Abstract
Oral cancer became a very common condition. WHO estimates that there are 4 cases of lip and oral cavity cancer for every 100,000 people worldwide. The early diagnosis of cancers is currently a top focus in the health sector. Recent systematic reviews and meta-analyses have identified promising biomarkers for early detection in several original research investigations. However, it is still unclear the quality of these evidence and which biomarker performs the best in terms of early detection. Therefore, the objective was, to map the methodological and reporting quality of available oral squamous cell carcinoma (OSCC) or head/neck squamous cell carcinoma (HNSCC) systematic reviews and meta-analysis. Secondly, to evaluate diagnostic accuracy of salivary biomarkers for common craniofacial cancers and to compare the diagnostic value of different salivary biomarkers.
PubMed, Scopus, Web of Science, Embase and Cochrane Library electronic databases were used to map the methodological and reporting quality of the systematic reviews and meta-analysis conducted on the HNSCC, OSCC using the AMSTAR-2 checklist. The inclusion criteria were systematic reviews and meta-analysis published in the topic of HNSCC and OSCC biomarkers. Exclusion criteria were no animal studies; original primary studies, due to limitation of competency in other languages articles with language other than English were excluded. The sensitivity and specificity were calculated for salivary biomarkers and ranked according to network meta-analysis principles.
A total of N = 5893 patients were included from four meta-analysis studies. All together, these included n = 37 primary studies. n = 94 biomarkers were pooled from these four meta-analyses and categorised into the stages at which they were detected (I-IV). In OSCC, Chemerin and MMP-9 displayed the highest sensitivity, registering 0.94 (95% CI 0.78, 1.00) and a balanced accuracy of 0.93. Phytosphingosine closely followed, with a sensitivity of 0.91 (95% CI 0.68, 0.99) and a balanced accuracy of 0.87.
For HNSCC, the top three biomarkers are Actin, IL-1β Singleplex, and IL-8 ELISA. Actin leads with a sensitivity of 0.91 (95% CI 0.68–0.99), a specificity of 0.67, and an overall accuracy of 0.79. Subsequently, IL-1β Singleplex exhibits a sensitivity of 0.62 (95% CI 0.30–0.88), a specificity of 0.89, and an accuracy of 0.75, followed by IL-8 ELISA with a sensitivity of 0.81 (95% CI 0.54–0.97), a specificity of 0.59, and an accuracy of 0.70.
In conclusion, there was highest sensitivity for MMP-9 and chemerin salivary biomarkers. There is need of further more studies to identify biomarkers for HNSCC and OSCC.
Keywords: Oral neoplasm, Oral cancer, Cancers, Mouth, Marker, Biological, Biomarker, Head and neck squamous cell carcinoma, HNSCC, OSCC, Network meta-analysis, NMA, Saliva
Highlights
-
•
Methodological and reporting quality of systematic reviews and meta-analysis was more than 50% based on AMSTAR-2 assessment.
-
•
UPLC-MS (31.79%) was the most used method to detect the salivary biomarkers followed by RT-qPCR (11.26%).
-
•
Chemerin & MMP-9 show top sensitivity in OSCC followed by Phytosphingosine.
-
•
Top 3 biomarkers for HNSCC are Actin, IL-1β Singleplex and IL-8 ELISA.
-
•
Top biomarkers in early OSCC I-II: chemerin, MMP-9, Phytosphingosine, Pipecolinic acid.
1. Introduction
Oral cancer is become a well-known malignant neoplasm with a high incidence globally [1], [2], [3] with an estimated 1401,931 cases worldwide in 2019 [4]. In 21st century, accurate and effective healthcare strategy is a need improve quality of life of HNSCC and OSCC patients [5], [6], [7], [8]. The TNM approach, which is staged based on radiological and pathological report, is the most popular methodology for lip and mouth cancer grading and severity and helps in identifying the prognosis of specific intervention. [9], [10], [11], [12], [13]. Early detection, staging of malignancies and prompt diagnosis could present a useful approach for a drastic reduction in mortality rate and invasive surgeries [14], [15], [16].
For such objective to be met, there is need of simple methodological approach with state-of-art investigations that orient in early detection of HNSCC and OSCC. Currently, this is widely practiced using body fluids such as blood, urine and saliva with increased sensitivity and specificity [17], [18], [19], [20], [21], [22]. Blood and urine have demonstrated significant potential as a source of detecting early malignancies, but these fluids have challenges in real life operatory settings [23].
Human saliva and gingival fluids would bridge the gap and accelerate the process of diagnosis with greater accuracy and bridging the gap that occur by using blood and urine as a marker. Saliva contain complex natural reservoir of enzymes and amylases, cytokines, hormones, immune-modulators, immunoglobulins, ions and glycoproteins [24], [25]. Therefore, new research reports being published widely using saliva biomarkers in detecting malignancies but there are uncertainties in the evidence as which saliva biomarker is best expressed in HNSCC and OSCC. Also, the quality of systematic reviews that being published that aimed to screen the saliva biomarkers from clinical studies. Hence, determining the landscape, quality of evidence and best salivary biomarkers in early detection of HNSCC and OSCC is paramount.
The aim of the current review was;.
-
1.
To map the methodological and reporting quality of available systematic reviews and meta-analysis.
-
2.
To evaluate diagnostic accuracy of biomarkers for common craniofacial cancers, oral squamous cell carcinoma (OSCC) or head/neck squamous cell carcinoma (HNSCC) by re-analysing the results of meta-analysis.
-
3.
To compare the diagnostic value of different biomarkers for various oral squamous cell carcinoma (OSCC) or head/neck squamous cell carcinoma (HNSCC) with network meta-analysis (NMA).
2. Material and methods
2.1. Design and registration
The aim of this review was to conduct methodological and reporting quality assessment, re-assess the meta-analysis results and rank the biomarkers based on their diagnostic outcomes in oral, head/neck cancer using previously available meta-analysis. The present research has been registered on the open science framework platform (OSF) and follows the Preferred Reporting Items for Systematic Reviews and Meta-analysis and the Network Meta-analysis statements.
2.2. Information sources
Data search was performed using the electronic databases using PubMed, Scopus, Web of Science, Embase and Cochrane Library till date. The scientific papers related to meta-analysis with no limitations on year of publication or the language of the manuscript was searched. The references of relevant systematic reviews were searched to identify additional potential studies. The searching terms used were: “Biomarker”, “Biomarkers”, “Diagnostics”, “Saliva”, Oral cancer, oral squamous cell carcinoma, head and neck squamous cell carcinoma, OSCC, HNSCC, “Meta-analysis”, “Systematic Review”. The search strategy includes (((biomarker)) OR (biomarkers)) OR (((saliva)) AND (((oral squamous cell carcinoma)) OR (OSCC)) OR (HNSCC)).
2.3. Types of studies
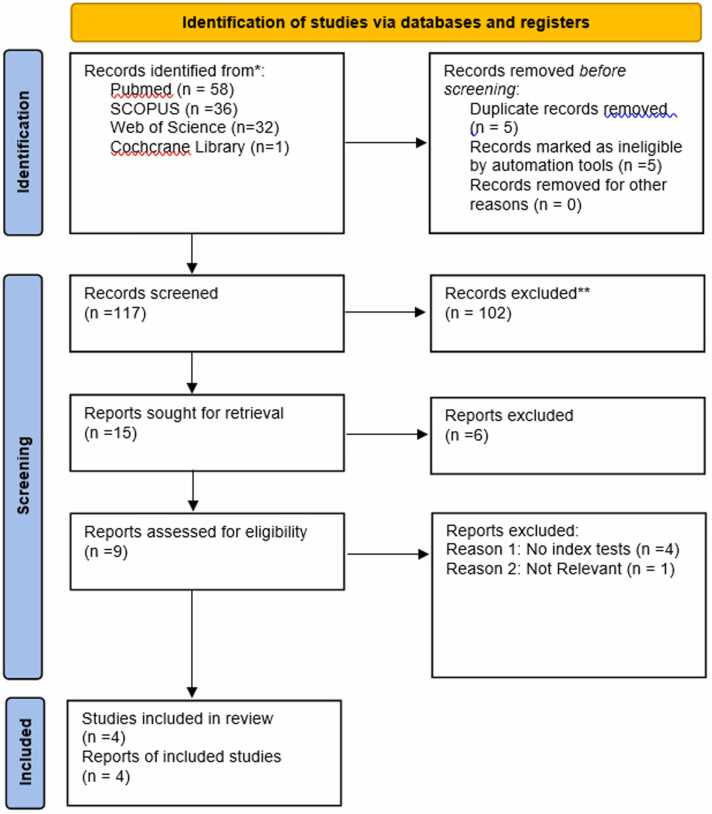
The selection of articles is demonstrated in the PRISMA flow chart (Fig. 1). We included meta-analysis reviews that considered randomized controlled trials, cross-sectional studies, case-control studies, and cohort studies in human subjects. There was no limitation on minimal quality, minimal sample size, or the number of patients. Exclusion criteria were narrative reviews, letters, personal opinions, book chapters, case reports, conference abstracts and meetings, duplicate publications and experimental in vitro and in vivo animal studies.
Fig. 1.
PRISMA Flow Chart for the selection of articles.
Then, full articles in which salivary biomarkers were used as potential diagnostic markers for OSCC or HNSCC was independently evaluated. The resulting manuscripts were initially screened by title and abstract, followed by a full-text analysis. Duplicates were checked manually and removed by two of the authors (SK and JY). Disagreements between the two authors have been discussed until a consensus has been reached and also communicated with third reviewer (MDF).
2.4. The selection was based on PICO criteria
2.4.1. Participants
Both healthy subjects and adults with suspected OSCC and HNSCC based on clinical symptoms and oral examination have been included. All participants received one or several index tests. There were no limitations for age or ethnicity.
2.4.2. Intervention(s), exposure(s)
Biomarkers include some common salivary biomarkers for tumor-specific biomarkers, and/or any type of biomarker is used to diagnose OSCC and HNSCC. The index test can be one biomarker, or one biomarker combines with other biomarkers. Blood and urine biomarkers were excluded as it was not the objective of this review.
2.4.3. Comparator(s)/control
Any type of biomarker is used to diagnose OSCC or HNSCC.
The reference standard included was placebo, control or other salivary biomarker of interest or standard of care with or without histological confirmation.
2.4.4. Context
Reviews excluded from the review that are (1) diagnostic tests of imaging modalities; (2) systematic reviews without meta-analysis; (3) review protocols and methodological articles.
2.4.5. Main outcome(s)
The primary outcomes were sensitivity (SEN), specificity (SPE), positive predictive value (PPV), negative predictive value (NPV), true positives (TP), true negative (TN), false positive (FP), false negative (FN), area under the curve (AUC), diagnostic odds ratio (DOR) and their respective 95% confidence intervals. The second outcomes were relative diagnostic estimates of different biomarkers.
2.4.6. Additional outcome(s)
Methodological quality and reporting quality. The relative diagnostic estimates of different biomarkers such as sensitivity, specificity, predictive positive value (PPV), negative predictive value (NPV), and diagnostic odds ratio (DOR).
2.4.7. Data extraction
Absolute sensitivity and specificity, Relative sensitivity and specificity, and relative DOR between different biomarkers have been calculated using STATA (17.0; Stata Corporation). Then, the relative diagnostic indices have been used to make the indirect comparison. When data were allowed, we conducted a network meta-analysis. NMA ranking has been done using superiority index (SI).
2.5. Data from meta-analysis reviews
For analysis purposes, following data variables have been extracted from studies included to facilitate rapid extraction of data: study id, author name, year, country, sponsor, gender (Control and Test), Age (Control and Test), Total sample size of the study (N), control sample size, test sample size, groups (test and control), pathological disease, diagnosis, diagnostic criteria, cut-off value, true positive (TP), true negative (TN), false positive (FP), false negative (FN), sensitivity, specificity, area under the curve value, diagnostic odds ratio (DOR), duration of months, anatomic tumour location, ethnicity, study design, Type of biomarker (Proteomic, Metabolomics, epigenomic, microbiomic, transcriptomic and genomic), fraction of saliva (Whole saliva, supernatant, pellet), unstimulated saliva or stimulated saliva, candidate biomarker selected for analysis, Index test (2 ×2 table contingency table), method of saliva collection, odds ratio, Source of MiRNA, Cancer spectrum, Type of Cancer, MiRNA profiling, conclusions and challenges were recorded.
2.6. Risk of bias (ROB)
The methodological quality of included systematic reviews was assessed using AMASAR-2 checklist, and the reporting quality was assessed using PRISMA-DTA checklist. Two review authors independently assessed the risk of bias in each study according to predefined criteria. Disagreements regarding by-item and overall rating of quality was resolved by consensus or third-party adjudication if consensus cannot be reached.
2.7. Strategy for data synthesis
The data for each OSCC and HNSCC was analysed separately, and these include following:
2.8. Evidence map
2.8.1. Map the biomarkers
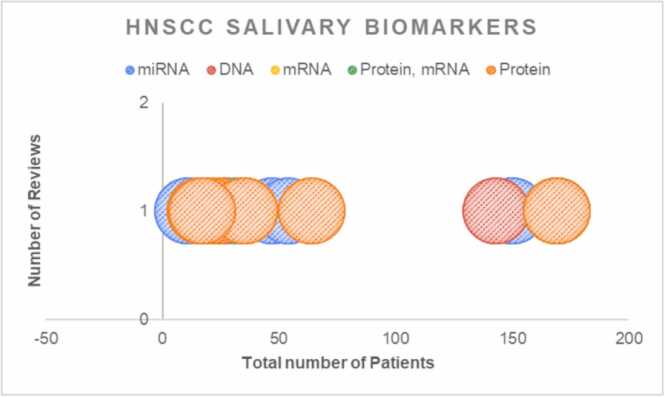
We created a bubble plot according to the biomarkers for all included meta-analysis. This map displays information in three dimensions (a) the bubble size represents the number of reviews, (b) the total number of participants included in the meta-analysis in the x-axis (c) the biomarkers in the y-axis. Mapping the quality. The bubble plot was created according to the methodological and reporting quality, where each bubble represents one meta-analysis. The information of three dimensions in the map is (a) the bubble size represents the number of primary studies included in the meta-analysis, (b) the methodological quality in the x-axis (c) the reporting quality in the y-axis.
2.9. Pairwise meta-analysis
We performed pairwise meta-analysis with the data of pooled sensitivity (SEN), specificity (SPE), diagnostic odds ratio (DOR), positive likelihood ratio (PLR), negative likelihood ratio (NLR) and their 95%CI lower limit, 95%CI upper limit using bivariate mixed-effects regression modelling with STATA V17.0. The heterogeneity between each study was estimated using the Q value and the inconsistency index (I² test). If the I² is≤ 50%, it suggests that there is negligible statistical heterogeneity, and the fixed effects model was employed. If the I² is > 50%, was explored sources of heterogeneity by subgroup analysis and meta-regression. If there is no clinical heterogeneity, the random effects model was used to perform the meta-analysis. Otherwise, clinical heterogeneity was explored through discussion with the review team.
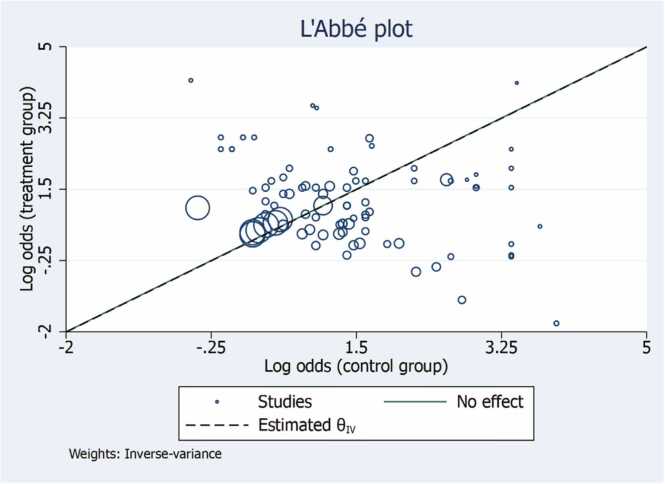
The L’Abbe plot explores between-study heterogeneity by comparing group-level summary outcome measures across studies. It can also be used to determine which type of effect size is more homogeneous across studies. Compared with other meta-analysis graphs, one important advantage of the L’Abbe´ plot is that it displays the data on individual studies for each of the two groups. Thus, in addition to identifying outlying studies, it can also identify the outlying groups within studies (Fig. 4).
Fig. 4.
L’Abbe Plot for treatment and control group assessing heterogeneity and comparing study specific event rates in the two groups. The circles with their sizes (areas) are proportional to study precision. The plot also contains a reference (diagonal) line, which indicates identical outcomes in the two groups and thus represents no effect, and the estimated overall effect-size.
2.10. Adjusted indirect comparisons
We calculated relative diagnostic outcomes between index tests including relative sensitivity, relative specificity, and relative DOR. Then, we conducted indirect comparisons using relative diagnostic outcomes. All analysis was performed using R version 4.1.3 software.
2.11. Analysis of subgroups or subsets
If sufficient data are available, we performed subgroup analysis based on the TNM staging, type of cancer and salivary biomarkers.
3. Results
A total of N = 4 meta-analysis reviews (Kang JW 2021; Guerra EN 2015; Liu D 2021; Hema Shree K 2019) [25], [26], [27], [28] worked on salivary biomarkers. In total, the meta-analysis reviews included n = 37 original studies with average mean age 56.77 ± 4.01. A total of n = 5893 patients were considered for analysis in this review (Fig. 6). In terms of patient data considered for NMA analysis, there were 2025 patients in the OSCC group and 3868 patients in the HNSCC group, resulting in a patient ratio of 0.53. There were three reviews on miRNA, two reviews on mRNA and protein combination, one review on metabolites (Fig. 6). The review considered oral squamous cell carcinoma (OSCC) and Head and neck carcinoma (HNSCC). Altogether there were n = 94 UPLC-MS (31.79%) was the most used method to detect the salivary biomarkers in the reviews selected followed by RT-qPCR (11.26%). Least percentage were with Gene expression array, multiplex assay, LC-MS;MS, ELISA, Immuno-blotting, MALDI-MS, Western blotting and MLPA (Supplementary file).
Fig. 6.
Bubble plot for OSCC salivary biomarker illustrating the mapping of reviews, number of patients and different class of biomarkers.
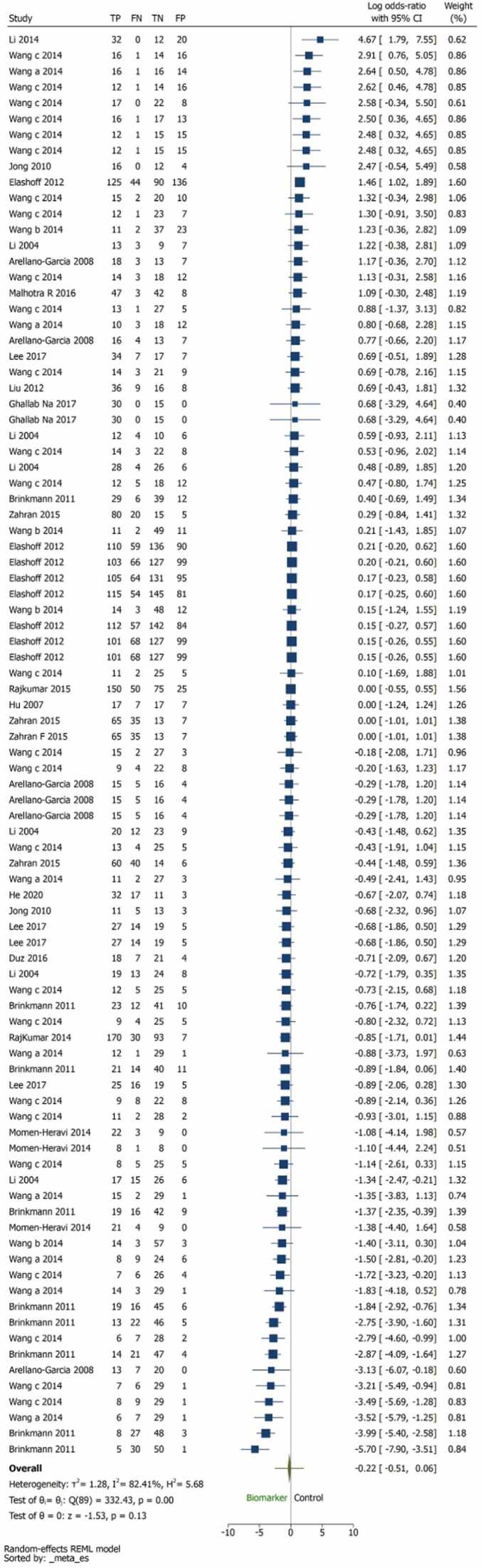
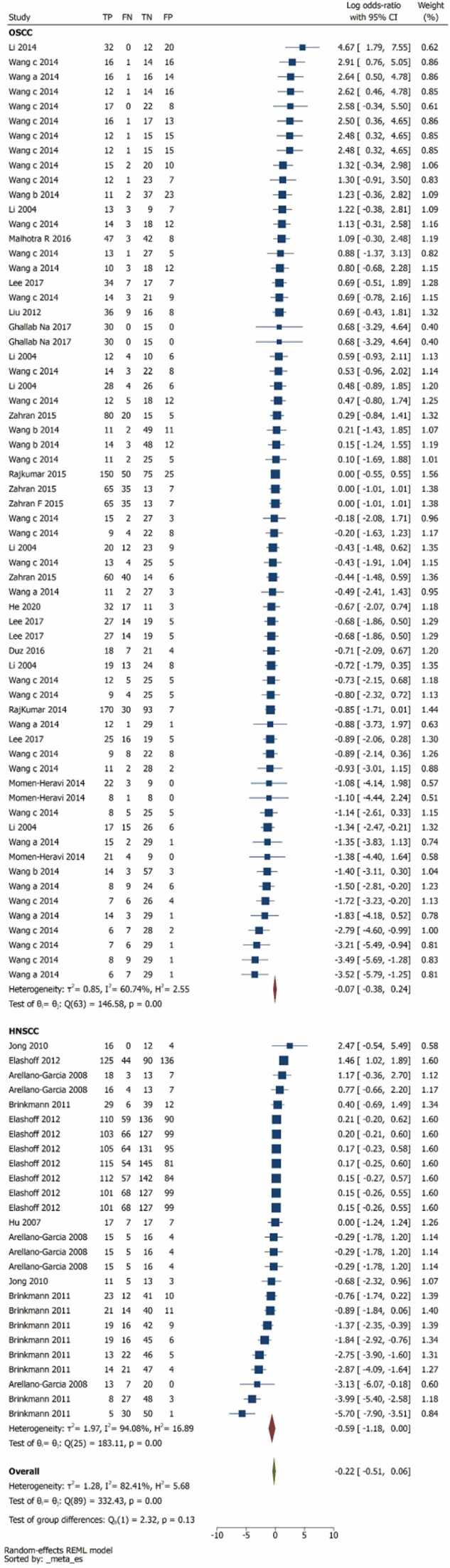
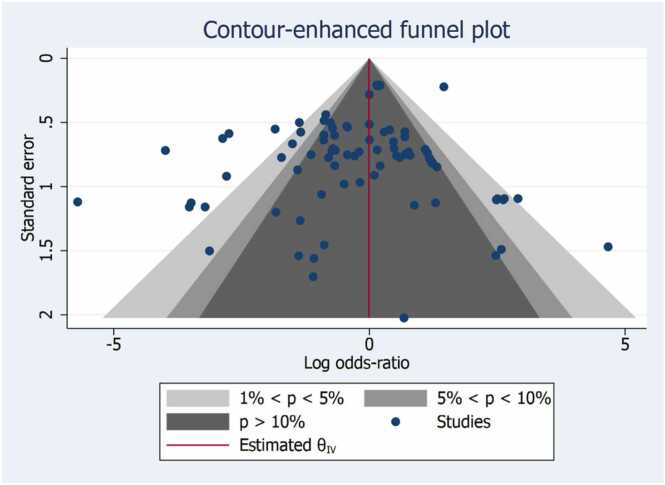
Pairwise comparison between index tests and biomarkers, control group was illustrated in Fig. 2. The overall log odds ratio was –0.22 (–0.51,0.06) favouring biomarkers (Fig. 2). The log odds ratio was for OSCC was –0.07 (–0.38,0.24) and HNSCC was –0.22(–0.51,0.06) (Fig. 3). 13 studies are unreliable that to be considered carefully for interpretation, as the studies are in the white area as illustrated in Fig. 5 of publication bias. The number of reviews and biomarker studies for HNSCC is illustrated in bubble plot Fig. 7. The three reviews were rated more than 50% in methodological and reporting quality except Hema Shree K 2019 (Supplementary Figure 1).
Fig. 2.
Pairwise comparison between biomarker and control group. The overall effect size was –0.22 (–0.51,0.06) favouring biomarkers.
Fig. 3.
Pairwise Forest plot of subgroup analysis OSCC and HNSCC.
Fig. 5.
Publication Bias. The studies should not be considered for interpretation as they are in white area and the null hypothesis of no effect should be rejected at 1% significance level (p < 0.01).
Fig. 7.
Bubble plot for HNSCC salivary biomarkers illustrating the mapping of reviews, number of patients and different class of biomarkers.
2. Network meta-analysis
2.1. Network plot
The network plot illustrates the number of comparison available between the biomarkers (thicker lines represents frequent comparisons available and the nodes represents the number of samples in the specific biomarkers). The network plot for biomarkers included in HNSCC and OSCC combined was illustrated in supplementary file Fig. 2. Similarly, the network plots for OSCC, HNSCC and OSCC TNM stage I-II was illustrated in supplementary file Fig. 3, Supplementary file Fig. 4 and Supplementary figure 8 respectively.
2.2. Forest plots
The forest plots for OSCC and HNSCC was illustrated in Supplementary File Fig. 5. Chemerin, MMP-9 and Phytosphingosine ranked higher in sensitivity.
In OSCC, Chemerin and MMP-9 took the lead by exhibiting the highest sensitivity, registering values of 0.94 (95% CI 0.78, 1.00) and achieving a balanced accuracy of 0.93. Following closely, Phytosphingosine demonstrated a sensitivity of 0.91 (95% CI 0.68, 0.99) and attained a balanced accuracy of 0.87 (Supplementary figure 6).
As for HNSCC, the top three biomarkers include Actin, IL-1β Singleplex, and IL-8 ELISA. Actin led the way with a sensitivity of 0.91 (95% CI 0.68–0.99), along with a specificity of 0.67 and an overall accuracy of 0.79. Subsequently, IL-1β Singleplex displayed a sensitivity of 0.62 (95% CI 0.30–0.88), a specificity of 0.89, and an accuracy of 0.75. Following that, IL-8 ELISA featured a sensitivity of 0.81 (95% CI 0.54–0.97), a specificity of 0.59, and an accuracy of 0.70 (Supplementary figure 7).
The biomarkers chemerin, MMP-9, Phytosphingosine, and Pipecolinic acid were found to be the most prominent in the early stages (TNM stage I-II) of oral squamous cell carcinoma (OSCC). (Supplementary figure 9).
5. Discussion
Several biomarkers have been studied for their potential use in detecting oral cancer, including mRNA, DUSP1, H3F3a, OAZ1, S100P, SAT, M2BP, MRP14, PMAIP1, PTPNI, DAPK1, and p16^INK4a. mRNA is a messenger ribosomal nucleic acid that plays a role in gene expression and is found to be overexpressed in oral cancer. DUSP1, a dual specificity phosphatase, regulates signaling pathways and is found to be downregulated in oral cancer. H3F3a, a histone protein, plays a role in gene regulation and is found to be overexpressed in oral cancer. OAZ1, an antizyme, regulates cell growth and is found to be downregulated in oral cancer. S100P, a calcium-binding protein, is found to be overexpressed in oral cancer and plays a role in tumor growth and invasion. SAT, a spermidine/spermine N1-acetyltransferase, regulates polyamine metabolism and is found to be downregulated in oral cancer. M2BP, a mac-2 binding protein, plays a role in inflammation and is found to be overexpressed in oral cancer. MRP14, a myeloid-related protein, is found to be overexpressed in oral cancer and is involved in tumor growth and invasion. PMAIP1, a protein induced by phorbol esters, plays a role in apoptosis and is found to be downregulated in oral cancer. PTPNI, a non-receptor type protein tyrosine phosphatase, regulates cell signaling and is found to be downregulated in oral cancer. DAPK1, a death-associated protein kinase, plays a role in apoptosis and is found to be downregulated in oral cancer. Finally, p16^INK4a, a cyclin-dependent kinase inhibitor, is found to be overexpressed in oral cancer and is used as a diagnostic biomarker.
Sahibzada et al., 2017 reported that the salivary expression of levels of different specific biomarkers IL-8, IL-6 and TNF-α have been correlated to the induction of the signalling pathways in the process of development of many different lesions and oral cancer through a semi-quantitative dose-dependent response [29]. Moreover, several authors indicated that also the circadian rhythm could produce a significant influence on the salivary flow rate, composition, cytokine and modulator expression [30], [31]. Li et al. supposed a multi-layered biomarkers and mRNA diagnostic approach, detecting an increased expression of markers such as DUSP1, H3F3A, IL1B, IL8, OAZ1, S100P, and SAT in subjects affected by oral squamous carcinoma with a high sensitivity and specificity [32]. This relationship was also correlated by the authors also the cancer staging reporting a sensitivity/specificity for T1-T2 of 0.67/ 0.96, and for T3-T4 of 0.82/0.84 [33]. Moreover, a binary methodology was proposed by Hamad et al. through a combinate detection of the specific inflammatory cytokines both in serum and salivary fluids, reporting an increase of the sensitivity of the in-chair IL-8, IL-6 tests [34].
The objective of this systematic review and meta‐analysis was to evaluate diagnostic accuracy of salivary biomarkers and to compare the diagnostic value of different salivary biomarkers for various OSCC or HNSCC with NMA principles. The first observation was that not all studies presented detection sensitivity and specificity values. The overall interpretation of the data must therefore take this into account when recommending salivary biomarkers.
The results reveal significant differences in biomarker sensitivity and accuracy between OSCC and HNSCC. In OSCC, Chemerin and MMP-9 emerged as the top performers with remarkable sensitivity values of 0.94 (95% CI 0.78, 1.00) and a balanced accuracy of 0.93. These findings are particularly significant because both Chemerin and MMP-9 are associated with key pathophysiological processes in oral squamous cell carcinoma (OSCC). Chemerin, a chemotactic cytokine, is involved in inflammation and immune responses, and its elevated sensitivity suggests its potential as a diagnostic marker for OSCC. Similarly, Matrix Metalloproteinase-9 (MMP-9) is an enzyme linked to tissue remodeling and cancer invasion. Its high sensitivity underscores its role as a potential indicator of OSCC progression.
Contrary, HNSCC displayed a different set of top biomarkers. Actin, with a sensitivity of 0.91 (95% CI 0.68–0.99), ranked highest, possibly due to its role as a structural protein involved in cell motility and its potential implication in cancer cell behavior. The specificity of 0.67 suggests some limitations in its ability to distinguish HNSCC from other conditions. IL-1β Singleplex, with a sensitivity of 0.62 (95% CI 0.30–0.88), is linked to inflammation, and its sensitivity could be attributed to the inflammatory nature of HNSCC. IL-8 ELISA, featuring a sensitivity of 0.81 (95% CI 0.54–0.97), indicates its role in immune responses and angiogenesis, which are often elevated in cancer. The lower specificity of 0.59 implies some overlap with non-cancerous conditions.
We found that the biomarkers chemerin, MMP-9, Phytosphingosine, and Pipecolinic acid were ranked higher as the most expressive during early stages (TNM stage I-II) of oral squamous cell carcinoma (OSCC) (Supplementary figure 8 and 9). However, due to insufficient studies within the HNSCC group, it was not possible to rank the biomarkers for early-stage detection. Therefore, further well-conducted clinical studies are needed to address this question.
The differences in biomarkers between OSCC and HNSCC may be attributed to variations in the underlying pathophysiology of these cancers. OSCC often originates in the oral cavity and may involve different molecular pathways and biomarkers compared to HNSCC, which encompasses a broader range of head and neck cancers. Additionally, variations in detection methods, such as ELISA and Singleplex assays, can contribute to differences in sensitivity and specificity.
Regarding patient data, the analysis considered 2025 patients in the OSCC group and 3868 patients in the HNSCC group, resulting in a patient ratio of 0.53. This discrepancy in patient numbers may reflect differences in the prevalence and incidence of these two types of cancer or variations in the availability of data for analysis. Further research is needed to explore the clinical implications of these findings and to optimize the use of these biomarkers in the diagnosis and management of OSCC and HNSCC.
Oral cancer and it remains a low 5-year survival rate for the past years [35]. It is reported that miRNAs may play an important role as promising salivary biomarkers, with the advantages of high diagnostic sensitivity and specificity and minimal invasiveness [36]. However, the estimation of miRNA-136 diagnostic performance in oral cancer remained inconsistent between studies. In one of such studies, Liu et al. reported the high accuracy of miRNA-136 in saliva for OSCC diagnostic: a sensitivity of 80.0%, miRNA-27B (77.0%) and miR-27b (78.0%) [37]. These percentages vary when specifically seen in individual groups i.e. OSCC and HNSCC.
Although mRNA biomarkers proved to be superior in our analyses as compared to cytokine biomarkers, in the early detection of OSCC and/or HNSCC, one must view these results in light of the relative differences in methodology and precision between the techniques used for the generation of both datasets. For salivary miRNAs, we observed that the studies published before 2015 showed higher sensitivity than after. Meanwhile, studies with larger sample size (n ≥ 100) showed lower accuracy than studies with small sample size (n < 100). Research have proven that the salivary biomarkers have good diagnostic potential in oral squamous cell carcinoma (OSCC). Guerra et al. [26] have evaluated the diagnostic value of salivary biological markers in the diagnosis of head and neck carcinoma. The article is an excellent concise of the relevant information on salivary biomarkers, where the authors have emphasized the point that combination of biomarkers have better accuracy than those tested individually [26].
The gold standard for the diagnosis of OSCC is a biopsy of the suspicious lesion but it is not always feasible for screening purposes for early oral cancer [38], [39], [40]. The discriminatory power of salivary biomarkers open up new avenues in oral cancer diagnosis. The overall log odds ratio of this review was –0.22 (–0.51,0.06) favouring biomarkers (Fig. 2). The log odds ratio was for OSCC was –0.07 (–0.38,0.24) and HNSCC was –0.22(–0.51,0.06).
Previous reviews have some limitations. First, miRNAs had low expression level in body fluids and only showed moderate diagnostic accuracy in detecting OSCC. Moreover, limited literatures reported the expression of blood or salivary miRNAs in OSCC, and some of the involved studies had small sample size with high specificity, resulting in the overestimating of the accuracy of the body fluid miRNAs in the diagnosis of OSCC. Second, some parameters including gender difference, age distribution, smoking habit, alcohol consumption were not detailed in some studies, which limited the subgroup analysis and meta regression. Third, the articles in this meta-analysis adopted diverse cut-off values and different controls for miRNA quantification, which may significantly affect the results of this analysis. Other limitations include, significant heterogeneity was encountered perhaps due to various regimens, doses, duration, center settings, populations enrolled etc. calling for cautious interpretation of the results. Many of the studies suffer from significant sources of bias.
The findings of this study have important practical and clinical implications for the early detection of oral cancers. The high sensitivity and accuracy of MMP-9 and chemerin salivary biomarkers suggest that they could be useful in screening and diagnosing oral cancers. The identification and evaluation of additional biomarkers could further improve early detection rates and patient outcomes. Incorporating these biomarkers into clinical practice could lead to earlier detection of oral cancers, which can increase the chances of successful treatment and improve survival rates.
6. Conclusion
Salivary biomarkers can represent a valuable tool for early screening and detection of head and neck squamous cell carcinoma and oral squamous cell carcinoma. The need for additional research to expand the panel of potential biomarkers for these cancers also emerged. The findings of this study are particularly important for public health, as early detection of oral cancers can significantly enhance patient outcomes and survival rates.
Source of funding
None.
Ethics approval statement
Not Applicable.
CRediT authorship contribution statement
S.K; J.Y; N.R; F.L; P.R;N.C; F.G; B.R;M.D.F and G.M.T conceived and planned the review. S.K; J.Y; N.R; P.R and MDF carried out the data search, synthesis, data extraction. NMW was performed by S.K and N.R. S.K; N.C; F.G; B.R and M.D.F. contributed to the interpretation of the results. S.K; F.G; B.R; F.L and P.R took the lead in writing the manuscript. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Conflict of interest
-
•
All authors have participated in (a) conception and design, or analysis and interpretation of the data; (b) drafting the article or revising it critically for important intellectual content; and (c) approval of the final version.
-
•
This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue.
-
•
The authors have no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript
-
•
The following authors have affiliations with organizations with direct or indirect financial interest in the subject matter discussed in the manuscript
Acknowledgement
None.
Footnotes
Registration: PROSPERO: n.345117
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jdsr.2023.10.003.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data availability
The data was extracted from the already published systematic reviews and meta-analysis and therefore the directly accessible through the included meta-analysis in this review.
References
- 1.Zygogianni A.G., Kyrgias G., Karakitsos P., Psyrri A., Kouvaris J., Kelekis N., et al. Oral squamous cell cancer: early detection and the role of alcohol and smoking. Head Neck Oncol. 2011;3 doi: 10.1186/1758-3284-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montero P.H., Patel S.G. Cancer of the oral cavity. Surg Oncol Clin N Am. 2015;24:491–508. doi: 10.1016/j.soc.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillon M., Smith D.J., Kanatas A. Oral cancer: another challenge for the dental team? Br Dent J. 2016;220:499–500. doi: 10.1038/sj.bdj.2016.357. [DOI] [PubMed] [Google Scholar]
- 4.Global Health Data Exchange | GHDx Available online: 〈http://ghdx.healthdata.org/〉 (accessed on 4 February 2022).
- 5.Khalili J. Oral cancer and role of general dental practitioners in its early detection. Lik Sprav. 2007:10–19. [PubMed] [Google Scholar]
- 6.Kujan O., Glenny A.M., Oliver R.J., Thakker N., Sloan P. Screening programmes for the early detection and prevention of oral cancer. Cochrane Database Syst Rev. 2006:CD004150. doi: 10.1002/14651858.CD004150.pub2. [DOI] [PubMed] [Google Scholar]
- 7.Van der Waal Isaäc, Remco de Bree Ruud Brakenhoff, Coebegh J.W. Early diagnosis in primary oral cancer: is it possible? Med Oral, Patol Oral Y cirugia Bucal. 2011;16(3):e300–e305. doi: 10.4317/medoral.16.e300. [DOI] [PubMed] [Google Scholar]
- 8.Mighell A.J., Gallagher J.E. Oral cancer - improving early detection and promoting prevention. Are you up to date? Br Dent J. 2012;213:297–299. doi: 10.1038/sj.bdj.2012.838. [DOI] [PubMed] [Google Scholar]
- 9.Ammann Yanic, Beutner Ulrich, Gian Vital Domenic, Morand Grgoire, Broglie Daeppen Martina A., Born Diana, et al. Impact of the new TNM staging system on oral tongue cancers. Swiss Med Wkly. 2021;151:w20493. doi: 10.4414/smw.2021.20493. [DOI] [PubMed] [Google Scholar]
- 10.Chiu K., Hosni A., Huang S.H., Tong L., Xu W., Lu L., et al. The potential impact and usability of the eighth edition TNM staging classification in oral cavity cancer. Clin Oncol. 2021;33(10):e442–e449. doi: 10.1016/j.clon.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Low Tsu‐Hui, Gao Kan, Elliott Michael, Clark Jonathan R. Tumor classification for early oral cancer: re‐evaluate the current TNM classification. Head neck. 2015;37(2):223–228. doi: 10.1002/hed.23581. [DOI] [PubMed] [Google Scholar]
- 12.Almulla Abdullah, Noel Christopher W., Lu Lin, Xu Wei, O’Sullivan Brian, Goldstein David P., et al. Radiologic-pathologic correlation of extranodal extension in patients with squamous cell carcinoma of the oral cavity: implications for future editions of the TNM classification. Int J Radiat Oncol* Biol* Phys. 2018;102(4):698–708. doi: 10.1016/j.ijrobp.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 13.Boeve Koos, Melchers Lieuwe J., Schuuring Ed, Roodenburg Jan L., Halmos Gyorgy B., van Dijk Boukje A., et al. Addition of tumour infiltration depth and extranodal extension improves the prognostic value of the pathological TNM classification for early‐stage oral squamous cell carcinoma. Histopathology. 2019;75(3):329–337. doi: 10.1111/his.13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fries Rudolf, Grabner Helmut, Leijhanec Josef, Wepner Friedrich, Kränzl Bernd, Krekeler Gisbert, et al. TNM-classification of carcinomas of the oral cavity—efficacy of clinically available data (TN) J Maxillofac Surg. 1976;4:231–238. doi: 10.1016/s0301-0503(76)80044-6. [DOI] [PubMed] [Google Scholar]
- 15.Mercante Giuseppe, Gaino Francesca, Giannitto Caterina, Ferreli Fabio, Virgilio Armando De, Franzese Ciro, et al. Discrepancies between UICC and AJCC TNM classifications for oral cavity tumors in the 8th editions and following versions. Eur Arch Oto-Rhino-Laryngol. 2022;279(1):527–531. doi: 10.1007/s00405-021-06964-6. [DOI] [PubMed] [Google Scholar]
- 16.Jones Daniel L. Oral cancer: diagnosis, treatment, and management of sequela. Tex Dent J. 2012;129(5):459. [PubMed] [Google Scholar]
- 17.Goldoni Riccardo, Scolaro Alessandra, Boccalari Elisa, Dolci Carolina, Scarano Antonio, Inchingolo Francesco, et al. Malignancies and biosensors: a focus on oral cancer detection through salivary biomarkers. Biosensors. 2021;11(10):396. doi: 10.3390/bios11100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tartaglia Gianluca Martino, Gagliano Nicoletta, Zarbin Luca, Tolomeo Giorgia, Sforza Chiarella. Antioxidant capacity of human saliva and periodontal screening assessment in healthy adults. Arch Oral Biol. 2017;78:34–38. doi: 10.1016/j.archoralbio.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Tartaglia A., Romasco T., D’Ovidio C., Rosato E., Ulusoy H.I., Furton K.G., et al. Determination of phenolic compounds in human saliva after oral administration of red wine by high performance liquid chromatography. J Pharm Biomed Anal. 2022;209 doi: 10.1016/j.jpba.2021.114486. [DOI] [PubMed] [Google Scholar]
- 20.Rapado-González Óscar, Martínez-Reglero Cristina, Salgado-Barreira Ángel, Takkouche Bahi, López-López Rafael, Suárez-Cunqueiro María Mercedes, et al. Salivary biomarkers for cancer diagnosis: a meta-analysis. Ann Med. 2020;52(3):131–144. doi: 10.1080/07853890.2020.1730431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Chen-Zi, Cheng Xing-Qun, Li Ji-Yao, Zhang Ping, Yi Ping, Xu Xin, et al. Saliva in the diagnosis of diseases. Int J Oral Sci. 2016;8(3):133–137. doi: 10.1038/ijos.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Bock Muriel, De Seny Dominique, Meuwis Marie-Alice, Chapelle Jean-Paul, Louis Edouard, Malaise Michel, et al. Challenges for biomarker discovery in body fluids using SELDI-TOF-MS. J Biomed Biotechnol. 2010;2009 doi: 10.1155/2010/906082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khurshid Zohaib, Zohaib Sana, Najeeb Shariq, Zafar Muhammad Sohail, Slowey Paul D., Almas Khalid. Human saliva collection devices for proteomics: an update. Int J Mol Sci. 2016;17(6):846. doi: 10.3390/ijms17060846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar Pavan, Singh Ashutosh, Kanaujia Surendra Kumar, Pradhan Asima. Human saliva for oral precancer detection: a comparison of fluorescence & stokes shift spectroscopy. J Fluoresc. 2018;28:419–426. doi: 10.1007/s10895-017-2203-2. [DOI] [PubMed] [Google Scholar]
- 25.Kang J.W., Eun Y.G., Lee Y.C. Diagnostic value of salivary miRNA in head and neck squamous cell cancer: systematic review and meta-analysis. Int J Mol Sci. 2021;22(13):7026. doi: 10.3390/ijms22137026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerra E.N., Acevedo A.C., Leite A.F., Gozal D., Chardin H., De Luca Canto G. Diagnostic capability of salivary biomarkers in the assessment of head and neck cancer: a systematic review and meta-analysis. Oral Oncol. 2015;51(9):805–818. doi: 10.1016/j.oraloncology.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Liu Dingshan, Xin Zhili, Guo Songsong, Li Sheng, Cheng Jie, Jiang Hongbing. Blood and salivary microRNAs for diagnosis of oral squamous cell carcinoma: a systematic review and meta-analysis. J Oral Maxillofac Surg. 2021;79(5):1082–e1. doi: 10.1016/j.joms.2020.12.043. [DOI] [PubMed] [Google Scholar]
- 28.Hema Shree K., Ramani Pratibha, Sherlin Herald, Sukumaran Gheena, Jeyaraj Gifrrina, Don K.R., et al. Saliva as a diagnostic tool in oral squamous cell carcinoma–a systematic review with meta analysis. Pathol Oncol Res. 2019;25:447–453. doi: 10.1007/s12253-019-00588-2. [DOI] [PubMed] [Google Scholar]
- 29.Sahibzada Haafsa Arshad, Khurshid Zohaib, Sannam Khan Rabia, Naseem Mustafa, Siddique Khalid Mahmood, Mali Maria, et al. Salivary IL-8, IL-6 and TNF-α as potential diagnostic biomarkers for oral cancer. Diagnostics. 2017;7(2):21. doi: 10.3390/diagnostics7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey Sue P., Russell T.Williamson. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85(2):162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 31.Tribble Gena D., Angelov Nikola, Weltman Robin, Wang Bing-Yan, Eswaran Sridhar V., Gay Isabel C., et al. Frequency of tongue cleaning impacts the human tongue microbiome composition and enterosalivary circulation of nitrate. Front Cell Infect Microbiol. 2019:39. doi: 10.3389/fcimb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Yang, Maie A.R.St. John, Zhou Xiaofeng, Kim Yong, Sinha Uttam, Jordan Richard C.K., et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10(24):8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 33.Roi Alexandra, Roi Ciprian Ioan, Negruțiu Meda Lavinia, Riviș Mircea, Sinescu Cosmin, Rusu Laura-Cristina. The challenges of OSCC diagnosis: salivary cytokines as potential biomarkers. J Clin Med. 2020;9(9):2866. doi: 10.3390/jcm9092866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Xiujuan, Chen Zhiying, Shi Shaomin, Wang Xianwen, Wang Wanli, Li Ning, et al. Clinical diagnostic implications of body fluid miRNA in oral squamous cell carcinoma: a meta-analysis. Medicine. 2015;94(37) doi: 10.1097/MD.0000000000001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C.‐J., Kao S.‐Y., Tu H.‐F., Tsai M.‐M., Chang K.‐W., Lin S.‐C. Increase of microRNA miR‐31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16(4):360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 36.Ranimol Prasanna, Anita Balan, Pillai M.Radhakrishna. Diagnostic capability of salivary biomarkers in the assessment of head and neck cancer. Oral Oncol. 2015;51(11) doi: 10.1016/j.oraloncology.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Liu C.J., Kao S.Y., Tu H.F., Tsai M.M., Chang K.W., Lin S.C. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16(4):360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 38.Brinkmann O., Kastratovic D.A., Dimitrijevic M.V., Konstantinovic V.S., Jelovac D.B., Antic J., et al. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol. 2011;47(1):51–55. doi: 10.1016/j.oraloncology.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S., Yang M., Li R., Bai J. Current advances in noninvasive methods for the diagnosis of oral squamous cell carcinoma: a review. Eur J Med Res. 2023;28(1):1–2. doi: 10.1186/s40001-022-00916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osman T.A., Costea D.E., Johannessen A.C. The use of salivary cytokines as a screening tool for oral squamous cell carcinoma: a review of the literature. J Oral Maxillofac Pathol. 2012;16(2):256. doi: 10.4103/0973-029X.99083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
The data was extracted from the already published systematic reviews and meta-analysis and therefore the directly accessible through the included meta-analysis in this review.