetamine noninjectors.16 Additionally, individuals with cocaine addiction had higher levels of IL-6 than healthy controls,17 and cocaine use has also been implicated in accelerating HIV disease progression.17 Nevertheless, detoxification of cocaine users led to cytokine levels realigning with levels detected in healthy individuals.18 Likewise, opioids have been linked to reduced gut motility, increased gut permeability, and potentially dysbiosis.11,19,20 PLWH with opioid use disorder displayed increased levels of sCD163, intermediate monocytes, and nonclassical monocytes compared to PLWH who did not use opioids.21 Further, PLWH with opioid use disorder showed a diminished ability to produce cytokines upon LPS stimulation ex vivo.21 Similarly, HCV-infected speedball users exhibited higher plasma cytokine levels than healthy controls but decreased lymphocyte proliferation.22 Therefore, IDU may demonstrate an additive or synergistic effect in inducing microbial translocation and persistent inflammation in PLWH, whether ART-treated or not.
Even though both IDU and HIV infection are individually associated with dysbiosis and inflammation, there has been limited information on comparative immune profiling in PWID with and without HIV infection. In this cross-sectional study, we investigated the potential synergistic impacts of chronic HIV infection and IDU on immune functions by comprehensively analyzing the inflammatory cytokines and mediators, as well as the immune cell population profiles between PLWH and PWID. From our analysis, we found remarkably similar patterns of immune dysfunction in individuals with HIV infection alone and PWID with and without HIV infection.
2. Materials and methods
2.1. Ethics statement
This study was approved by the institutional review boards of the University of Nebraska–Lincoln, Universidad Central del Caribe, and Louisiana State University Health Sciences Center–New Orleans. Written informed consent was obtained from all study participants, and all study data were deidentified before analysis and publication.
2.2. Patient recruitment and sample collection
Respondent-driven sampling (RDS) was used to recruit PWID and controls who do not inject drugs in San Juan, Puerto Rico, for this cross-sectional study.6,23 RDS, a type of chain referral recruitment, begins by selecting a small number of participants to take a survey. After completion, each participant is given recruitment coupons that they can pass out to people they know who qualify for the study. Those who are recruited through coupons are then given more coupons and recruitment spreads through the local community. Participants were eligible for an incentive for completing the survey and for each recruitment coupon that resulted in a new participant. Overall, most eligible participants who responded to solicitation (i.e. had an RDS coupon) did participate by attempting to schedule an appointment and enroll in the study. We were unable to estimate the proportion of coupons that were not passed on to potential new participants (i.e. the current participant is discreet about their status, the homeless participants may lose coupons, etc.), and thus the response rate could not be evaluated. At the study site after obtaining written informed consent and conducting the survey, we performed HIV rapid antibody tests (cat. 90-1019; BioLytical) to confirm each participant’s self-reported HIV status and collected urine samples for rapid drug use tests (CLIA-IDTC-14-BUPa; CLIAwaved, Inc.). Whole blood was collected in EDTA tubes and transferred to Universidad Central del Caribe (UCC) for processing. The plasma was separated by centrifugation (545 × g for 15 min), aliquoted, and stored at −80 °C. A portion of whole blood was reserved for mass cytometry staining.
2.3. Mass cytometry
The Maxpar Direct Immune Profiling Assay (MDIPA; cat. 201325; Fluidigm) was used per the manufacturer’s guidelines to quantify the levels of immune cell populations in whole blood (Fig. 1). Fresh whole blood was stained prior to stabilization and storage at −80 °C as recommended by the manufacturer. The Cytodelics whole-blood preservation kit was used to stabilize the stained whole blood for storage and shipping (cat. hC001-1000; Cytodelics) per the manufacturer’s guidelines. The processing of the sample was performed per manufacturer recommendations and Fluidigm’s application note, “Impact of Cryopreservation on Performance of the Maxpar Direct Immune Profiling System.” This is the recommended protocol by both manufacturers when sample collection, processing, and analysis are performed at separate study sites. Importantly, our study performed the staining of fresh whole blood prior to stabilization, freezing, and shipping in order to avoid any negative staining effects.
Fig. 1.
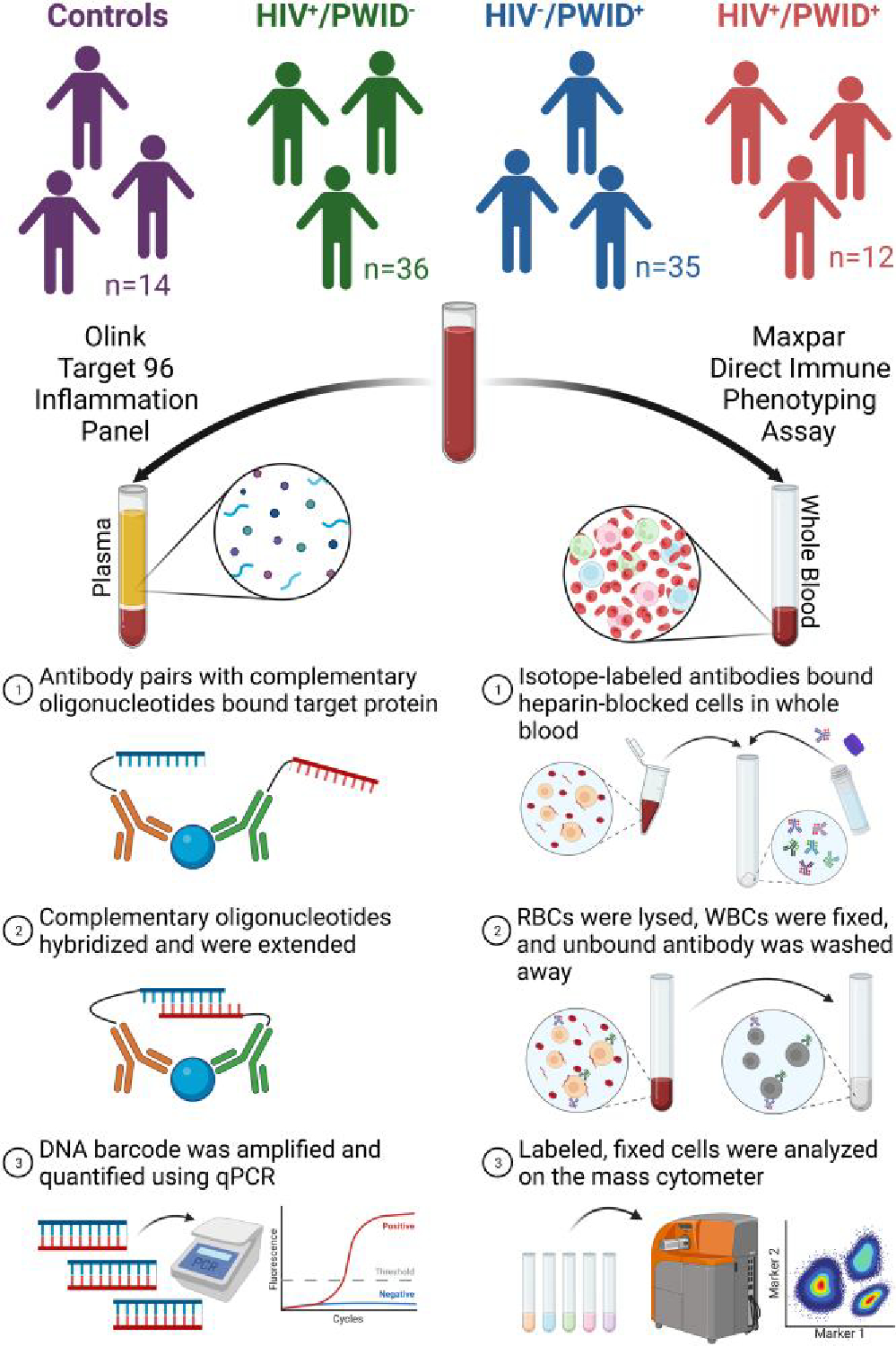
Graphical representation of study design and methodology. Whole-blood samples were collected and partitioned into plasma while reserving some whole blood. The plasma was subjected to the Olink proximity extension assay with the Olink Target 96 inflammation panel. The reserved whole blood was heparin blocked and isotope labeled before stabilization, fixation, washing, and quantification on the mass cytometer at OMRF. Created with BioRender.com. Abbreviations: RBCs, red blood cells; WBC, white blood cells.
Briefly, at UCC, 300 μL of whole blood was mixed with 3 μL of 10 kU/mL heparin solution and incubated at room temperature for 20 min to reduce nonspecific antibody binding. Next, 270 μL of heparin-blocked whole blood was transferred to 1 MDIPA tube. MDIPA contains a standard panel of 30 antibodies that can identify 37 immune cell populations. Then, 3 μL of each antibody in the T-cell expansion pack 1 (cat. 201405; Fluidigm) was added to the heparin-blocked whole blood in the MDIPA tube. Adding the T-cell expansion pack 1 allows further characterization of T-cell activation and exhaustion. The MDIPA tube was gently vortexed and incubated at room temperature for 30 min. Cytodelics Stabilizer was added to 2-mL cryovials (1:1 ratio with whole blood) and equilibrated to room temperature for 5 to 10 min. Finally, the stained whole blood was transferred from the MDIPA tube to the cryovials containing the Cytodelics Stabilizer, mixed by inverting 10 to 15 times, and incubated at room temperature for 10 min. The stabilized, stained whole blood was stored at −80 °C until shipment to the University of Nebraska–Lincoln (UNL).
At UNL, the ≈630 μL of stabilized, stained whole blood was thawed and mixed with 3 mL of 1× Cytodelics Fixation Buffer and incubated at room temperature for 15 min (vortexed multiple times throughout). Then, 12 mL of 1× Cytodelics Lysis Buffer was added and incubated for 15 to 20 min. Once the solution was crystal clear red, the cells were pelleted by centrifugation (300 × g for 5 min), and the supernatant was aspirated. The cells were fixed with fresh 1.6% formaldehyde for 10 min at room temperature. Finally, the cells were stained with Cell-ID Intercalater-Ir in Fix Perm Buffer overnight at 4 °C before storing at −80 °C until shipment to the Oklahoma Medical Research Foundation (OMRF) for analysis.
On the day of acquisition at OMRF, the samples were thawed, washed with Maxpar Cell Staining Buffer, counted, and resuspended in Maxpar Cell Acquisition Solution at 1.0 × 106 cells/mL with 0.1× EQ beads. Data were acquired on a Helios mass cytometer and normalized using CyTOF Software v6.7. The normalized FCS files were used for downstream analyses.
The normalized FCS files were imported into FlowJo v10.8 Software (BD). The immune cell populations were gated as described in Fluidigm’s technical note, “Approach to Bivariate Analysis of Data Acquired Using the Maxpar Direct Immune Profiling Assay,” and Maffei et al.24 The gating strategy (Supplementary Table 1 and Fig. 1), the antibody clone, mass isotope label, and catalog number (Supplementary Table 2) and the cell counts in the final gates (Supplementary Data 2) are provided in the supplementary materials. Cell populations containing fewer than 100 cells in the final gate for >20% of individuals were excluded from downstream analysis. The percentage of lineage parent was calculated for each cell population and used for downstream analysis (i.e. CD4+ T cells/total T cells). GraphPad Prism v9.3.1 was used to analyze and graph the cell populations. Additionally, the live, singlet population was exported from FlowJo Software into FCS files and analyzed further using the ImmunoCluster program (https://github.com/kordastilab/ImmunoCluster)25 in RStudio v2021.9.2.382 (R v4.1.2). Uniform manifold approximation and projection (UMAP) was used as a dimensionality reduction algorithm, and self-organizing maps followed by consensus clustering were used to identify k = 8 clusters of cells based on the 30 markers included in MDIPA.
2.4. Olink immunoproteomics
Inflammatory mediators were quantified using the Olink Target 96 Inflammation Panel, which quantifies 92 inflammation-related proteins simultaneously using Olink’s proximity extension assay technology (Olink) (Fig. 1). Samples were randomized, and 50 μL of plasma was added to each well of a fully skirted 96-well plate. The 96-well plate was sealed, frozen, and shipped to Olink on dry ice. The output of Olink’s proximity extension assay is a relative quantification, referred to as a normalized expression value (NPX), for each protein in the panel. Per the manufacturer’s guidelines, 8 “bridge” samples were run on both plates and used to combine the data from the 2 plates. The NPXs from each plate of samples were bridge normalized using the 8 bridge samples and the Olink Analyze R package (https://github.com/Olink-Proteomics/OlinkRPackage) in RStudio v2021.9.2.382 (R v4.1.2). GraphPad Prism v9.3.1 was used to analyze and graph the bridged NPX values. Gene Ontology (GO) Enrichment Analysis was performed on subsets of inflammatory mediators (http://geneontology.org) with the following conditions: analysis type—PANTHER Overrepresentation Test (released 20221013), annotation version—PANTHER version 17.0 (released 2022-02-22), reference list—Homo sapiens (all genes in database), annotation data set—PANTHER GO-slim Biological Process, test type—Fisher’s exact, and correction—false discovery rate.26–28 The most specific subclass of the hierarchically sorted results table was recorded. The Bridged NPXs are provided in the supplementary materials (Supplementary Data 2).
2.5. HIV-1 plasma viral load
HIV-1 plasma viral load was quantified as described previously.29 Briefly, viral RNA was extracted using the QIAamp viral RNA mini kit with on-column DNase I treatment (cat. 52904; Qiagen). Quantitative PCR for HIV-1 LTR was performed using AcroMetrix HIV-1 High Control samples (cat. 964001; Thermo Scientific). QuantStudio Design and Analysis Software (ThermoFisher) calculated HIV-1 copies/mL from the standard curve. Statistical analysis was performed in GraphPad Prism v9.3.1.
2.6. Additional statistical analysis
Demographics, drug use characteristics, and HIV characteristics were compared between groups using the Mann–Whitney test for continuous variables and Fisher’s exact test for categorical variables where appropriate. Principal component analysis (PCA) was performed in GraphPad Prism v9.3.1 with the default parameters (standardized data, select PCs based on Monte Carlo simulations with auto seed, 1,000 simulations, and 95% percentile).
Next, the inflammatory mediators and immune cell populations were compared among the 3 IDU/HIV groups and controls (HIV−/PWID−, HIV+/PWID−, HIV−/PWID+, HIV+/PWID+). Adjusted pairwise comparisons were performed by fitting a multivariable linear regression with dummy variables for the 3 of the 4 IDU/HIV groups (1 baseline) and inclusion of the following patient potential confounding variables: age, homeless in 12 mo or now, male identity, married, employed, and some college experience. This is an adjusted ANOVA (i.e. ANCOVA). Bonferroni adjusted P values were computed for testing each pair of IDU/HIV groups through Wald-based P values, using the estimated difference in postadjusted group means and the estimated differential standard error. These analyses were performed in R, and results are provided in tabular format in the supplemental materials (Supplementary Table 3).
Finally, multivariable linear regression with main effects for HIV status, IDU status, and their interaction was performed with adjustment for the potential confounding variables discussed above. This is an adjusted 2-factor factorial ANOVA. Due to the use of all observations in the study for these comparisons, the assumption of residual normality is more likely to hold due to the central limit theorem, whereas this was not the case in unadjusted pairwise comparisons. We did not correct the P values here for studying multiple outcomes since this analysis was exploratory in nature, which may limit the power to detect observed differences in outcomes as was done in previous studies.30–33 Instead, we adjusted our P values due to multiple comparisons within a given outcome. These analyses were performed in R, and results are provided in tabular format in the supplemental materials (Supplementary Table 3).
3. Results
3.1. Cohort summary and drug use characteristics
To compare the immune profiles in people with and without HIV who are either using injection drugs or not, we recruited 97 individuals spread across 4 groups: HIV−/PWID− (controls; 14), HIV+/PWID− (36), HIV−/PWID+ (35), and HIV+/PWID+ (12) (Table 1). Most participants were born in Puerto Rico and reported residence in the Rio Piedras, Santurce, Puerto Nuevo, and Hato Rey “barrios” of metro San Juan, Puerto Rico. The gender of non-injectors was 38% male, while the injectors were primarily male (89%); the age of injectors was not significantly different from non-injectors (Table 1). Notably, there were more PLWH in the non-injector group compared to the injector group; however, all but 3 of the PLWH were currently taking ART, resulting in comparable viral loads across the relevant groups (Table 1). Empirical HIV-1 plasma viral load quantities confirmed self-reported ART adherence. Previous work has established that the prevalence of HCV in Puerto Rican PWID is >75%, and infection is rapidly acquired upon initiation of IDU.34 Therefore, it was not surprising that injectors self-reported having previously tested positive for HCV more than non-injectors (Table 1).
Table 1.
Cohort demographics (n = 97).
| Variable | Non-injectors (n = 50) |
Injectors (n = 47) |
Injectors vs Non-injectors | ||||
|---|---|---|---|---|---|---|---|
| HIV− (n = 14) | HIV+(n = 36) | P | HIV− (n = 35) | HIV+ (n = 12) | P | P | |
|
| |||||||
| Demographic | |||||||
| Age (ys) | 43.5 (5.5) | 48 (10.25) | 0.0288 | 44 (12) | 49 (12) | 0.1678 | 0.6884 |
| Male gender | 4 (29%) | 15 (42%) | 0.5218 | 31 (89%) | 11 (92%) | >0.9999 | <0.0001 |
| Married currently | 7 (50%) | 5 (14%) | 0.0225 | 4 (11%) | 6 (50%) | 0.0104 | 0.8114 |
| Attended some college | 13 (93%) | 15 (42%) | 0.0012 | 8 (23%) | 4 (33%) | 0.4708 | 0.0036 |
| Employed | 11 (79%) | 10 (28%) | 0.0016 | 3 (9%) | 2 (17%) | 0.5904 | 0.0005 |
| Homeless in the past year | 0 (0%) | 6 (17%) | 0.1670 | 24 (69%) | 4 (33%) | 0.0445 | <0.0001 |
| Homeless currently | 0 (0%) | 3 (8%) | 0.5500 | 13 (37%) | 0 (0%) | 0.0209 | 0.0055 |
| Sleep/night (h) | 7 (0.88) | 7.5 (2.13) | 0.4804 | 5.5 (2.75) | 6.5 (3.5) | 0.2022 | 0.0003 |
| Exercise/wk (d) | 2 (2) | 1 (3) | 0.4505 | 2 (4.5) | 3 (3.5) | 0.5421 | 0.2201 |
| HCV | |||||||
| Self-report past testing | 10 (71%) | 28 (78%) | 0.7060 | 32 (91%) | 12 (100%) | 0.5597 | 0.0233 |
| Ever tested positive | 0 (0%) | 8 (29%) | 0.0786 | 23 (72%) | 12 (100%) | 0.0821 | <0.0001 |
| HIV | |||||||
| Viral load (cps/mL) | ... | 1,600 (2 × 104) | ... | ... | 1,900 (1 × 104) | ... | 0.4000 |
| Undetectablea | ... | 33 (92%) | ... | ... | 8 (67%) | ... | 0.0552 |
| ART current | ... | 34 (94%) | ... | ... | 11 (92%) | ... | >0.9999 |
| Always adhering | ... | 31 (91%) | ... | ... | 6 (55%) | ... | 0.0141 |
Continuous variables are shown as median with interquartile range and compared using Mann-Whitney tests. Categorical data are shown as counts with percentages (%) and compared using Fisher’s exact tests. The P values are italicized and significant P values are bolded.
Undetectable HIV-1 plasma viral load is defined as <70 copies/mL.
The self-reported non-injection drug use (NIDU) characteristics, treatment program history, and the results of urine toxicology tests performed at the time of blood sample collection were also compared between the groups (Table 2). Injectors were more likely to have engaged in NIDU in the past 12 mo and to have participated in drug treatment programs than non-injectors (Table 2). While HIV+ non-injectors were more likely to test positive for drug use in their urine samples than controls, injectors were more prone to polysubstance abuse (testing positive for >1 drug) than non-injectors (Table 2). However, there were no significant differences in self-reported IDU characteristics (age at first injection, injection frequency, types of drugs injected, number of drugs injected, and frequency of sterile needle use) between the HIV− and HIV+ injectors (Table 3).
Table 2.
Self-reported non-injection drug use characteristics, drug treatment program history, and urine toxicology results (n = 97).
| Non-injectors (n = 50) |
Injectors (n = 47) |
Injectors vs Non-injectors | |||||
|---|---|---|---|---|---|---|---|
| Variable | HIV− (n = 14) | HIV+ (n = 36) | P | HIV− (n = 35) | HIV+ (n= 12) | P | P |
|
| |||||||
| Self-reported NID use | |||||||
| NIDs used in the past year | 6 (43%) | 14 (39%) | 0.7466 | 27 (77%) | 7 (58%) | 0.2687 | 0.0020 |
| Alcohol | 6 (l00%) | 13 (93%) | >0.9999 | 20 (74%) | 5 (71%) | >0.9999 | 0.0719 |
| Marijuana | 0 (0%) | 4 (29%) | 0.2675 | 9 (33%) | 1 (14%) | 0.6445 | 0.5326 |
| Crystal meth | 0 (0%) | 4 (29%) | 0.2675 | 13 (48%) | 2 (29%) | 0.4263 | 0.0862 |
| Cocaine | 1 (17%) | 4 (29%) | >0.9999 | 13 (48%) | 3 (43%) | >0.9999 | 0.1511 |
| Downers | 0 (0%) | 0 (0%) | >0.9999 | 5 (19%) | 1 (14%) | >0.9999 | 0.0740 |
| MDMA/ecstasy | 0 (0%) | 0 (0%) | >0.9999 | 7 (26%) | 1 (14%) | >0.9999 | 0.0201 |
| Heroin | 1 (7%) | 0 (0%) | 0.3000 | 1 (4%) | 1 (14%) | 0.3845 | >0.9999 |
| Xylazine | 0 (0%) | 0 (0%) | >0.9999 | 4 (15%) | 0 (0%) | 0.5585 | 0.2845 |
| Buprenorphine | 6 (100%) | 5 (36%) | 0.0141 | 9 (33%) | 2 (29%) | >0.9999 | 0.1521 |
| # NIDs used | 2 (0) | 2 (1.75) | 0.4989 | 3 (2) | 2 (2) | 0.3515 | 0.2407 |
| Drug treatment history | |||||||
| Ever | 0 (0%) | 12 (33%) | 0.0215 | 30 (86%) | 11 (92%) | >0.9999 | <0.0001 |
| In the past year | 0 (0%) | 3 (25%) | >0.9999 | 18 (60%) | 9 (82%) | 0.2753 | 0.0195 |
| Urine toxicology | |||||||
| Urine test positive | 2 (14%) | 28 (78%) | <0.0001 | 34 (97%) | 11 (92%) | 0.4496 | <0.0001 |
| Multidrug positive | 0 (0%) | 15 (54%) | 0.4828 | 28 (82%) | 7 (64%) | 0.2279 | 0.0233 |
| Fentanyl | 0 (0%) | 0 (0%) | >0.9999 | 21 (62%) | 4 (36%) | 0.1757 | <0.0001 |
| Cocaine | 0 (0%) | 6 (21%) | >0.9999 | 19 (56%) | 5 (45%) | 0.7302 | 0.0044 |
| Opiates | 0 (0%) | 0 (0%) | >0.9999 | 15 (44%) | 4 (36%) | 0.7363 | <0.0001 |
| Benzodiazepines | 0 (0%) | 13 (46%) | 0.4920 | 10 (29%) | 2 (18%) | 0.6991 | 0.1444 |
| Methadone | 0 (0%) | 2 (7%) | >0.9999 | 18 (53%) | 7 (64%) | 0.7295 | <0.0001 |
| Marijuana | 2 (100%) | 13 (46%) | 0.4828 | 13 (38%) | 3 (27%) | 0.7202 | 0.2393 |
| TCAs | 0 (0%) | 5 (18%) | >0.9999 | 6 (18%) | 0 (0%) | 0.3111 | 0.7461 |
| Amphetamine | 0 (0%) | 2 (7%) | >0.9999 | 5 (15%) | 0 (0%) | 0.3131 | 0.6952 |
| Oxycodone | 0 (0%) | 1 (4%) | >0.9999 | 0 (0%) | 0 (0%) | >0.9999 | 0.4000 |
| Barbiturates | 0 (0%) | 1 (4%) | >0.9999 | 0 (0%) | 0 (0%) | >0.9999 | 0.4000 |
| Buprenorphine | 0 (0%) | 1 (4%) | >0.9999 | 0 (0%) | 4 (36%) | 0.0022 | 0.6424 |
| # positive drugs in urine | 0 (0) | 1 (1) | <0.0001 | 3 (2) | 2 (2.5) | 0.2390 | <0.0001 |
Continuous variables are shown as median with interquartile range and compared using Mann-Whitney tests. Categorical data are shown as counts with percentages (%) and compared using Fisher’s exact tests. The P values are italicized and significant P values are bolded.
NID, non-injection drug; TCA, tricyclic antidepressant.
Table 3.
Self-reported injection drug use characteristics (n = 47).
| Self-Reported IDU | HIV− Injectors (n = 35) | HIV+ Injectors (n = 11a) | P |
|---|---|---|---|
|
| |||
| Age at first injection | 24 (12.75) | 19 (8) | 0.2048 |
| Injecting daily | 27 (77%) | 6 (55%) | 0.2479 |
| Drugs used in past year | |||
| Speedball | 30 (86%) | 8 (73%) | 0.3744 |
| Heroin alone | 17 (49%) | 6 (55%) | >0.9999 |
| Cocaine alone | 10 (29%) | 2 (18%) | 0.7006 |
| Xylazine alone | 1 (3%) | 0 (0%) | >0.9999 |
| Crystal meth | 1 (3%) | 0 (0%) | >0.9999 |
| Prescription opioids | 0 (0%) | 2 (18%) | 0.0531 |
| Buprenorphine | 2 (6%) | 1 (9%) | >0.9999 |
| # injection drugs used | 1 (1) | 1 (1) | 0.9285 |
| Rarely used sterile needles | 13 (37%) | 1 (9%) | 0.1331 |
Continuous variables are shown as median with interquartile range and compared using Mann-Whitney tests. Categorical data are shown as counts with percentages (%) and compared using Fisher’s exact tests. The P values are italicized and significant P values are bolded.
The 12th HIV+ injector did not participate in the IDU portion of the suwey.
Finally, the self-reported use of multiple non-injection and injection drugs was analyzed for common combinations of drugs and visualized using heatmaps of the row percentages (Supplementary Fig. 2). For example, 1 of 38 speedball injectors (2.6%) reported also injecting xylazine (Supplementary Fig. 2B, see row 1), whereas the single xylazine injector also reported injecting speedball (100%; Supplementary Fig. 2B, see row 3). The most frequently reported non-injection drug was alcohol (Supplementary Fig. 2A), while the most frequently reported injection drug was “speedball,” a combination of cocaine and heroin (Supplementary Fig. 2B). Buprenorphine and alcohol were the most frequently co-occurring substances; of the 22 individuals who reported using buprenorphine through non-injection routes, 95.5% reported also drinking alcohol within the past 12 mo (Supplementary Fig. 2A).
3.2. Exploratory analysis of high-throughput immunoproteomics and immunophenotyping
Drug abuse has been shown to affect the levels of isolated cytokines in various studies,16–18,21,22 but no study in humans has comprehensively investigated the extent to which IDU affects peripheral immune soluble inflammatory mediators. To evaluate differentials in plasma inflammatory profiles between injectors and non-injectors, we employed an established Olink proximity extension assay (Target 96 Inflammation Panel; Olink Bioscience AB) (Fig. 1). Consistent with anticipated assay performance, 74 of 92 target proteins were detected in >90% of study subjects. PCA was performed to identify any outlier samples in the data set and discover any underlying patterns or linear segregation of the 4 groups (HIV−/PWID−, HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+). PCA revealed slight segregation of the controls (highlighted by the purple ellipse) from the 3 HIV/IDU groups; however, the 3 HIV/IDU groups were inseparable (Fig. 2A). Finally, outlier samples were not detected.
Fig. 2.
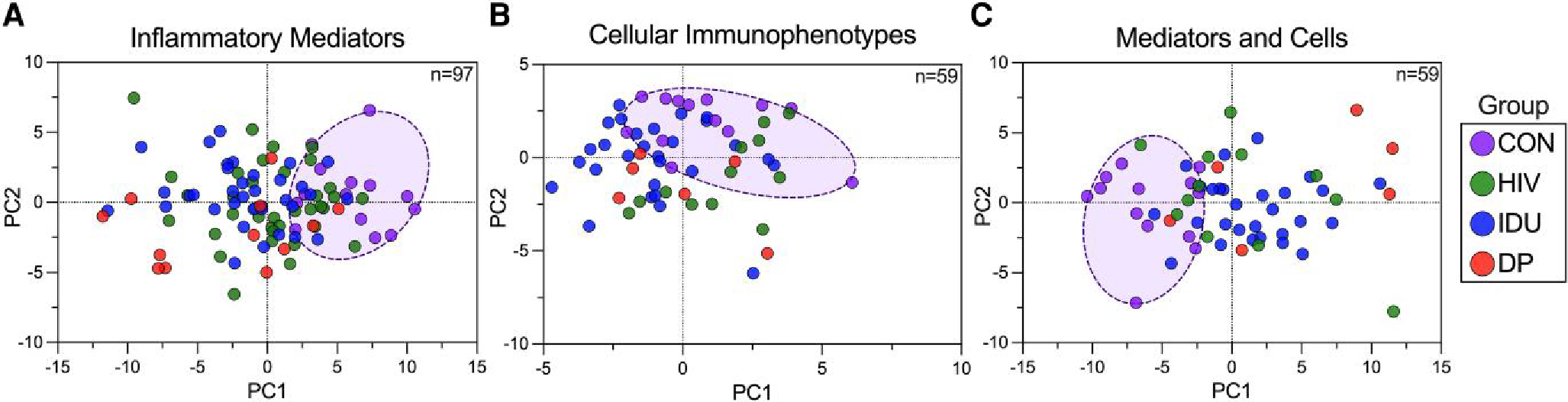
Exploratory PCA of inflammatory mediators and cellular immunophenotypes. A) Bridge normalized NPX values from Olink Target 96 Inflammation Panel, B) percentage of lineage parent population for each of 37 immune cell types, or C) the combined data sets were the inputs for PCA. In each case, the input data were standardized and centered. The individuals are colored by group (HIV: HIV+/PWID−, IDU: HIV−/PWID+, DP: HIV+/PWID+), and the control group (CON: HIV−/PWID−) was circled.
Studies to define the impact of IDU on the immune cell populations have focused on limited cell lineages. To develop a more comprehensive appreciation of how IDU alters peripheral immune cell phenotypes, we employed a mass cytometry panel of 36 isotopically defined mass markers to detect 47 immune cell populations, including granulocytes, in a single assay (MDIPA; Fluidigm) (Fig. 1). Of the 97 individuals included in inflammatory mediator analyses, cell phenotypes were analyzed in parallel for the 59 individuals as follows: 13 controls, 12 HIV+/PWID−, 28 HIV−/PWID+, and 6 HIV+/PWID+. We collected an average of 214,174 events/sample (range: 73,273 to 500,000), where each group average was between 199,505 and 224,565 events/sample. Nine of 47 immune cell populations did not meet the detection threshold in >80% of participants (>100 cells in the final gate), and those populations were excluded from downstream analyses (see Materials and Methods and Supplementary Table 1). Surprisingly, this group included the exhausted T-cell subsets. Notably, 2 of the excluded populations, exhausted CD8+ T cells and activated, senescent CD8+ T cells, were detectable in 89% of HIV+ participants yet were only detectable in 49% of the HIV− participants, which rendered them excluded from downstream analyses. This HIV association was not observed in the remaining excluded populations. We calculated the percentage of lineage parent (i.e. CD4 T cells/total T cells * 100) for each of the 37 detectable immune cell populations with detectable lineage parent populations (Supplementary Table 1). Then, we applied PCA, and outliers were not detected (Fig. 2B). The highlighted cluster of controls overlapped with the 3 HIV/IDU groups to a greater extent than did the inflammatory mediators (Fig. 2B).
Data sets for both inflammatory mediators and cellular immunophenotypes were merged for 59 participants, and PCA was applied (Fig. 2C). This merged analysis revealed the best segregation of the control group (highlighted) from the 3 HIV/IDU groups that, in turn, were indistinguishable. Since the merged data set provided the best group segregation, we projected the PCA results in 3 dimensions (Supplementary Fig. 3). In the 3-dimensional projection, the controls were more demonstrably separable from the HIV+/PWID+ group by a hyperplane, and the 3 HIV/IDU groups mostly overlapped (Supplementary Fig. 3).
Overall, PCA suggested that the control group was more distinguishable from the 3 HIV/IDU groups than the 3 HIV/IDU groups from each other. High variation between samples within groups and lack of tight clustering highlight the heterogeneity intrinsic to hard-to-reach human populations in a non-clinical setting. To ensure that potentially confounding demographic differences between groups (Table 1) were not immunologically confounding factors, adjustment for age, homelessness, gender, marital status, employment status, and education was performed as described in the Materials and Methods and applied to all downstream analyses. The lack of clear separation between the 3 HIV/IDU groups suggests that the overall inflammatory mediator and cellular immunophenotype profiles are not able to discriminate the groups. Nevertheless, pairwise comparisons are needed to elucidate finer resolution differences between the groups and identify any potential additivity or synergy between HIV infection and IDU.
3.3. Similar inflammatory mediator profiles among the PLWH, PWID, and HIV+ PWID
To determine if specific inflammatory mediators are affected by IDU, HIV infection, or the combination of the two, we stratified the participants by HIV/IDU statuses and conducted pairwise comparisons of the inflammatory profiles between all 4 groups. In this confounder-adjusted analysis, 34 of 74 detectable inflammatory mediators were significant in at least 1 comparison. Of the 34 mediators, 18, 9, and 24 were elevated in HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+ compared to controls (HIV−/PWID−), respectively (Fig. 3A). Differences were readily detected between each of the 3 IDU/HIV groups and controls, but very few significant differences were found between the mono- and dual-affected individuals (Fig. 3A). These results suggest limited, if any, additivity or synergy between HIV infection and IDU.
Fig. 3.
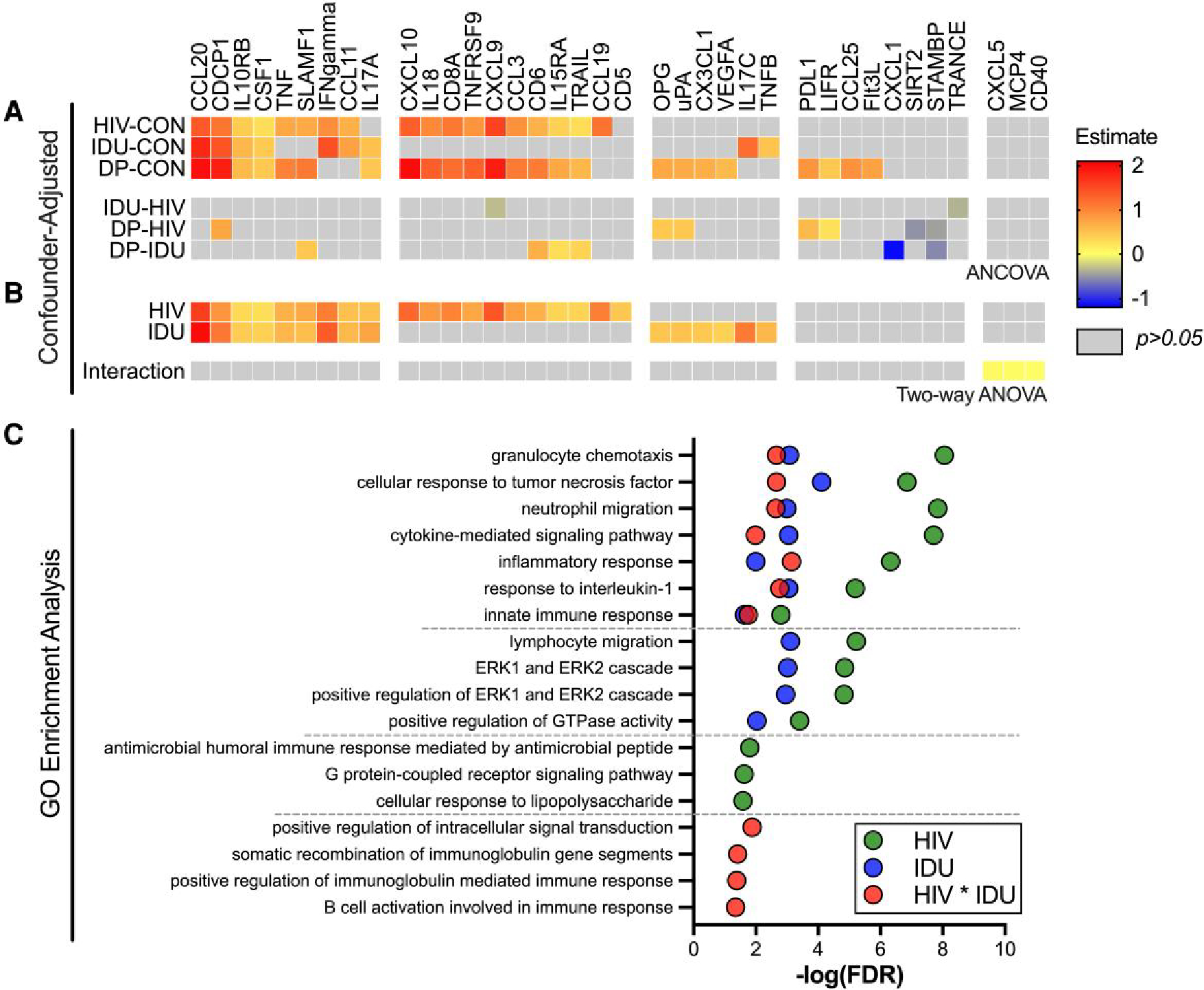
Pairwise comparison of the inflammatory mediators in HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+. Bridge normalized NPX values from Olink Target 96 Inflammation Panel were used to compare levels of 74 inflammatory mediators between HIV−/PWID− (controls; CON), HIV+/PWID− (HIV), HIV−/PWID+ (IDU), and HIV+/PWID+ (DP) A) post-adjusting for age, homeless in the past year or now, male identity, married, employed, and some college experience (ANCOVA). B) The adjusted data were also subjected to 2-way ANOVA to identify HIV infection, injection drug use, and interaction (synergy) effects. C) GO enrichment analysis was performed for the inflammatory mediators with significant interaction effects in B, and the most specific subclass of the hierarchical ontology cluster is shown. The estimate difference (adjusted mean difference) (A), coefficient estimates (effect size) (B), and −log(false discovery rate) (C) are shown when significant (P < 0.05).
To further gauge whether HIV infection and IDU synergistically affected the immune system, we tested for the interaction effect between the two using 2-way ANOVA (Fig. 3B). Twenty inflammatory mediators were significantly associated with HIV infection (see row HIV, Fig. 3B), 15 were significantly associated with IDU (see row IDU, Fig. 3B), and 9 were shared between HIV infection and IDU. However, none of these 26 upregulated inflammatory mediators evinced significant synergistic effects, meaning they were not more elevated in the HIV+ injectors than would be predicted by adding up the effects of HIV infection or IDU alone (see row Interaction, Fig. 3B). Interestingly, 3 unique inflammatory mediators—CXCL5, MCP-4, and CD40—did demonstrate significant synergy between HIV infection and IDU (see row Interaction, Fig. 3B). To investigate the functional roles of these cytokines, we performed GO enrichment analysis (Fig. 3C). While CXCL5, MCP-4, and CD40 are components of similar biological processes as the cytokines significantly associated with HIV infection or IDU alone, they are also components of B-cell processes (Fig. 3C). While these GO enrichments and interaction effects are significant, they have small coefficient estimates (i.e. effect sizes), reinforcing the subtlety of these synergistic effects. We would therefore caution against overinterpretation. In summary, plasma inflammatory profiling revealed differentiation between HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+ from controls. The unanticipated similarities between the plasma inflammatory profiles of PLWH and PWID resulted in limited evidence of additivity or synergy between HIV infection and IDU.
Because it was anticipated that different types of drugs abused and the frequency of use could differentially affect inflammatory profiles, we also investigated the associations of inflammatory mediators with drug use parameters collected during the study surveys (Table 2, Table 3). Nine inflammatory mediators correlated with the extent of polysubstance abuse evident in the urine test (Supplementary Fig. 4A). These results indicate that the number of drugs abused can additionally influence plasma inflammatory mediator profiles.
3.4. The peripheral immune cell profiles of HIV+ PWID resemble those of PLWH and PWID
To examine the potential additive effects of HIV infection and IDU on the immune cell populations, we again stratified our cohort into 4 groups based on HIV and IDU status. First, we compared HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+ to controls post-adjustment for age, homelessness, gender, marital status, employment status, and education (Fig. 4). As expected in HIV infection, the CD4 T cell population was depleted, resulting in a larger proportion of CD8 T cells compared to controls (see row HIV-CON, Fig. 4A). Although our PLWH have been treated, ART does not always lead to rapid or full reconstitution of the CD4+ T-cell population. Additionally, mucosal-associated invariant T or natural killer T (MAIT/NKT) cells were reduced compared to controls, and natural killer (NK) cells shifted toward a late phenotype (CD57+) (see row HIV-CON, Fig. 4A). Notably, none of these 5 cell populations were significantly different between HIV−/PWID+ and controls (see row IDU-CON, Fig. 4A), reinforcing that these effects are unique to HIV infection and are not associated with IDU. Only 2 cell populations were significantly elevated in HIV− injectors compared to controls, CD4+ terminal effectors, and IgD-negative memory B cells (see row IDU-CON, Fig. 4A).
Fig. 4.
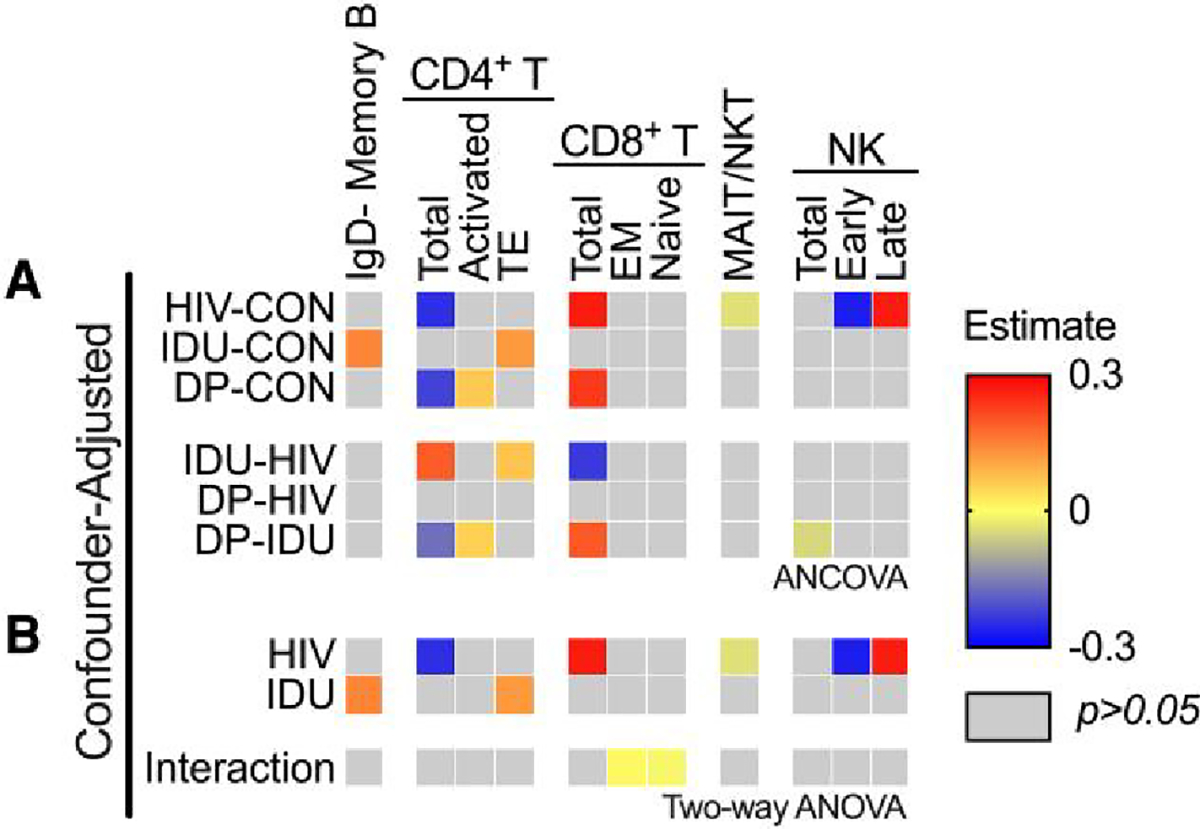
Pairwise comparison of the cellular immunophenotypes in HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+. Percentage of lineage parent population was calculated for each of 37 immune cell types and compared between HIV−/PWID− (controls; CON), HIV+/PWID− (HIV), HIV−/PWID+ (IDU), and HIV+/PWID+ (DP) A) post-adjusting for age, homeless in the past year or now, male identity, married, employed, and some college experience (ANCOVA). B) The adjusted data were also subjected to 2-way ANOVA to identify HIV infection, injection drug use, and interaction effects. The estimate difference (adjusted mean difference) (A) and coefficient estimates (effect size) (B) are shown when significant (P < 0.05). EM, effector memory; TE, terminal effector.
Surprisingly, significant differences in the neutrophil and monocyte populations were not detected, contrary to what was expected since injectors would likely have increased exposure to pathogens. Thus, we employed an unbiased clustering algorithm of our live, singlet population. We performed UMAP dimensionality reduction with FlowSOM clustering (k = 8). No global cell population differences were detected between our cohorts using this strategy (Supplementary Fig. 5), validating the results of our bivariate gating strategy. Further, no cell populations had a statistically significant correlation with the number of positive drugs in urine tests (Supplementary Fig. 4B).
While differences in immune cell populations between HIV+/PWID−, HIV−/PWID+, and HIV+/PWID+ compared to controls were evident, few differences between HIV+/PWID+ and HIV+/PWID− or HIV−/PWID+ were detected, indicating that there is not a notable additive effect of HIV infection and IDU on peripheral immune cell populations. To ensure subtle changes were not overlooked, we tested for significant effects associated with HIV infection, IDU, and the interaction between them using 2-way ANOVA (Fig. 4B). Consistent with the pairwise comparisons, HIV had a significant effect on the ratios of CD4/CD8 T cells and early/late NK cells, as well as MAIT/NKT cells (see row HIV, Fig. 4B), and IDU had a significant effect on CD4+ terminal effector T cells and IgD-negative memory B cells (see row IDU, Fig. 4B). While synergistic effects were not evident from the analysis of the aforementioned immune cell populations, significant synergy of HIV infection and IDU on CD8+ effector memory (elevated) and naive T cells (reduced) was detected, indicating these CD8+ T cell populations were more altered in the HIV+/PWID+ than would be predicted by summing the effects of HIV infection or IDU alone (see row Interaction, Fig. 4B). Like the soluble mediators, there were significant interaction effects, but the coefficient estimates (i.e. effect sizes) were extremely small. Thus, the biological significance of these subtle synergies is unclear. Overall, these data suggest that HIV has a larger effect on the peripheral immune cell populations than IDU but only a marginal synergistic effect in the context of IDU.
4. Discussion
Although previous studies have analyzed aspects of the effects of HIV and IDU on the immune system, this study, to our knowledge, provides the most comprehensive peripheral immune profiling data set available on PWID. Furthermore, this study focused on a unique population of Puerto Rican PWID and PLWH for whom the potential impacts of both insults on immune dysfunction have not been previously analyzed. Our results revealed similar inflammation and immune dysfunction patterns in PLWH, PWID, and HIV+ PWID. There was limited evidence for a pattern of cytokines that could differentiate PLWH from PWID, suggesting that both HIV infection and IDU dysregulate the immune system, resulting in similar cytokine and immune cell profiles.
Furthermore, only signs of subtle synergy between HIV infection and IDU were evident. Twenty-six inflammatory mediators and 7 immune cell populations were significantly associated with HIV infection and/or IDU, while 3 distinct inflammatory mediators and 2 immune cell populations exhibited significant synergy between HIV infection and IDU (Fig. 5). Despite their significance, these interaction effects were accompanied by weak coefficient estimates, reinforcing the subtlety of any potential synergy. Moreover, the considerable differences in inflammatory mediators between PLWH and PWID vs controls did not translate into notable changes in cell phenotypes, and only weak to moderate correlations were noted between cell phenotypes and their associated inflammatory mediators (Supplementary Fig. 6), suggesting a disconnect between the cytokine signaling and cell phenotypes in the blood. This is further reinforced by the lack of concordance between inflammatory mediators and cell populations with significant HIV, IDU, or interaction effects (Fig. 5). The inflammatory mediators may be originating from tissue-resident cells (i.e. gut epithelium), but further studies are needed to understand this relationship.
Fig. 5.
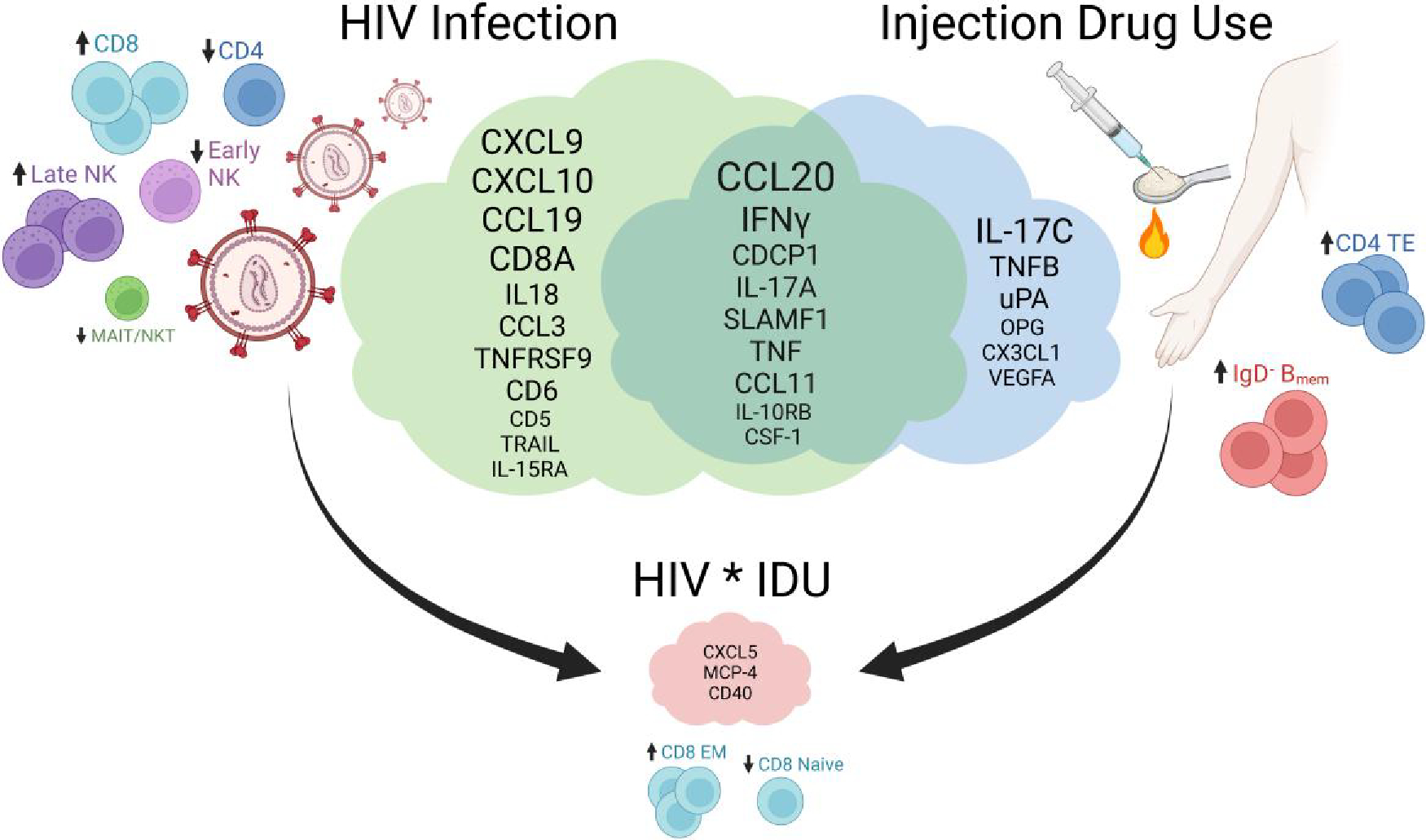
The effects of HIV infection and injection drug use on the peripheral immune repertoire. The upregulated cytokines are shown in the cloud Venn diagram for HIV infection, injection drug use, and those that are in common. The dysregulated immune cell populations are also displayed with arrows indicating an increasing effect (“up” arrow) or decreasing effect (“down” arrow). The upregulated cytokines and dysregulated cell populations with significant interaction effects (synergy) between HIV infection and injection drug use are shown. The smaller text size indicates a less significant effect, while the larger text size indicates a more significant effect. Created with BioRender.com.
Albeit through potentially different mechanisms, both drug abuse and HIV infection have been associated with gut dysbiosis and microbial translocation. Such translocation could explain the similarity in peripheral immune profiles. Microbial translocation can result from gut dysbiosis, weakening of the mucosal barrier, or an immune deficiency35 and has been implicated in systemic inflammation.36 HIV-associated microbial translocation is generally thought to result from gut dysbiosis induced by HIV infection and depletion of the CD4+ Th17 cells in the gut-associated lymphoid tissue.37–39 Additionally, opioid use has been associated with decreased gut motility through interaction with the enteric nervous system and gut dysbiosis,40,41 while cocaine has been associated with changes in the gut barrier, gut microbiome, and production of short-chain fatty acids,42 and Tomescu et al.33 demonstrated elevations in plasma levels of both sCD14 and sCD163 in PWID. Thus, both IDU and HIV infection have the potential to induce microbial translocation, resulting in the similarity in detected patterns of systemic inflammation in PWID and PLWH.
To our knowledge, this study is the first to apply Olink technology and mass cytometry to the study of immune dysfunction in PWID. The results are concordant with published studies using other assays in the context of drug abuse. In a study of speedball users, TNF, IL-6, IFN-γ, IL-2, IL-4, IL-10, and IL-12 were increased compared to healthy controls.22 Further, when PBMCs were stimulated with a pan-T-cell activator or biological response modifier, TNF, IL-6, and IFN-γ were consistently detected at higher levels in speedball users than in healthy controls upon stimulation.22 Additionally, Tomescu et al.33 reported increased activation of NK and CD4+ T cells in PWID compared to controls. Importantly, this did not translate into any functional innate or adaptive differences,32 suggesting that although we detect differential cell population numbers, these are unlikely to translate into functional changes in the immune response. Further, in the study of abstinent cocaine users seeking treatment, Araos et al.43 found TNF-α to be reduced in abstinent cocaine users compared to controls. These findings support our study identifying TNF as associated with HIV infection and IDU because the participants were actively using various drugs of abuse.
Given the anticipation of high HCV prevalence and the aforementioned rates of self-reported HCV infection, we were unable to control for HCV infection status in our study and therefore did not seek to quantify HCV viral load. Additionally, other studies have found HCV co-infection in PLWH not to be a factor confounding cytokine or cell phenotypes.44,45 For example, in a study of HIV+/HBV+/HCV+ PWID, the authors found CD4+ T cells were reduced and CD8+ T cells were elevated compared to HIV−/HBV+/HCV+ and HIV−/HBV−/HCV− PWID.44 These findings support our results that both HIV infection and IDU alter T-cell populations and suggest that HCV and hepatitis B virus (HBV) are unlikely to further affect the immunophenotypes. Similarly for cytokines, IL-18, CXCL10, IFN-γ, and IL-15 were significantly elevated in HIV/HCV co-infection compared to mono-infection.45 We also found elevations of these cytokines in our HIV/IDU groups compared to controls. This supports the notion that our PWID have already been infected by HCV, which was likely not a confounding factor in the cytokine dysregulation detected in our study.
This cross-sectional study has described the most comprehensive and contemporary data set on comparative immune profiling of PWID with and without HIV infection. While cause and effect cannot be established, this study has generated preliminary results using novel contemporary approaches upon which future studies can build. Importantly, the results also support those from several other studies focused on HIV and the changes in various cellular factors associated with infection,46–49 and our mass cytometry data demonstrated a depletion of CD4 T cells and an increased proportion of CD8 T cells, along with a decrease in MAIT/NKT cells in our HIV+ groups compared to HIV− groups, all of which are hallmarks of HIV infection.50 Studies of cocaine use in PLWH have demonstrated more severe HIV disease in cocaine users compared to non-users but also found a small number of cytokines that are associated with cocaine use in PLWH.51 Importantly, our cohort predominantly consisted of aviremic PLWH who self-reported always adhering to their ART, which may differ from other cohorts. Finally, other recent studies have reported minimal and complex synergistic effects between HIV infection and heroin use.30,31 This comprehensive study bolsters those findings and highlights the intricate relationship between HIV and IDU.
As a function of non-targeted recruitment, this study had uneven and small sample sizes between the subcohorts, with controls and HIV+/PWID+ being about half the size of the HIV+/PWID− and HIV−/PWID+ groups. However, the disproportionality is representative of the natural populations. Puerto Rican PLWH tend to move to the continental United States in search of HIV treatment, and the remaining HIV+/PWID+ population is hard to reach due to social stigmas and lack of contact information.52 We employed RDS, which our team has demonstrated to be effective in past studies.6 However, the HIV+/PWID+ population proved especially hard to reach and was heterogeneous, as expected. Additionally, it is important to note that the demographics of San Juan are not necessarily representative of the rest of Puerto Rico or the continental United States. It would be important for future studies to obtain a large sample from diverse regions and poverty levels, allowing the study to be more generalizable and evaluate the subtle differences between groups that may prove biologically or therapeutically relevant. There was also a higher proportion of males in our injectors group than non-injectors. Again, this is representative of the natural populations6 and was accounted for in the confounder-adjusted analyses. Last, we had a diverse drug user population, and 31% of our non-injector population self-reported NIDU (excluding prescriptions, alcohol, and marijuana). However, when we removed the non-injectors with evidence of NIDU, our results remained largely the same.
In summary, we report that the inflammation and immune cell profiles of HIV+ PWID resemble those of HIV+ non-injectors and HIV− PWID. While many inflammatory mediators were significantly different between the mono- and dual-affected individuals and controls, there were very few differences between the mono- and dual-affected individuals themselves. We also identified subtle synergistic effects between HIV infection and IDU, but these effects may not be biologically relevant and did not translate into differences in peripheral immune cell phenotypes. Future studies with larger sample sizes and a focus on single drug-using populations would better elucidate how each drug of abuse affects the peripheral immune profile, and analysis of the gut microbiome, microbial translocation, and functional analysis of immune cells would paint a complete picture of how IDU and HIV infection, in combination, affect the immune system.
Supplementary Material
Acknowledgments
The authors thank all the study participants for their generosity and participation; Anjelica Rivera and Gabriel Cruz Rodriguez for their roles in patient recruitment and collection of samples; Danielle Shea for her indispensable support of the study and training on laboratory techniques; and the Arthritis & Clinical Immunology Human Phenotyping Core at the OMRF for CyTOF data acquisition and initial data analysis. This work was supported by National Institutes of Health (NIH) grants (R01 DA047823, T32 AI125207). The group at OMRF was supported by NIH grants (P30 AR073750, U54 GM104938), Presbyterian Health Foundation, and Oklahoma Center for Adult Stem Cell Research funds (JMG) for this work.
Abbreviations:
- ART
antiretroviral therapy
- GO
Gene Ontology
- HCV
hepatitis C virus
- IDU
injection drug use
- LPS
lipopolysaccharide
- MAIT/NKT
mucosal-associated invariant T or natural killer T cells
- MDIPA
Maxpar Direct Immune Profiling Assay
- NIDU
non-injection drug use
- NPX
normalized expression unit
- PBMC
peripheral blood mononuclear cell
- PLWH
people living with HIV
- PWID
people who inject drugs
- RDS
respondent-driven sampling
- sCD163
soluble CD163
- UMAP
uniform manifold approximation and projection
Footnotes
Conflict of interest statement. None declared.
Supplementary material
Supplementary materials are available at Journal of Leukocyte Biology online.
References
- 1.Colon HM, Finlinson HA, Robles RR, Deren S, Andía J, Kang SY, Oliver-Vélez D. Joint drug purchases and drug preparation risk behaviors among Puerto Rican injection drug users. AIDS Behav. 2001:5(1):85–96. 10.1023/A:1009515723223 [DOI] [Google Scholar]
- 2.Colón HM, Robles RR, Deren S, Sahai H, Finlinson HA, Andía J, Cruz MA, Kang SY, Oliver-Vélez D. Between-city variation in frequency of injection among Puerto Rican injection drug users: east Harlem, New York, and Bayamon, Puerto Rico. J Acquir Immune Defic Syndr. 2001:27(4):405–413. 10.1097/00126334-200108010-00012 [DOI] [PubMed] [Google Scholar]
- 3.Gelpí-Acosta C, Rodríguez-Díaz CE, Aponte-Meléndez Y, Abadie R. Puerto Rican syndemics: opiates, overdoses, HIV, and the hepatitis C virus in a context of ongoing crises. Am J Public Health. 2020:110(2):176–177. 10.2105/AJPH.2019.305487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Handel MM, Rose CE, Hallisey EJ, Kolling JL, Zibbell JE, Lewis B, Bohm MK, Jones CM, Flanagan BE, Siddiqi AE, et al. County-level vulnerability assessment for rapid dissemination of HIV or HCV infections among persons who inject drugs, United States. J Acquir Immune Defic Syndr. 2016:73(3):323–331. 10.1097/QAI.0000000000001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mino M, Deren S, Colón HM. HIV and drug use in Puerto Rico: findings from the ARIBBA study. J Int Assoc Physicians AIDS Care (Chic). 2011:10(4):248–259. 10.1177/1545109710397768 [DOI] [PubMed] [Google Scholar]
- 6.Hautala D, Abadie R, Khan B, Dombrowski K. Rural and urban comparisons of polysubstance use profiles and associated injection behaviors among people who inject drugs in Puerto Rico. Drug Alcohol Depend. 2017:181:186–193. 10.1016/j.drugalcdep.2017.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006:12(12):1365–1371. 10.1038/nm1511 [DOI] [PubMed] [Google Scholar]
- 8.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003:77(21):11708–11717. 10.1128/JVI.77.21.11708-11717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013:21(1): 6–13. 10.1016/j.tim.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klase Z, Ortiz A, Deleage C, Mudd JC, Quiñones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015:8(5):1009–1020. 10.1038/mi.2014.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng J, Sindberg GM, Roy S. Disruption of gut homeostasis by opioids accelerates HIV disease progression. Front Microbiol. 2015:6:643. 10.3389/fmicb.2015.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz IT, Maughan-Brown B. Improved life expectancy of people living with HIV: who is left behind? Lancet HIV. 2017:4(8): e324–e326. 10.1016/S2352-3018(17)30086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodés B, Cadiñanos J, Esteban-Cantos A, Rodríguez-Centeno J, Arribas JR. Ageing with HIV: challenges and biomarkers. EBioMedicine. 2022:77:103896. 10.1016/j.ebiom.2022.103896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chichetto NE, Polanka BM, So-Armah KA, Sung M, Stewart JC, Koethe JR, Edelman EJ, Tindle HA, Freiberg MS. Contribution of behavioral health factors to non-AIDS-related comorbidities: an updated review. Curr HIV/AIDS Rep. 2020:17(4):354–372. 10.1007/s11904-020-00498-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiao EY, Coghill A, Kizub D, Fink V, Ndlovu N, Mazul A, Sigel K. The effect of non-AIDS-defining cancers on people living with HIV. Lancet Oncol. 2021:22(6):e240–e253. 10.1016/S1470-2045(21)00137-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller M, Lee JY, Fulcher JA, Roach ME, Dilworth SE, Chahine A, Pallikkuth S, Fuchs D, Pahwa S, Carrico AW. Getting to the point: methamphetamine injection is associated with biomarkers relevant to HIV pathogenesis. Drug Alcohol Depend. 2020:213:108133. 10.1016/j.drugalcdep.2020.108133 [DOI] [PubMed] [Google Scholar]
- 17.Ersche KD, Döffinger R. Inflammation and infection in human cocaine addiction. Curr Opin Behav Sci. 2017:13:203–209. 10.1016/j.cobeha.2016.12.007 [DOI] [Google Scholar]
- 18.Levandowski ML, Viola TW, Prado CH, Wieck A, Bauer ME, Brietzke E, Grassi-Oliveira R. Distinct behavioral and immunoendocrine parameters during crack cocaine abstinence in women reporting childhood abuse and neglect. Drug Alcohol Depend. 2016:167:140–148. 10.1016/j.drugalcdep.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Meng J, Zhang L, Johnson T, Chen C, Roy S. Morphine induces changes in the gut microbiome and metabolome in a morphine dependence model. Sci Rep. 2018:8(1):3596. 10.1038/s41598-018-21915-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng J, Banerjee S, Zhang L, Sindberg G, Moidunny S, Li B, Robbins DJ, Girotra M, Segura B, Ramakrishnan S, et al. Opioids impair intestinal epithelial repair in HIV-infected humanized mice. Front Immunol. 2020:10:2999. 10.3389/fimmu.2019.02999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underwood ML, Nguyen T, Uebelhoer LS, Kunkel LE, Korthuis PT, Lancioni CL. Altered monocyte phenotype and dysregulated innate cytokine responses among people living with HIV and opioid-use disorder. AIDS. 2020:34(2):177–188. 10.1097/QAD.0000000000002416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ríos-Olivares E, Vilá LM, Reyes JC, Rodríguez JW, Colón JH, Pagán NO, Marrero A, Ríos-Orraca ZM, Boukli NM, Shapshak P, et al. Impaired cytokine production and suppressed lymphocyte proliferation activity in HCV-infected cocaine and heroin (“speedball”) users. Drug Alcohol Depend. 2006:85(3):236–243. 10.1016/j.drugalcdep.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 23.Abadie R, Habecker P, Carrasco KG, Chiou KS, Fernando S, Bennett SJ, Valentin-Acevedo A, Dombrowski K, West JT, Wood C. Employing respondent driven sampling (RDS) to recruit people who inject drugs (PWID) and other hard-to-reach populations during COVID-19: lessons learned. Front Psychiatry. 2022:13: 990055. 10.3389/fpsyt.2022.990055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maffei VJ, Siggins RW, Luo M, Brashear MM, Mercante DE, Taylor CM, Molina P, Welsh DA. Alcohol use is associated with intestinal dysbiosis and dysfunctional CD8+ T-cell phenotypes in persons with human immunodeficiency virus. J Infect Dis. 2021:223(6): 1029–1039. 10.1093/infdis/jiaa461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opzoomer JW, Timms JA, Blighe K, Mourikis TP, Chapuis N, Bekoe R, Kareemaghay S, Nocerino P, Apollonio B, Ramsay AG, Tavassoli M, et al. Immunocluster provides a computational framework for the nonspecialist to profile high-dimensional cytometry data. Elife. 2021:10:e62915. 10.7554/eLife.62915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000:25(1):25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021:49(D1):D325–D334. 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER Version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019:47(D1):D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tso FY, Kossenkov AV, Lidenge SJ, Ngalamika O, Ngowi JR, Mwaiselage J, Wickramasinghe J, Kwon EH, West JT, Lieberman PM, et al. RNA-Seq of Kaposi’s sarcoma reveals alterations in glucose and lipid metabolism. PLoS Pathog. 2018:14(1):e1006844. 10.1371/journal.ppat.1006844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hileman CO, Bowman ER, Gabriel J,Kettelhut A, Labbato D, Smith C, Avery A, Parran T, Funderburg N, McComsey GA. Impact of heroin and HIV on gut integrity and immune activation. J Acquir Immune Defic Syndr. 2022:89(5):519–526. 10.1097/QAI.0000000000002893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hileman CO, Durieux JC, Janus SE, Bowman E, Kettelhut A, Nguyen TT, Avery AK, Funderburg N, Sullivan C, McComsey GA. Heroin use is associated with vascular inflammation in human immunodeficiency virus. Clin Infect Dis. 2023:76(3):375–381. 10.1093/cid/ciac812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azzoni L, Giron LB, Vadrevu S, Zhao L, Lalley-Chareczko L, Hiserodt E, Fair M, Lynn K, Trooskin S, Mounzer K, et al. Methadone use is associated with increased levels of sCD14, immune activation, and inflammation during suppressed HIV infection. J Leukoc Biol. 2022:112(4):733–744. 10.1002/JLB.4A1221-678RR [DOI] [PubMed] [Google Scholar]
- 33.Tomescu C, Colon K, Smith P, Taylor M, Azzoni L, Metzger DS, Montaner LJ. Persons who inject drugs (PWID) retain functional NK cells, dendritic cell stimulation, and adaptive immune recall responses despite prolonged opioid use. J Leukoc Biol. 2021:110(2): 385–396. 10.1002/JLB.5A0920-604R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habecker P, Abadie R, Welch-Lazoritz M, Reyes JC, Khan B, Dombrowski K. Injection partners, HCV, and HIV status among rural persons who inject drugs in Puerto Rico. Subst Use Misuse. 2018:53(7):1128–1138. 10.1080/10826084.2017.1400562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaishnavi C Translocation of gut flora and its role in sepsis. Indian J Med Microbiol. 2013:31(4):334–342. 10.4103/0255-0857.118870 [DOI] [PubMed] [Google Scholar]
- 36.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012:30(1):149–173. 10.1146/annurev-immunol-020711-075001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012:10(9):655–666. 10.1038/nrmicro2848 [DOI] [PubMed] [Google Scholar]
- 38.Ramendra R, Isnard S, Mehraj V, Chen J, Zhang Y, Finkelman M, Routy JP. Circulating LPS and (1→3)-β-D-glucan: a folie à deux contributing to HIV-associated immune activation. Front Immunol. 2019:10:465. 10.3389/fimmu.2019.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luján JA, Rugeles MT, Taborda NA. Contribution of the microbiota to intestinal homeostasis and its role in the pathogenesis of HIV-1 infection. Curr HIV Res. 2019:17(1):13–25. 10.2174/1570162X17666190311114808 [DOI] [PubMed] [Google Scholar]
- 40.Jalodia R, Abu YF, Oppenheimer MR, Herlihy B, Meng J, Chupikova I, Tao J, Ghosh N, Dutta RK, Kolli U, et al. Opioid use, gut dysbiosis, inflammation, and the nervous system. J Neuroimmune Pharmacol. 2022:17(1–2):76–93. 10.1007/s11481-021-10046-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbarali HI, Dewey WL. Gastrointestinal motility, dysbiosis and opioid-induced tolerance: is there a link? Nat Rev Gastroenterol Hepatol. 2019:16(6):323–324. 10.1038/s41575-019-0150-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chivero ET, Ahmad R, Thangaraj A, Periyasamy P, Kumar B, Kroeger E, Feng D, Guo ML, Roy S, Dhawan P, et al. Cocaine induces inflammatory gut milieu by compromising the mucosal barrier integrity and altering the gut microbiota colonization. Sci Rep. 2019:9(1):12187. 10.1038/s41598-019-48428-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Araos P, Pedraz M, Serrano A, Lucena M, Barrios V, García-Marchena N, Campos-Cloute R, Ruiz JJ, Romero P, Suárez J, et al. Plasma profile of pro-inflammatory cytokines and chemokines in cocaine users under outpatient treatment: influence of cocaine symptom severity and psychiatric co-morbidity. Addict Biol. 2015:20(4):756–772. 10.1111/adb.12156 [DOI] [PubMed] [Google Scholar]
- 44.Kallas E, Huik K, Türk S, Pauskar M, Jõgeda EL, Šunina M, Karki T, Des Jarlais D, Uusküla A, Avi R, et al. T cell distribution in relation to HIV/HBV/HCV coinfections and intravenous drug use. Viral Immunol. 2016:29(8):464–470. 10.1089/vim.2016.0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veenhuis RT, Astemborski J, Chattergoon MA, Greenwood P, Jarosinski M, Moore RD, Mehta SH, Cox AL. Systemic elevation of proinflammatory interleukin 18 in HIV/HCV coinfection versus HIV or HCV monoinfection. Clin Infect Dis. 2017:64(5): 589–596. 10.1093/cid/ciw771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vos AG, Dodd CN, Delemarre EM, Nierkens S, Serenata C, Grobbee DE, Klipstein-Grobusch K, Venter WDF. Patterns of immune activation in HIV and non HIV subjects and its relation to cardiovascular disease risk. Front Immunol. 2021:12:647805. 10.3389/fimmu.2021.647805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babu H, Ambikan AT, Gabriel EE, Svensson Akusjärvi S, Palaniappan AN, Sundaraj V, Mupanni NR, Sperk M, Cheedarla N, Sridhar R, et al. Systemic inflammation and the increased risk of inflamm-aging and age-associated diseases in people living with HIV on long term suppressive antiretroviral therapy. Front Immunol. 2019:10:1965. 10.3389/fimmu.2019.01965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babu H, Sperk M, Ambikan AT, Rachel G, Viswanathan VK, Tripathy SP, Nowak P, Hanna LE, Neogi U. Plasma metabolic signature and abnormalities in HIV-infected individuals on long-term successful antiretroviral therapy. Metabolites. 2019:9(10): 210. 10.3390/metabo9100210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.deFilippi C, Toribio M, Wong LP, Sadreyev R, Grundberg I, Fitch KV, Zanni MV, Lo J, Sponseller CA, Sprecher E, et al. Differential plasma protein regulation and statin effects in human immunodeficiency virus (HIV)-infected and non-HIV-infected patients utilizing a proteomics approach. J Infect Dis. 2020:222(6): 929–939. 10.1093/infdis/jiaa196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamat A, Misra V, Cassol E, Ancuta P, Yan Z, Li C, Morgello S, Gabuzda D. A plasma biomarker signature of immune activation in HIV patients on antiretroviral therapy. PLoS One. 2012:7(2): e30881. 10.1371/journal.pone.0030881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parikh N, Dampier W, Feng R, Passic SR, Zhong W, Frantz B, Blakey B, Aiamkitsumrit B, Pirrone V, Nonnemacher MR, et al. Cocaine alters cytokine profiles in HIV-1–infected African American individuals in the DrexelMed HIV/AIDS genetic analysis cohort. J Acquir Immune Defic Syndr. 2014:66(3):256–264. 10.1097/QAI.0000000000000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abadie R, Goldenberg S, Welch-Lazoritz M, Fisher CB. Establishing trust in HIV/HCV research among people who inject drugs (PWID): insights from empirical research. PLoS One. 2018:13(12): e0208410. 10.1371/journal.pone.0208410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


