Abstract
The dcuA and dcuB genes of Escherichia coli encode homologous proteins that appear to function as independent and mutually redundant C4-dicarboxylate transporters during anaerobiosis. The dcuA gene is 117 bp downstream of, and has the same polarity as, the aspartase gene (aspA), while dcuB is 77 bp upstream of, and has the same polarity as, the anaerobic fumarase gene (fumB). To learn more about the respective roles of the dcu genes, the environmental and regulatory factors influencing their expression were investigated by generating and analyzing single-copy dcuA- and dcuB-lacZ transcriptional fusions. The results show that dcuA is constitutively expressed whereas dcuB expression is highly regulated. The dcuB gene is strongly activated anaerobically by FNR, repressed in the presence of nitrate by NarL, and subject to cyclic AMP receptor protein (CRP)-mediated catabolite repression. In addition, dcuB is strongly induced by C4-dicarboxylates, suggesting that dcuB is under the control of an uncharacterized C4-dicarboxylate-responsive gene regulator. Northern blotting confirmed that dcuA (and aspA) is expressed under both aerobic and anaerobic conditions and that dcuB (and fumB) is induced anaerobically. Major monocistronic transcripts were identified for aspA and dcuA, as well as a minor species possibly corresponding to an aspA-dcuA cotranscript. Five major transcripts were observed for dcuB and fumB: monocistronic transcripts for both fumB and dcuB; a dcuB-fumB cotranscript; and two transcripts, possibly corresponding to dcuB-fumB and fumB mRNA degradation products. Primer extension analysis revealed independent promoters for aspA, dcuA, and dcuB, but surprisingly no primer extension product could be detected for fumB. The expression of dcuB is entirely consistent with a primary role for DcuB in mediating C4-dicarboxylate transport during anaerobic fumarate respiration. The precise physiological purpose of DcuA remains unclear.
Escherichia coli can utilize C4-dicarboxylates for energy generation under both aerobic and anaerobic conditions (8). Under aerobic conditions, the uptake of C4-dicarboxylates (fumarate, malate, and succinate) and l-aspartate is mediated by a secondary transporter and/or a binding protein-dependent system, designated Dct (19, 22). Three mutations (cbt at 16.2 min, dctA at 79.2 min, and dctB at 16.1 min) have been reported to result in the inactivation of components involved in aerobic C4-dicarboxylate transport (23). The dctA gene has been sequenced, and the role of its product (DctA) in the utilization of C4-dicarboxylates (and the cyclic monocarboxylate orotate) is supported by complementation studies of Salmonella typhimurium dctA or outA mutants (2, 37). The dctB and cbt genes are predicted to encode inner membrane and periplasmic binding proteins, respectively (21), but the genes have yet to be identified in the E. coli genome sequence.
Uptake, exchange, and efflux of C4-dicarboxylic acids under anaerobic conditions are mediated by the Dcu systems (Km for fumarate uptake, 50 to 400 μM), which are genetically distinct from the aerobic Dct system (8, 9, 45). Transport studies suggest that the Dcu systems are expressed exclusively under anaerobic conditions, activated by the anaerobic activator protein FNR, and repressed in the presence of nitrate. Three independent Dcu systems have been identified, DcuA, DcuB, and DcuC (36, 45). DcuA and DcuB are homologous proteins (36% identical), whereas DcuC is only 22 to 24% identical to DcuA and DcuB. The DcuA protein is encoded by a gene (dcuA, at 94.0 min) located 117 bp downstream of the anaerobically expressed aspartase gene aspA (see Fig. 1). DcuB is encoded by dcuB (at 93.5 min), which is 77 bp upstream of the gene (fumB) encoding the anaerobic fumarase, FumB (see Fig. 1). DcuC is encoded by the dcuC gene at 14.1 min.
FIG. 1.
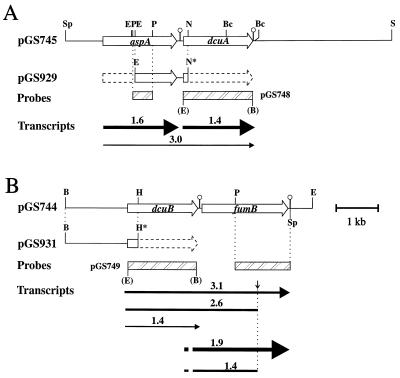
Restriction maps and transcriptional organization of the aspA-dcuA (A) and dcuB-fumB (B) regions. The inserts of plasmids pGS745, pGS748, and pGS929 (A) and of plasmids pGS744, pGS749, and pGS931 (B) are shown. Restriction site abbreviations: B, BamHI; Bc, BclI; E, EcoRI; H, HindIII; N, NarI; P, PstI; S, SalI; Sp, SphI. Restriction sites in parentheses were introduced during PCR amplification. Asterisks indicate restriction sites that have been inactivated by digestion with the corresponding restriction enzyme followed by filling in with the Klenow fragment of DNA polymerase I. Open arrows indicate the positions and polarities of the aspA, dcuA, dcuB, and fumB structural genes. Closed arrows indicate primary transcripts and putative processed transcripts (sizes in kilonucleotides) as detected by Northern hybridization experiments. The relative abundance of each transcript is indicated by arrow thickness. The small downward arrow indicates a potential endoribonucleolytic processing site. Hatched bars represent DNA fragments used as hybridization probes, and strongly predicted stem-loop structures are indicated.
Growth tests and transport studies using dcuA, dcuB, and dcuC single, double, and triple mutants have shown that DcuA, DcuB, and DcuC each mediate exchange as well as uptake (36, 45). The triple mutant was almost completely devoid of Dcu activity and was unable to use fumarate for anaerobic respiratory growth, but growth could be supported by fermentation. The single mutants exhibited no phenotype, but the dcuA dcuB double mutant was markedly defective in both C4-dicarboxylate transport and fumarate respiratory growth, suggesting that DcuA and DcuB have analogous and mutually complementary transport functions in the anaerobic uptake of C4-dicarboxylates (36, 45). The affinities of DcuA and DcuB for C4-dicarboxylates are similar, except for the lower affinity of DcuA for malate (45).
Expression of the aspA gene is repressed by glucose (∼10-fold) under aerobic conditions and enhanced anaerobically (∼10-fold) in a manner which is partially (∼2-fold) FNR dependent (16, 44). Expression of fumB is also induced anaerobically (1.5- or 5-fold) in a manner which is dependent on both FNR and ArcA (41, 44). The anaerobic expression of fumB is reduced by ∼10- or 2-fold in an fnr background, and unlike aspA expression, fumB expression is not subject to glucose-mediated repression (41, 44). The anaerobic induction of aspA and fumB is appropriate since both aspartase and fumarase B are thought to function in the generation of fumarate for utilization as an anaerobic electron acceptor. Predicted FNR and cyclic AMP receptor protein (CRP) sites at suitable distances from putative promoter sequences are located immediately upstream of aspA and dcuB (36, 44). However, a previous analysis of the aspA-dcuA and dcuB-fumB intergenic regions showed no potential CRP sites and the putative FNR sites were poorly placed, suggesting that the dcuB-fumB and aspA-dcuA gene pairs are cotranscribed (36).
This paper describes studies on the transcriptional regulation and organization of the dcuA and dcuB genes that provide further information concerning the roles of DcuA and DcuB. The results show that the dcuB gene is expressed exclusively under anaerobic conditions in a manner that is FNR dependent and that it is repressed by NarL in the presence of nitrate and is subject to CRP-mediated catabolite repression. Furthermore, dcuB is strongly induced by C4-dicarboxylates presumably mediated by an uncharacterized C4-dicarboxylate-responsive gene regulator. In contrast, the dcuA gene shows little variation in expression under the growth conditions investigated. These findings suggest that DcuB functions primarily as a C4-dicarboxylate transporter during fumarate respiration, whereas DcuA has a general role in anaerobic C4-dicarboxylate transport.
MATERIALS AND METHODS
Strains and plasmids.
All E. coli strains, plasmids, and phages used in this study are listed in Table 1. A plasmid containing the aspA-dcuA region was obtained by subcloning the 6.2-kb SphI-SalI fragment of pGS73 (13) into the corresponding sites of pBR322 to generate pGS745 (Fig. 1A). A dcuA′-lacZYA transcriptional fusion was produced by subjecting pGS745 to digestion with NarI, treatment with DNA polymerase I (Klenow fragment) to generate blunt-ended fragments, and digestion with EcoRI and then subcloning the resulting 1-kb EcoRI-NarI dcuA′-containing fragment into EcoRI- and SmaI-digested pRS415 (35) to generate pGS929 (Fig. 1A).
TABLE 1.
Strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Δ(argF-lac)U169 (φ80ΔlacZM15) recA | 30 |
| JRG1999 | MC1000 ΔcrpT8 | S. T. Cole, Paris, France |
| JRG1728 | MC1000 Δ(tyrR-fnr-rac-trg) 17 zdd-230::Tn9 | 39 |
| JRG3834 | MC4100 [λRS45::dcuA′-lacZYA] | This work |
| JRG3835 | MC4100 [λRS45::dcuB′-lacZYA] | This work |
| JRG3836 | MC1000 [λRS45::dcuA′-lacZYA] | This work |
| JRG3837 | MC1000 [λRS45::dcuB′-lacZYA] | This work |
| JRG3838 | JRG3834fnr | This work |
| JRG3839 | JRG3835fnr | This work |
| JRG3840 | JRG3834arc | This work |
| JRG3841 | JRG3835arc | This work |
| JRG3842 | JRG3835narL | This work |
| JRG3843 | JRG3835narP | This work |
| JRG3845 | JRG1999 [λRS45::dcuA′-lacZYA] | This work |
| JRG3846 | JRG1999 [λRS45::dcuB′-lacZYA] | This work |
| MC1000 | ΔlacX74 Δ(araABC-leu) | 33 |
| MC4100 | Δ(argF-lac)U169 rpsL | 33 |
| RKP3580 | narL215::Tn10 | R. K. Poole, Sheffield, United Kingdom |
| RKP3655 | narP258::Tn10d | R. K. Poole, Sheffield, United Kingdom |
| RM315 | MC4100 ΔarcA1 zjj::Tn10 | 31 |
| Plasmids | ||
| pGS73 | aspA dcuA | 13 |
| pGS78 | dcuB fumB | 12 |
| pGS744 | pBR322 + 4.7-kb BamHI-EcoRI dcuB-fumB fragment | This work |
| pGS745 | pBR322 + 6.2-kb SphI-SalI aspA-dcuA fragment | This work |
| pGS748 | pUC118 + 1.3-kb dcuA fragment | This work |
| pGS749 | pUC119 + 1.3-kb dcuB fragment | This work |
| pGS929 | pRS415 + dcuA′-lacZYA | This work |
| pGS931 | pRS528 + dcuB′-lacZYA | This work |
| pRS415/528 | lacZYA transcriptional fusion vectors | 35 |
| Phages | ||
| P1vir1 | 26 | |
| λRS45 | 35 |
A plasmid containing the dcuB-fumB region was made by subcloning the 4.7-kb BamHI-EcoRI fragment of pGS78 (12) into BamHI- and EcoRI-digested pBR322 to generate pGS744 (Fig. 1B). A dcuB′-lacZYA transcriptional fusion was generated by subjecting pGS744 to digestion with HindIII, treatment with DNA polymerase I (Klenow), and then digestion with BamHI and then subcloning the resulting 1-kb BamHI-HindIII dcuB′ fragment into BamHI- and SmaI-digested pRS528 (35) to generate pGS931 (Fig. 1B). The dcuA and dcuB-lacZ fusions were transferred to the phage λRS45 (35) by homologous recombination in vivo as described by Simons et al. (35), and the resulting Lac+-conferring phages, λRS45(dcuA′-lacZYA) and λRS45(dcuB′-lacZYA), were used to produce monolysogenic derivatives of appropriate strains of E. coli (Table 1).
A 1.3-kb dcuA fragment, suitable for use as a hybridization probe in Northern blot analysis, was PCR amplified by using the plasmid pGS73 (13) as a template, Pfu DNA polymerase (Stratagene), and the following two primers: DCUA1, 5′-ccgaattcat2129ATGCTAGTTGTAGAACTC2146-3′; and DCUA4, 5′-ccggatcc3436TGATCATTACAGCATGAAG3418-3′ (where underlining indicates the dcuA start codon, small capitals indicate mismatches, boldface type indicates EcoRI and BamHI sites, italics indicate NdeI site, and coordinates are from the work of Six et al. [36]; primers were designed to introduce flanking EcoRI-NdeI and BamHI sites). The EcoRI- and BamHI-digested PCR product was subcloned into plasmid pUC118 (42) to generate the plasmid pGS748 (Fig. 1A). A similar strategy was adopted for subcloning the dcuB gene in pUC119 (42), except that a dcuB-containing template, pGS78 (12), and different PCR primers were used; the primers were DCUB1, 5′-ccgaattcat170ATGTTATTTACTATCCAAC188-3′, and DCUB2, 5′-ccggatcc1516GTGCATTTATAAGAACCCG1497-3′. The 1.3-kb dcuB-containing fragment was subcloned in pUC119, generating plasmid pGS749 (Fig. 1B).
To investigate the effects of the global regulators Fnr, Arc, NarL, and NarP on dcuA and dcuB expression, the fnr, arc, narL, and narP deletions were transferred from the corresponding donor strains, JRG1728, RM315, RKP3580, and RKP3655, by P1vir-mediated transduction to the dcuA- and dcuB-lacZ fusion strains, JRG3834 and JRG3835 (Table 1). To study the effects of CRP on dcuA and dcuB expression, a pGS279 (crp+) transformant of strain JRG1999 (Δcrp) was infected with λRS45(dcuA′-lacZYA) and λRS45(dcuB′-lacZYA) to generate the monolysogens JRG3845(pGS279) and JRG3846(pGS279), respectively, which were subsequently cured of the plasmid by propagation of the strains under nonselective conditions.
Growth media and conditions.
Cultures were grown at 37°C, either aerobically in shaking 250-ml conical flasks or anaerobically in 10-ml bijou bottles (for β-galactosidase measurements) or in 300-ml medical flats (for the preparation of total RNA). Bacteria were usually grown aerobically in L broth for DNA manipulation and in L broth supplemented with glycerol and fumarate for the extraction of total RNA. For measurements of β-galactosidase activity, strains were grown in M9 minimal salts medium (Sigma), unless otherwise stated, with glucose (0.4%) or glycerol (0.4%) as a carbon source, and supplements of 0.5% Casamino Acids, 1 mM MgSO4, 0.1 mM CaCl2, and 0.5 mg of vitamin B1 per ml. When used, fumarate, nitrate, and trimethylamine N-oxide (TMAO) were present at 50 mM.
β-Galactosidase measurements.
Samples with an optical density at 650 nm (OD650) of 0.5 were withdrawn from cultures grown in duplicate at regular intervals during the growth cycle. The samples were cooled on ice, and the bacteria were pelleted by centrifugation, resuspended in ice-cold saline, and resedimented at 4°C. After complete removal of the supernatant by aspiration, the bacteria were frozen at −20°C and used within 3 days.
Cell extracts were prepared from thawed cells and assayed for β-galactosidase activity and protein content by using an iEMS Reader MF microtiter plate spectrophotometer (Labsystems) by the method of Phillips-Jones et al. (27). β-Galactosidase specific activities (micromoles of ONPG [o-nitrophenyl-β-d-galactopyranoside] per minute per milligram of protein) were averaged from samples taken from two independent cultures. Each of the two samples was assayed in duplicate.
Northern hybridization and primer extension analysis.
Total RNA present in MC4100 was extracted by using a method based upon that described by Aiba et al. (1). Cultures (500 ml) were harvested by centrifugation and resuspended in 3 to 5 ml of a solution containing 20 mM sodium acetate (pH 5.5), 0.5% SDS, and 1 mM EDTA at 4°C. An equal volume of acid-phenol (phenol equilibrated with 20 mM sodium acetate [pH 5.5]), was added, and the mixture was incubated for 5 min at 65°C. After centrifugation, the aqueous layer was removed, extracted twice more with acid-phenol, and then extracted once with chloroform and once with a mixture of phenol-CHCl3-isoamyl alcohol (25:24:1; equilibrated with 0.1 M Tris-Cl [pH 8.0]). RNA in the aqueous layer was precipitated with 3 volumes of ethanol and dissolved in ∼1 ml of water. The RNA content was determined spectroscopically by assuming that an OD260 of 1 corresponds to ∼40 μg of RNA per ml.
Northern hybridization was performed by fractionating total RNA, along with RNA molecular weight standards (Gibco-BRL), in a 1% agarose gel containing 2.2 M formaldehyde. Denatured RNA was transferred to nitrocellulose membranes and hybridized with DNA probes radiolabeled with [α-32P]dCTP by using the Ready to Go DNA Labeling Kit (−dCTP) (Pharmacia). Hybridization was performed at 65°C as described by Sambrook et al. (30). The hybridization probes used were the 1.3-kb EcoRI-BamHI dcuA fragment of pGS748, the 1.3-kb EcoRI-BamHI dcuB fragment of pGS749, the 0.375-kb PstI aspA fragment of pGS745 (10), and the 1-kb PstI-SphI fumB fragment of pGS744 (Fig. 1). DNA-RNA hybrids were detected by autoradiography using BioMax MS (Kodak) autoradiography film.
Reverse transcriptase-mediated primer extension analysis was performed as described by Quail et al. (28) by using two oligonucleotide primers for each promoter. The primers used and the corresponding base pair coordinates (2, 36) were as follows: for dcuA, P1dcuA, 5′-2235GCAAGAACCAGCACCCCCAATCCGCCTGCA2206-3′, and P2dcuA, 5′-2211CCTGCAAAACCAATACCTATTCCCCCCAAT2182-3′; for dcuB, P1dcuB, 5′-294AGGTGGAAGACGAAGACCAGAATGACCAGA265-3′, and P2dcuB, 270ACCAGACCGATACCGCCTAATAAACCCAGC241-3′; for aspA, P1aspA, 693TTGTTGTTGCTGATATAGAAGTTTACAATC664-3′, and P2aspA, 5′-669ACAATCGCTCTCAGAGTGTGAACACCATAG640-3′; and for fumB, P1fumB, 5′-1336GGTTTCGCCGTCGAAGTCGGCAACGCTAAC1307-3′, and P2fumB, 5′-1309AACGTAATCGGAAGTGAGTAGATAGTA1283-3′. Sequence ladders were generated by using the T7 Sequenase DNA sequencing kit (Amersham), together with plasmids pGS744 and pGS745 as templates, and the primers described above. Primer extension products and sequencing ladders were visualized by autoradiography. Autoradiographic images were digitized by use of a model GS-690 imaging densitometer (Bio-Rad) linked to a computer equipped with Molecular Analyst (Bio-Rad) software. Potential FNR, CRP, and NarL binding sites were identified by using the score matrix searching option of the xnip program (40) and score matrices derived from 22 FNR, 25 CRP, and 8 NarL experimentally determined binding sites.
RESULTS
Effects of growth conditions on the expression of single-copy dcuA- and dcuB-lacZ transcriptional fusions.
The expression of the dcuA and dcuB genes was investigated by using derivatives of MC4100 (ΔlacZ) containing single copies of the corresponding lacZ transcriptional gene fusion (see Materials and Methods). The fusions were transferred from plasmids to λRS45 via homologous recombination for use in constructing the monolysogens JRG3834 (MC4100 [λRS45::dcuA′-lacZYA]) and JRG3835 (MC4100 [λRS45::dcuB′-lacZYA]). In order to include the entire dcuA and dcuB operator-promoter regions, 1.1- and 1.4-kb segments of DNA located immediately upstream of the respective structural genes were fused to the lacZ reporter gene. Both fusions were active (see below), indicating that dcuA and dcuB each possess independent promoters and that dcuA transcription is not dependent on the upstream aspA gene.
The β-galactosidase specific activities of JRG3834 (dcuA-lacZ) and JRG3835 (dcuB-lacZ) were determined after aerobic and anaerobic growth in minimal medium containing glycerol plus fumarate and Casamino Acids (Fig. 2). Expression of the dcuA-lacZ fusion under aerobic conditions (∼0.5 μmol/min/mg) increased by only two- to threefold anaerobically (0.9 to 1.5 μmol/min/mg). In contrast, aerobic expression of the dcuB-lacZ fusion was very low (∼0.02 μmol/min/mg) but was up to 150-fold higher (3 μmol/min/mg) anaerobically (Fig. 2). Thus, dcuB expression is strongly induced during anaerobiosis whereas dcuA expression is only slightly increased by lack of oxygen. The expression of dcuA and dcuB increased only slightly (up to ca. twofold) in the transition from exponential to postexponential growth, indicating that expression is weakly affected by the growth phase (Fig. 2). For this reason, all subsequent data (Fig. 3 to 7) relate to samples taken during the mid-logarithmic to late logarithmic phase.
FIG. 2.
Expression of dcuA-lacZ and dcuB-lacZ transcriptional fusions during aerobic (A) and anaerobic (B) growth at 37°C in M9 minimal salts medium containing 0.4% glycerol, 50 mM fumarate, and 0.05% Casamino Acids. The strains used were JRG3834 (dcuA-lacZ) (• and ○) and JRG3835 (dcuB-lacZ) (■, □). Growth (open symbols) and β-galactosidase activities (closed symbols) are shown.
FIG. 3.
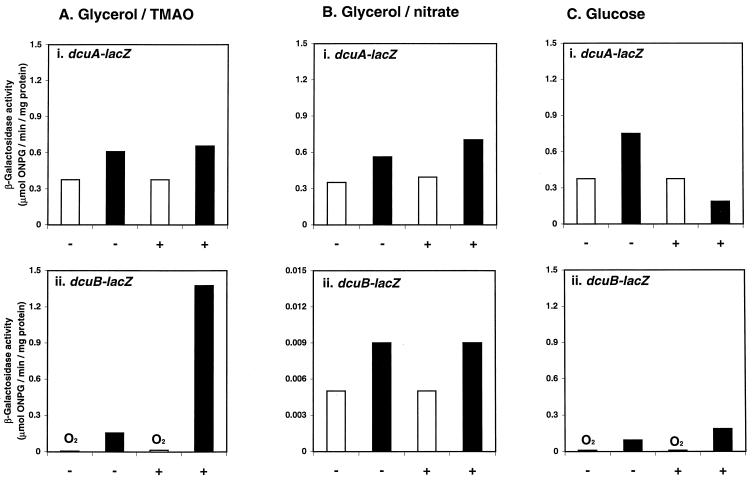
Expression of the dcuA- and dcuB-lacZ fusions during aerobic (open bars) and anaerobic (closed bars) growth at 37°C in M9 minimal salts medium containing 0.05% Casamino Acids plus either 0.4% glycerol and 50 mM TMAO (A), 0.4% glycerol with 50 mM nitrate (B), or 0.4% glucose (C). The plus and minus signs indicate the presence and absence of 50 mM fumarate, respectively. β-Galactosidase activities were assayed in samples of mid-logarithmic to late logarithmic cultures of JRG3834 (dcuA-lacZ) and JRG3835 (dcuB-lacZ).
FIG. 7.
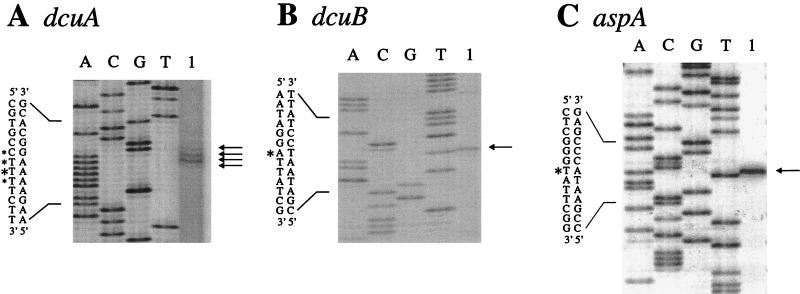
Determination of the dcuA (A), dcuB (B), and aspA (C) transcriptional start sites by reverse transcriptase-mediated primer extension. Lanes 1 indicate primer extension products. Sequencing ladders (lanes A, C, G, and T) were generated by using the primers used for the reverse transcriptase reaction. The sequencing ladders and primer extension products shown in panels A, B, and C were generated by using, respectively, primers P1dcuA, P2dcuB, and P2aspA (see Materials and Methods). Similar results were obtained with primers P2dcuA, P1dcuB, and P1aspA (data not shown). The sequence, its complement, and the transcriptional start sites (indicated by asterisks) are shown. Horizontal arrows indicate primer extension products.
Although dcuB was strongly expressed during anaerobic respiratory growth with fumarate as the terminal electron acceptor (∼1.3 μmol/min/mg), expression was 8- and 144-fold lower when TMAO and nitrate (0.16 and 0.009 μmol/min/mg), respectively, were used as sole electron acceptors (Fig. 2B, 3Aii, and 3Bii). However, these values are still 30- and 2-fold higher than the corresponding aerobic activities (0.05 and 0.005 μmol/min/mg) (Fig. 3Aii and Bii). Addition of fumarate to minimal medium containing glycerol plus TMAO and Casamino Acids resulted in a ninefold increase in dcuB expression under anaerobic conditions (Fig. 3Aii), but addition of fumarate to minimal medium containing glycerol plus nitrate and Casamino Acids had no effect on dcuB expression (Fig. 3Bii). These findings indicate that the anaerobic expression of dcuB is strongly repressed by nitrate, and in the absence of nitrate and oxygen, it is strongly induced by fumarate. In contrast, expression of dcuA is only slightly (less than twofold) affected by nitrate and fumarate (Fig. 3Ai and Bi).
Expression of the dcuB-lacZ fusion was moderate (0.09 μmol/min/mg) during anaerobic fermentative growth in glucose minimal medium plus Casamino Acids and was increased only twofold (to 0.18 μmol/min/mg) by the inclusion of fumarate (Fig. 3Cii). The latter value is sevenfold lower than that (1.3 μmol/min/mg) observed when glucose was replaced by glycerol (Fig. 2B), indicating that dcuB transcription is subject to catabolite repression. During aerobic growth, dcuB was very weakly expressed (0.005 to 0.014 μmol/min/mg) under all conditions employed (Fig. 3). Expression of the dcuA-lacZ fusion (0.35 to 0.75 μmol/min/mg) was only slightly affected by the growth conditions, indicating that dcuA is expressed constitutively.
The studies described above show that dcuB is strongly repressed by oxygen and nitrate and is moderately repressed by glucose. In the absence of these repressing substrates, dcuB is strongly induced by fumarate. To determine whether dcuB expression is induced by C4-dicarboxylic acids other than fumarate, expression was measured in minimal medium containing glycerol plus TMAO and either aspartate, fumarate, malate, maleate, or succinate (Fig. 4A). Casamino Acids was omitted from the media used in these experiments to eliminate any stimulation caused by its aspartate residue content. The expression of dcuB was lowered ninefold (from 0.16 to 0.018 μmol/min/mg) by omitting Casamino Acids (Fig. 4A), indicating that the addition of Casamino Acids does indeed induce dcuB expression. In the absence of Casamino Acids, dcuB expression was induced ∼40- to 70-fold by the five C4-dicarboxylic acids tested (Fig. 4A). However, the carboxylic acids (pyruvate, acetate, and lactate) had no effect (Fig. 4A), indicating that dcuB induction is specific for C4-dicarboxylates as is Dcu transport (8). The degree of dcuB induction by C4-dicarboxylates decreased in the order fumarate (1.2 μmol/min/mg) ≈ malate (1.1) ≈ aspartate (1.0) > succinate (0.8) > maleate (0.6). This order differs from the order of substrate preference for DcuB measured as a function of competitive inhibition of succinate antiport activity in a dcuA mutant (36), suggesting that there is no direct link between the substrate binding specificity of DcuB and the regulation of dcuB by C4-dicarboxylates.
FIG. 4.
Effects of carboxylates on expression of the dcuB-lacZ fusion during anaerobic growth in M9 salts medium containing 0.4% glycerol and 50 mM TMAO but lacking Casamino Acids (except where indicated). The alternative carboxylates (50 mM) or 0.5% Casamino Acids (CA) (A) and the fumarate concentrations (B) are indicated on the x axis. Other details are as described for Fig. 3.
The activation of dcuB expression by fumarate was directly related to the initial concentration of fumarate in the medium over the range of 1 to 50 mM (Fig. 4B). Fumarate concentrations of ≥250 mM inhibited growth, whereas those of ≤0.1 mM were too low to affect dcuB expression (Fig. 4B). The apparent Km for fumarate of the C4-dicarboxylate-responsive dcuB-regulatory system can be estimated to be 4 to 9 mM. The latter value is 10- to 100-fold greater than the Km of the Dcu transporters (50 to 400 μM) but is appropriate to ensure induction of DcuB when substrate concentrations are at levels that could be saturating for the DcuA and DcuC transporters. Thus, anaerobic induction of dcuB by C4-dicarboxylates would be expected to result in increased Dcu transport activity, which is indeed what has been observed (8).
Effect of global regulators on dcuA- and dcuB-lacZ expression.
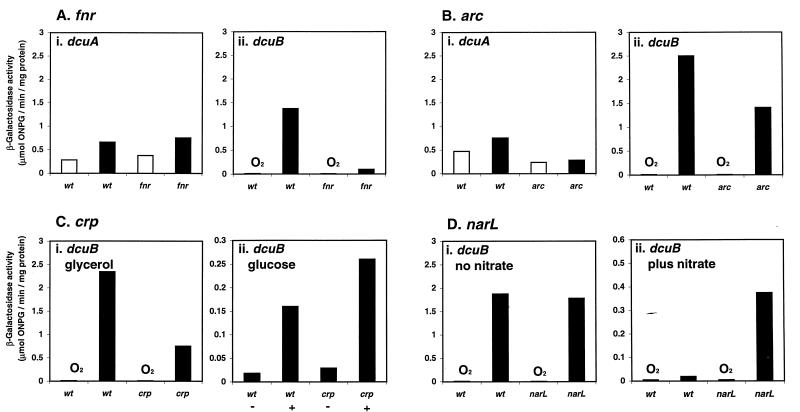
The glucose, oxygen, and nitrate repression of dcuB expression suggests that dcuB could be subject to regulation by the global transcriptional regulatory proteins FNR, CRP, ArcA, NarP, and NarL. This was tested by measuring dcuB-lacZ (and dcuA-lacZ) expression in the appropriate regulatory mutants (Fig. 5). The anaerobic expression of dcuB in minimal medium containing glycerol plus TMAO and fumarate was 15-fold lower in a Δfnr strain, JRG3839, than in the parental strain, JRG3835 (Fig. 5Aii). However, even in the absence of FNR, the anaerobic expression level was still 19-fold higher (0.094 μmol/min/mg) than the aerobic level (0.005 μmol/min/mg). These observations suggest that the anaerobic activation of dcuB transcription is mediated by FNR-dependent and FNR-independent mechanisms. Aerobic dcuB expression was only twofold lower in the Δfnr strain (0.005 μmol/min/mg) than in the parental strain (0.009 μmol/min/mg), indicating that the FNR activation of dcuB expression is mainly an anaerobic process (Fig. 5A). Both the aerobic and anaerobic expression levels of dcuB were only ca. twofold lower in the arcA mutant (JRG3841), indicating that ArcA plays no more than a minor role in regulating dcuB expression in response to oxygen (Fig. 5B) and that ArcA is not responsible for the FNR-independent mechanism of anaerobic activation of dcuB transcription. The activation of dcuB expression was lowered ca. twofold by TMAO (from 2.5 to 1.4 μmol/min/mg Fig. 5Aii and Bii), although dcuA expression was unaffected by TMAO (Fig. 5Ai and Bi), suggesting that TMAO represses dcuB expression.
FIG. 5.
Effect of fnr (A), arcA (B), crp (C), and narL (D) on dcuA and/or dcuB expression. Growth was performed under both aerobic (open bars) and anaerobic (closed bars) conditions in M9 minimal medium containing either 0.4% glycerol, 50 mM TMAO, 50 mM fumarate, and 0.05% Casamino Acids (A and D); 0.4% glycerol and 50 mM fumarate (B and Ci); and 0.4% glucose with (+) or without (−) 50 mM fumarate (Cii). The strains used were JRG3834 (Ai and Bi), JRG3835 (Aii, Bii, and D), JRG3837 and JRG3846 (C), JRG3838 (Ai), JRG3839 (Aii), JRG3840 (Bi), JRG3841 (Bii), and JRG3842 (D). Other details are as described for Fig. 3. wt, wild type.
During anaerobic growth in glycerol and fumarate, the activity of the dcuB-lacZ fusion (0.75 μmol/min/mg) in the Δcrp strain, JRG3846, was threefold lower than in the parental strain, JRG3837 (2.3 μmol/min/mg) (Fig. 5Ci). This indicates that the cyclic AMP (cAMP)-CRP complex weakly activates dcuB expression in the absence of glucose. In the absence of TMAO and in the presence of fumarate, the anaerobic expression of dcuB was repressed 14-fold by glucose (Fig. 5Ci and Cii), and this repression was relieved only slightly (to ∼9-fold repression) by the crp mutation (Fig. 5Cii). These findings indicate that some other factor accounts for most of the 14-fold repression of dcuB by glucose.
The effect of NarL (and NarP) on the expression of the dcuB-lacZ fusion was measured in the presence and absence of nitrate (Fig. 5D). Although the narL mutation had no effect on dcuB expression in the absence of nitrate (Fig. 5Di), the strong nitrate repression of dcuB expression in the narL+ parental strain, JRG3835, was reduced 20-fold in the ΔnarL strain, JRG3842. This demonstrates that NarL has the major role in nitrate-induced repression of dcuB expression (Fig. 5Dii). However, even in the absence of NarL, nitrate still caused a fourfold repression in dcuB expression. The reason for this is unclear, but since nitrate repression of dcuB expression was unaffected by a narP null mutation (data not shown), it appears that NarP is not involved in mediating the nitrate-induced repression of dcuB.
The twofold anaerobic induction of the dcuA-lacZ fusion was unaffected by the fnr deletion (data not shown), revealing that FNR has no role in dcuA expression. The ΔarcA mutation caused a ca. twofold reduction in dcuA expression, both aerobically and anaerobically (data not shown), suggesting a role for ArcA in the constitutive expression of dcuA. It is uncertain whether the minor effects of ArcA on dcuA and dcuB expression reflect a metabolic consequence of deregulation of the arcA regulon or a direct interaction of the dcuA and dcuB genes with the ArcA protein. The fnr, crp, and narL mutations had no effect on dcuA expression (data not shown).
Northern blot analysis of the dcuA, dcuB, aspA, and fumB transcripts.
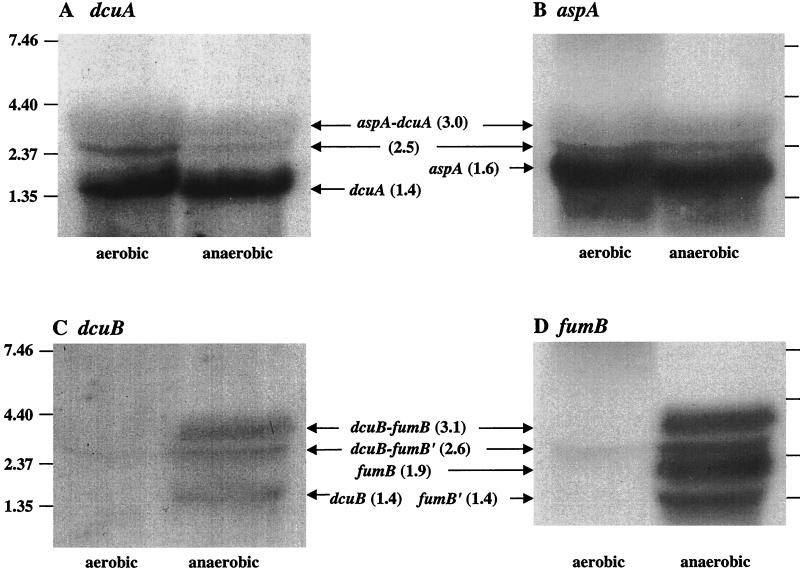
To determine whether the lacZ fusion analyses reflect transcript abundance and to examine the transcriptional organization of the aspA-dcuA and dcuB-fumB genes, Northern hybridization experiments were performed on the dcuA, dcuB, aspA, and fumB gene transcripts. Total RNA was extracted from MC4100 grown aerobically and anaerobically to mid-logarithmic phase in L broth containing glycerol plus fumarate. RNA was hybridized with labeled dcuA, dcuB, aspA, and fumB gene fragments (Fig. 1 and 6).
FIG. 6.
Northern hybridization analysis of the dcuA (A), aspA (B), dcuB (C), and fumB (D) transcripts. Total RNA was extracted from MC4100 grown aerobically and anaerobically in L broth containing 0.4% glycerol and 50 mM fumarate and, after electrophoresis and capillary transfer, was hybridized with labeled dcuA, aspA, dcuB, and fumB gene fragments (see Materials and Methods). The positions of RNA molecular weight standards (in kilonucleotides) are indicated. The arrows mark hybridizing transcripts (sizes in kilonucleotides) and a minor band (2.5 kilonucleotides) possibly corresponding to nonspecifically hybridizing 23s rRNA.
A major dcuA-hybridizing band, corresponding to a transcript of 1,400 nucleotides (nt), was detected under both aerobic and anaerobic conditions (Fig. 6A). Its size corresponds to that predicted for a dcuA monocistronic transcript initiating at the promoter defined in the aspA-dcuA intergenic region (bp 2060; see below) and terminating at an inverted repeat (bp 3440 to 3459) positioned just 10 bp downstream from the dcuA stop codon (bp 3430) (36). The transcript appears to be equally abundant in cells grown in aerobic and anaerobic conditions. The minor transcript of 3,000 nt corresponds in size to an aspA-dcuA cotranscript, whereas the minor 2,500-nt transcript could either be a degradation product of the aspA-dcuA cotranscript or a nonspecifically hybridizing species (such as 23s rRNA). The aspA hybridization revealed a major transcript of 1,600 nt, equally abundant in the aerobic and anaerobic samples (Fig. 6B). The size of this transcript matches that predicted for the aspA monocistronic transcript which initiates from the aspA promoter (bp 470; see below) and is presumed to terminate at an inverted repeat immediately downstream of aspA (bp 2063) (43). The minor 2,500- and 3,000-nt bands, which were detected by using the dcuA hybridization probe, were also detected with the aspA probe (Fig. 6A). This indicates that these transcripts correspond to cross-hybridizing aspA-dcuA cotranscripts, as suggested above. The dcuA hybridization results are consistent with the dcuA-lacZ fusion data and so confirm that dcuA is transcribed in both the presence and absence of oxygen. The aspA gene is, likewise, transcribed both aerobically and anaerobically. This supports a previous study showing that aspA is strongly expressed anaerobically and, in the absence of glucose, is also strongly expressed aerobically (44), but contrasts with the increased aspartase activity under anaerobiosis reported by Jerlström et al. (16). This difference may relate to posttranslational effects on aspartase activity or to differences in the growth conditions used. Despite the obvious potential for cotranscription, the dcuA and aspA genes seem to be predominantly transcribed independently, at least under the conditions tested, as indicated by the hybridization data and the expression of the dcuA-lacZ fusion.
Three dcuB-hybridizing transcripts of 1,400, 2,600, and 3,100 nt were observed at a ratio of approximately 1:1:2 in RNA isolated from anaerobic cells (Fig. 6C). No major transcripts were detected in the aerobic samples. The predominant 3,100-nt transcript is likely to correspond to a dcuB-fumB cotranscript. This would be expected to initiate at bp 151 (see below) and terminate at an inverted repeat immediately downstream of fumB (bp 3251 to 3270) (3, 36), giving an overall size of ∼3,100 nt. The 1,400-nt transcript is the expected size for a dcuB monocistronic transcript that initiates at bp 151 (see below) and presumably terminates at the stem-loop structure (bp 1520 to 1547) between the dcuB and fumB genes (36). The 2,600-nt transcript could be a dcuB-fumB degradation product lacking ∼500 nt at the 3′ end (see below). Four anaerobic transcripts were detected with the fumB hybridization probe, but as for dcuB, none were present in the aerobic sample (Fig. 6D). The four transcripts were 1,400, 1,900, 2,600, and 3,100 nt in length and were present at a ratio of approximately 2:4:1:2 (Fig. 6D). The 3,100-nt transcript is likely to be identical to the dcuB-fumB cotranscript detected with the dcuB probe (Fig. 6C). The predominant 1,900-nt RNA probably corresponds to a fumB transcript, which should be at least 1,700 nt if it initiates at a fumB promoter located in the dcuB-fumB intergenic region (bp 1511 to 1587) and terminates at the inverted repeat (bp 3251 to 3270) downstream of fumB (36). The 1,400- and 2,600-nt RNA species could be derived from the 1,900-nt fumB and 3,100-nt dcuB-fumB transcripts, respectively, through endonucleolytic cleavage at a site ∼500 nt from the 3′ ends of the corresponding transcripts (Fig. 1).
The dcuB Northern blotting results are consistent with the dcuB-lacZ expression data reported above and thus confirm that dcuB transcription is repressed by oxygen. Also, the absence of a fumB transcript in the aerobic sample is consistent with previous studies showing anaerobic induction of fumB-lacZ fusions of up to 5-fold and 3- to 10-fold reductions in anaerobic expression in the absence of FNR and/or ArcA (41, 44). The relative abundance of the dcuB and fumB transcripts is approximately 1:2, indicating that fumB is twofold more highly expressed than dcuB.
Determination of the transcriptional start sites for the dcuA, dcuB, and aspA genes.
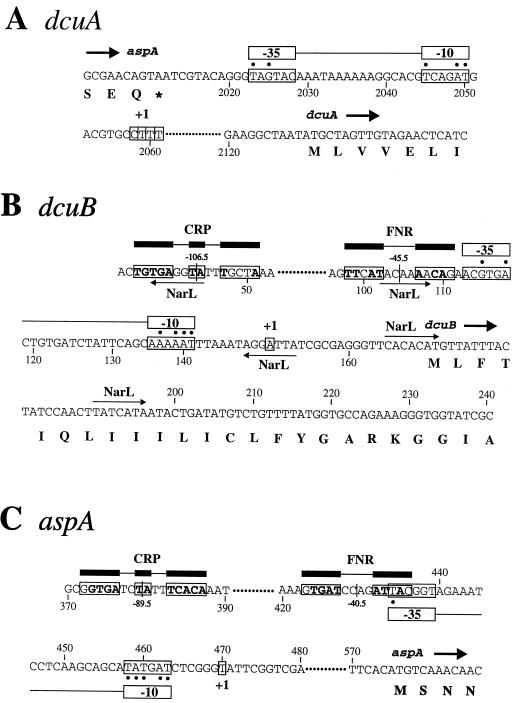
The transcriptional start sites of the dcuA, dcuB, and aspA genes were determined by primer extension analysis (Fig. 7). Four primer extension products were obtained for the dcuA transcript; they correspond to transcriptional start sites at C-2058, T-2059, T-2060, and T-2061 (Fig. 7A and 8A). These sites are approximately 30 bp upstream of the +1 site predicted by Six et al. (36). The relative abundances of the four cDNA species were 1:3:6:1, respectively, indicating that of the four alternative transcriptional start sites, T-2060 is preferred. The latter is 70 bp upstream of the dcuA initiation codon, and suitably positioned and well predicted −10 and −35 sequences are centered at bp 2047.5 and bp 2025.5, respectively. No potential FNR or CRP binding site sequences were detected directly upstream of the dcuA promoter, which is consistent with the expression studies.
FIG. 8.
Nucleotide sequence of the dcuA (A), dcuB (B), and aspA (C) promoter regions. Coordinates for the aspA gene are from the work of Woods et al. (43), and coordinates for the dcuA and dcuB gene are from the work of Six et al. (36). The experimentally determined +1 sites are boxed and labeled, as are the deduced −35 and −10 sites and predicted CRP and FNR sites. Putative NarL binding sites are indicated by horizontal arrows: rightward arrows indicate that the binding site is on the coding strand, while leftward arrows indicate that the binding site is on the noncoding strand. Residues matching the corresponding consensus sequence residues for the FNR and CRP sites are in boldface type. Closed circles indicate residues matching the corresponding consensus sequence residues for the −35 and −10 sites. The positions of the predicted FNR and CRP sites with respect to the +1 sites are indicated.
A single, moderately abundant cDNA product corresponding to an initiation site at A-151, 20 bp upstream of the dcuB initiation codon (Fig. 7B and 8B) and just 3 bp upstream from that predicted by Six et al. (36), was observed for the dcuB transcript. A well-predicted −10 site is appropriately positioned at bp 138.5, but the associated −35 site at bp 115.5 is very poor. A well-predicted FNR site and a moderately predicted CRP site are centered at bp 103.5 (−45.5) and bp 43.5 bp (−106.5), respectively (Fig. 8B). These sites were identified by virtue of the high or moderate probability values obtained in a score matrix-based search (see Materials and Methods) and correspond to those previously predicted (36). The predicted FNR site matches the consensus FNR binding site at 7 of the 10 conserved positions, and the predicted CRP site matches the CRP binding site consensus sequence at 9 of a possible 12 positions. FNR sites are normally found just upstream of the −35 site, centered at around −41.5 (11, 20). The FNR site of the FNR-activated fumarate reductase (Frd) operon (frdABCD) is also centered at −45.5 (or −46.5), indicating that the position of the predicted dcuB-FNR site is appropriate to allow FNR-dependent transcriptional activation at the dcuB promoter (11). FNR-dependent promoters often possess weak −35 sites and good −10 sites, and in such cases the FNR site is compensatory in allowing FNR-dependent expression of otherwise weakly transcribed genes. This model for FNR induction is likely to apply to dcuB also. CRP is known to activate transcription by binding to sites located at various positions upstream of the −35 site (from −41.5 to −103) (11). The location of the CRP site at −106.5 is therefore appropriate to permit CRP-dependent activation. Several genes are known to be subject to dual FNR- and CRP-dependent activation (11). These include the ansB (asparaginase II) gene which contains FNR and CRP sites (at −41.5 and −91.5, respectively) with an organization resembling that of dcuB (15). Thus, like the ansB promoter, the dcuB promoter can be classified as class III (32).
Five sequences with similarity to the NarL or NarP binding site consensus sequence (7) were identified by using the score matrix approach. These heptameric sequences are centered at −109, −45, +1, +19, and +44 and are suitably positioned to interfere with the binding of FNR, CRP, and RNA polymerase and/or transcript extension. NarL or NarP sites are normally located, at multiple positions, upstream of the −35 site (7). The NarP heptameric binding sites possess a 7-2-7 organization, whereas NarL can recognize heptamers in various arrangements (7). The organization of the putative NarL sites in the dcuB operator-promoter region clearly does not conform to the preferred 7-2-7 arrangement, which is consistent with the observation that NarL, but not NarP, represses dcuB expression.
A single, relatively abundant cDNA product was detected in the primer extension analysis of the aspA transcript (Fig. 7C). The size of this product indicates a transcriptional initiation site at T-470, 105 bp upstream of the aspA initiation codon (Fig. 8C). A good −10 site (bp 461.5) and a weak −35 site (bp 436.5) are correctly positioned upstream of the aspA +1 site (Fig. 8C). A weakly predicted FNR site and a strongly predicted CRP binding site are centered at −40.5 and −89.5, respectively. Since aspA expression is reported to be induced 10- to 15-fold anaerobically by FNR (16, 44), a good FNR site would be expected. However, the Northern blot analysis showed no anaerobic induction of aspA expression under the growth conditions used here, and this is consistent with a weak FNR site. The strong CRP binding site is correctly positioned to function in aspA transcriptional activation by the cAMP-CRP complex. Such an interaction could compensate for the weak −35 site and could also account for the fivefold repression of aspA by glucose (44).
No primer extension products were observed in the analysis of the fumB transcript. The reason for this is uncertain. The Northern blot analysis indicated a ∼1,900-nt fumB transcript. If fumB transcription terminates, as expected, at the stem-loop structure at bp 3250, then this would place the transcriptional initiation site ∼250 bp upstream of the fumB initiation codon, well within the dcuB structural gene. This is far further upstream than anticipated (36) and might explain the failure to detect a primer extension product for the fumB transcript. Previous analyses of fumB expression utilizing fumB′-lacZ fusions suggest that the fumB gene possesses an independent promoter and associated operator region located within the 800-bp region directly upstream of the fumB initiation codon (41, 44).
DISCUSSION
This study shows that the dcuA and dcuB genes of E. coli are differentially regulated. The dcuB gene is expressed exclusively under anaerobic conditions in a manner that is largely FNR dependent, is repressed by nitrate through a mechanism that is mostly NarL mediated, and is strongly induced by C4-dicarboxylates anaerobically. Expression of dcuB is also repressed by glucose, is slightly repressed by TMAO, and in the absence of glucose is threefold induced by CRP. In agreement with the expression data, the dcuB promoter region contains well-predicted binding sites for FNR, NarL, and CRP (Fig. 8). ArcA has no significant role in dcuB regulation, although it appears that a factor other than FNR also contributes to anaerobic induction of dcuB. Furthermore, in the absence of NarL, dcuB is repressed fourfold by nitrate in a manner that appears to be NarP independent, and, in addition, dcuB expression is reduced by glucose in a fashion that is mostly CRP independent. Thus, factors other than FNR, NarL, and CRP also appear to contribute to the regulation of dcuB in response to oxygen, nitrate, and glucose. In contrast to the strong regulation of dcuB, expression of the dcuA gene is virtually unaffected by the environmental and regulatory factors tested. The dcuB gene is ca. fourfold more strongly expressed than dcuA during fumarate respiration (Fig. 3A, 5Bii, and 5Ci). This supports the results of transport experiments with dcu mutants showing that DcuB is the dominant Dcu carrier during fumarate respiration (45). However, the dcuA gene is more strongly expressed than dcuB under most of the other growth conditions examined, and this is concordant with the better codon usage of dcuA relative to that of dcuB (36).
The expression and transport data are consistent with each other insofar as Dcu transport activity and the combined dcuA- and dcuB-lacZ activities are activated anaerobically by FNR, repressed anaerobically in the presence of nitrate by NarL, increased by fumarate or succinate, and weakly affected by ArcA (8, 9). The factors affecting Dcu transport activity described above clearly reflect those influencing dcuB expression. This is because dcuB expression, but not dcuA expression, is strongly modulated by these factors and also because dcuB expression exceeds that of dcuA during fumarate respiration. The slight repression effect (greater than twofold) of TMAO on dcuB expression is consistent with the lack of effect of TMAO on Dcu transport activity. It is possible that the TMAO effect on dcuB expression is mediated by the TMAO-responsive two-component sensor-regulator system, TorS-TorR (18, 34), although this has not been tested.
Surprisingly, the correspondence between transport and expression activities is not maintained under all conditions examined. Previous studies showed that Dcu activity is only slightly reduced by glucose and is unaffected by cAMP or a cya mutation, suggesting that CRP does not regulate Dcu synthesis (8, 9, 45). However, clear glucose and CRP effects on dcuB expression were observed in the studies reported here, although the CRP effect was relatively weak. The reason for these discrepancies is unclear, but they could reflect posttranscriptional effects or a lack of specificity when transport activity mediated by at least three alternative systems is measured.
The 70-fold induction of dcuB expression by C4-dicarboxylates (Fig. 4) is likely to be mediated by an undefined C4-dicarboxylate-dependent regulator able to sense and respond to exogenous C4-dicarboxylates. A good candidate for such a regulator is a putative two-component regulatory system composed of a response regulator and a membrane-associated histidine kinase sensor encoded by the yjdHG genes located just 570 bp upstream of dcuB. The corresponding proteins have strong sequence similarity to the Klebsiella CitB and CitA proteins involved in the regulation of anaerobic citrate metabolism (5, 25). The possible involvement of these genes in C4-dicarboxylate-responsive gene regulation is being investigated. Other E. coli genes are also known to be induced, albeit weakly, by C4-dicarboxylates. These are the frdABCD and nuo operons which are 1.5-fold induced by fumarate and ∼2.5-fold induced by fumarate or succinate, respectively (4, 17). The mechanism governing this regulation is unknown.
The pattern of dcuA and dcuB expression provides important clues for the likely physiological functions of the homologous and functionally related DcuA and DcuB proteins. The profile for dcuB expression is consistent with a role for DcuB in the provision of substrate and export of product for the reaction catalyzed by Frd during fumarate respiration. Indeed, the expression profile of dcuB resembles that of the frdABCD operon. Appropriately, both are anaerobically induced by FNR and repressed by NarL in response to nitrate (14, 17), thus ensuring that oxygen and nitrate are utilized in preference to fumarate as terminal electron acceptors. The fumB gene, which is adjacent to dcuB, encodes the enzyme fumarase B, which acts as a malate dehydratase in the conversion of malate to fumarate. Fumarase B thus provides substrate for Frd during anaerobic fumarate respiration. Therefore, DcuB and fumarase B are both involved in feeding substrate to Frd and constitute consecutive steps in the anaerobic transport and metabolism of malate. The dcuB and fumB genes would therefore be expected to be expressed in a coordinated fashion, as is suggested by the Northern blotting analysis showing strong anaerobic induction and partial cotranscription (Fig. 6).
The expression of dcuA under both aerobic and anaerobic conditions is inconsistent with the previously proposed, anaerobic function for DcuA (36, 45) and suggests that DcuA has an aerobic function in addition to contributing to anaerobic C4-dicarboxylate uptake. However, transport studies failed to demonstrate any Dcu activity, attributable to DcuA, under aerobic conditions, possibly because the Dcu systems are inactivated by oxygen (8, 9). Therefore, it is possible that the aerobic expression of dcuA produces an inactive DcuA protein. However, inactivation of Dcu activity by transient exposure to oxygen (or other oxidants) can be reversed by subsequent treatment with reducing agents (8). This offers the possibility that constitutive expression of dcuA allows E. coli to respond rapidly to transitions from aerobic to anaerobic conditions through the activation of presynthesized DcuA.
Aspartase, the product of the aspA gene located upstream of dcuA, converts l-aspartate to fumarate. Together with the constitutive aspartate aminotransferase (encoded by aspC), it is thought to provide an alternative mechanism for converting oxaloacetate to fumarate as part of the reductive branch of the noncyclic form of the citric acid cycle (6). A previous lacZ fusion analysis (44) and the Northern blot analysis reported above suggest that, like aspC and dcuA, the aspA gene is well expressed under both aerobic and anaerobic conditions, indicating that DcuA and aspartase have aerobic as well as anaerobic functions. However, it should be stressed that this is inconsistent with aspartase activity measurements that show that aspartase is anaerobically induced (16). Aspartase may also have a role, together with the anaerobically induced periplasmic asparaginase II, in the utilization of exogenous asparagine (24) and in the degradation of aspartate for use as a carbon source (29). Furthermore, aspartase is required for regenerating oxaloacetate in the aerobic and anaerobic utilization of glutamate (38). Whether the DcuA protein assists in any of these processes is uncertain, but the colocations of the aspA and dcuA genes and the related substrate specificities of their products are certainly suggestive of linked functions.
ACKNOWLEDGMENTS
We thank the BBSRC for a project grant (S.C.A. and J.R.G.) and for an Advanced Fellowship (S.C.A.).
ADDENDUM IN PROOF
A recent publication (E. Zientz, J. Bongaerts, and G. Unden, J. Bacteriol. 180:5421–5425, 1998) as well as our own unpublished results show that the yjdHG (dcuSR) genes do indeed encode a sensor-regulator system responsible for the C4-dicarboxylate-dependent regulation of dcuB (and other genes).
REFERENCES
- 1.Aiba H, Adkya S, de Crombrugghe B. Evidence for a functional gal promoter in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Baker K E, Ditullio K P, Neuhard J, Kelln R A. Utilization of orotate as a pyrimidine source by Salmonella typhimurium and Escherichia coli requires the dicarboxylate transport protein encoded by dctA. J Bacteriol. 1996;178:7099–7105. doi: 10.1128/jb.178.24.7099-7105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell J B, Andrews S A, Sivak M N, Guest J R. Nucleotide sequence of the FNR-regulated fumarase gene (fumB) of Escherichia coli K-12. J Bacteriol. 1989;171:3494–3503. doi: 10.1128/jb.171.6.3494-3503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;6:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 5.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 6.Cronan J E, Jr, LaPorte D. Tricarboxylic acid cycle and glyoxylate bypass. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecther M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 206–216. [Google Scholar]
- 7.Darwin A J, Tyson K L, Busby S J W, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 8.Engel P, Kramer R, Unden G. Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from aerobic dicarboxylate uptake. J Bacteriol. 1992;174:5533–5539. doi: 10.1128/jb.174.17.5533-5539.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel P, Kramer R, Unden G. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli: energetics and mechanism of uptake and efflux. Eur J Biochem. 1994;222:605–614. doi: 10.1111/j.1432-1033.1994.tb18903.x. [DOI] [PubMed] [Google Scholar]
- 10.Golby P, Kelly D J, Guest J R, Andrews S. Topological analysis of DcuA, an anaerobic C4-dicarboxylate transporter of Escherichia coli. J Bacteriol. 1998;180:4821–4827. doi: 10.1128/jb.180.18.4821-4827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guest J R, Green J, Irvine A S, Spiro S. The FNR modulon and FNR-regulated gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. Washington, D.C: Landes Co. and Chapman & Hall; 1996. pp. 317–342. [Google Scholar]
- 12.Guest J R, Miles J S, Roberts R E, Woods S A. The fumarase genes of Escherichia coli: location of the fumB gene and discovery of a new gene (fumC) J Gen Microbiol. 1985;131:2971–2984. doi: 10.1099/00221287-131-11-2971. [DOI] [PubMed] [Google Scholar]
- 13.Guest J R, Roberts R E, Wilde R J. Cloning of the aspartase gene (aspA) of Escherichia coli. J Gen Microbiol. 1984;130:1271–1278. doi: 10.1099/00221287-130-5-1271. [DOI] [PubMed] [Google Scholar]
- 14.Iuchi S, Lin E C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase TMAO operons in Escherichia coli. Proc Natl Acad Science USA. 1987;84:3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings M P, Beacham I R. Co-dependent positive regulation of the ansB promoter of Escherichia coli by CRP and the FNR protein—a molecular analysis. Mol Microbiol. 1993;1:155–164. doi: 10.1111/j.1365-2958.1993.tb01677.x. [DOI] [PubMed] [Google Scholar]
- 16.Jerlström P G, Liu J, Beacham I R. Regulation of Escherichia colil-asparaginase II and l-aspartase by the fnr gene-product. FEMS Microbiol Lett. 1987;41:127–130. [Google Scholar]
- 17.Jones H M, Gunsalus R P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J Bacteriol. 1987;169:3340–3349. doi: 10.1128/jb.169.7.3340-3349.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jourlin C, Bengrine A, Chippaux M, Mejean V. An unorthodox sensor protein (TorS) mediates the induction of the Tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol Microbiol. 1996;20:1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 19.Kay W W, Kornberg H L. The uptake of C4 dicarboxylic acids by Escherichia coli. Eur J Biochem. 1971;18:274–281. doi: 10.1111/j.1432-1033.1971.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 20.Kolb A, Busby S J W. The Cap Modulon. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. Washington, D.C: Landes Co. and Chapman & Hall; 1996. pp. 255–280. [Google Scholar]
- 21.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecther M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 307–342. [Google Scholar]
- 22.Lo T C Y. The molecular mechanism of dicarboxylic acid transport in Escherichia coli K12. J Supramol Struct. 1977;7:463–480. doi: 10.1002/jss.400070316. [DOI] [PubMed] [Google Scholar]
- 23.Lo T C Y, Rayman K, Sanwal B D. Transport of succinate in Escherichia coli K12. J Biol Chem. 1972;247:6323–6331. [PubMed] [Google Scholar]
- 24.McFall E, Newman E B. Amino acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 358–379. [Google Scholar]
- 25.Meyer M, Dimroth P, Bott M. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A+T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J Mol Biol. 1997;269:719–731. doi: 10.1006/jmbi.1997.1076. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for E. coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 27.Phillips-Jones M K, Watson F J, Martin R. The 3′ codon context effect on UAG suppressor tRNA is different in Escherichia coli and human cells. J Mol Biol. 1993;233:1–6. doi: 10.1006/jmbi.1993.1479. [DOI] [PubMed] [Google Scholar]
- 28.Quail M A, Haydon D J, Guest J R. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol Microbiol. 1994;12:95–104. doi: 10.1111/j.1365-2958.1994.tb00998.x. [DOI] [PubMed] [Google Scholar]
- 29.Reizer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sawers G, Suppmann B. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by ArcA and FNR proteins. J Bacteriol. 1992;174:3474–3478. doi: 10.1128/jb.174.11.3474-3478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott S, Busby S, Beacham I. Transcriptional co-activation at the ansB promoters: involvement of the activating regions of CRP and FNR when bound in tandem. Mol Microbiol. 1995;18:521–531. doi: 10.1111/j.1365-2958.1995.mmi_18030521.x. [DOI] [PubMed] [Google Scholar]
- 33.Silhavy T J, Barman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 34.Simon G, Mejean V, Jourlin C, Chippaux M, Pascal M C. The TorR gene of Escherichia coli encodes a response regulator protein involved in the expression of trimethylamine N-oxide reductase genes. J Bacteriol. 1994;176:5601–5606. doi: 10.1128/jb.176.18.5601-5606.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 36.Six S, Andrews S C, Unden G, Guest J R. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct) J Bacteriol. 1994;176:6470–6478. doi: 10.1128/jb.176.21.6470-6478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sofia H J, Burland V, Daniels D L, Plunkett III G, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spencer M E, Lebeter V M, Guest J R. Location of the aspartase gene (aspA) on the linkage map of Escherichia coli K12. J Gen Microbiol. 1976;97:73–82. doi: 10.1099/00221287-97-1-73. [DOI] [PubMed] [Google Scholar]
- 39.Spiro S, Guest J R. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol Microbiol. 1988;2:701–707. doi: 10.1111/j.1365-2958.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 40.Staden R. The Staden sequence-analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 41.Tseng C-P. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol Lett. 1997;157:67–72. doi: 10.1111/j.1574-6968.1997.tb12754.x. [DOI] [PubMed] [Google Scholar]
- 42.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 43.Woods S A, Miles J S, Roberts R E, Guest J R. Structural and functional relationships between fumarase and aspartase. Nucleotide sequence of the fumarase (fumC) and aspartase (aspA) genes of Escherichia coli K12. Biochem J. 1986;237:547–557. doi: 10.1042/bj2370547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woods S A, Guest J R. Differential roles of the Escherichia coli fumarases and fnr-dependent expression of fumarase B and aspartase. FEMS Microbiol Lett. 1987;48:219–224. [Google Scholar]
- 45.Zientz E, Six S, Unden G. Identification of a third secondary carrier (DcuC) for anaerobic C4-dicarboxylate transport in Escherichia coli: roles of the three Dcu carriers in uptake and exchange. J Bacteriol. 1996;178:7241–7247. doi: 10.1128/jb.178.24.7241-7247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]