Highlights
-
•
PRRSV reduces the formation of PML nuclear bodies in which PML protein is a major component.
-
•
PML NBs were significantly downregulated by PRRSV nsp1β.
-
•
Viral suppression of PML NBs was common in the family Arteriviridae.
Keywords: PRRSV, TRIM19, PML, nsp1, Immune evasion, IFN antagonism, Promyelocytic leukemia protein
Abstract
Tripartite motif (TRIM)-containing proteins are a family of regulatory proteins that can participate in the induction of antiviral cytokines and antagonize viral replication. Promyelocytic leukemia (PML) protein is known as TRIM19 and is a major scaffold protein organizing the PML nuclear bodies (NBs). PML NBs are membrane-less organelles in the nucleus and play a diverse role in maintaining cellular homeostasis including antiviral response. Porcine reproductive and respiratory syndrome virus (PRRSV), a member virus of the family Arteriviridae, inhibits type I interferon (IFN) response during infection, and nonstructural protein 1 (nsp1) of the virus has been identified as a potent IFN antagonist. We report that the numbers of PML NBs per nucleus were significantly downregulated during infection of PRRSV. The overexpression of all six isoforms of PML suppressed the PRRSV replication, and conversely, the silencing of PML gene expression enhanced the PRRSV replication. The suppression of PML NBs by the nsp1 protein was common in other member viruses of the family, represented by equine arteritis virus, lactate dehydrogenase elevating virus of mice, and simian hemorrhagic fever virus. Our study unveils a conserved viral strategy in arteriviruses for innate immune evasion.
1. Introduction
The innate immune system functions as an early defense to prevent the establishment of viral infections. Innate immunity plays a pivotal role in sensing and limiting invading viruses and enhancing adaptive immune response that eventually controls infections. The sensing of invading viruses begins with the recognition of viral pathogen-associated molecular patterns (PAMPs) by host cell pattern-recognition receptors (PRRs). Successful employment of the innate immune system relies on the activation of relevant host factors to create an antiviral state of the cell. If inflammatory responses remain unchecked, certain antiviral mechanisms may become detrimental to the host because of the induction of apoptosis or tissue damage. Post-translational modifications (PTMs) are cellular processes to attach or detach small molecules to proteins that can function as on-off switches. PTMs play a vital role in regulating the host-cell innate immunity. Conversely, viruses have evolved to develop mechanisms to modify PTMs and circumvent cellular antiviral response to establish productive infections.
TRIM proteins. Tripartite motif (TRIM)-containing proteins are a family of PTMs and are widely distributed in mammalian cells. TRIM proteins have a conserved structural arrangement of three domains (RBCC) in their N-terminal region: RING (R) domain, B-box (B) domain, and coiled-coil (CC) domain (van Gent et al., 2018). The RING (really interesting new gene) domain is found in most TRIM proteins and contains a zinc finger motif. The RING domain also possesses an E3-ubiquitin ligase activity which can conjugate different polyubiquitin linkage types that determine the fate of the target protein. Lysine (K) 48-linked ubiquitination induces proteasomal degradation of substrates, whereas K63-linked polyubiquitin modulates the activity, subcellular localization, or interaction with other proteins of substrates (Davis and Gack, 2015). In addition to conjugating ubiquitin to their targets, TRIM proteins can also conjugate ubiquitin-like molecules such as ISG15 (interferon-stimulated gene 15) and SUMO (small ubiquitin-like modifier). The RING domain is crucial for the antiviral activity of TRIM proteins by allowing them to modulate the function of a wide range of both host and viral substrates via ubiquitin or ubiquitin-like modifications. TRIM proteins also contain one or two B-box domains containing cysteine-histidine-rich and zinc-binding motifs (Meroni, 2012). The B-box domains are thought to modulate self-assembly, E3-ligase activity, or protein-protein interactions. The alpha-helical CC domain mediates the homomeric or heteromeric assembly of TRIM proteins, which is also crucial for their antiviral activities.
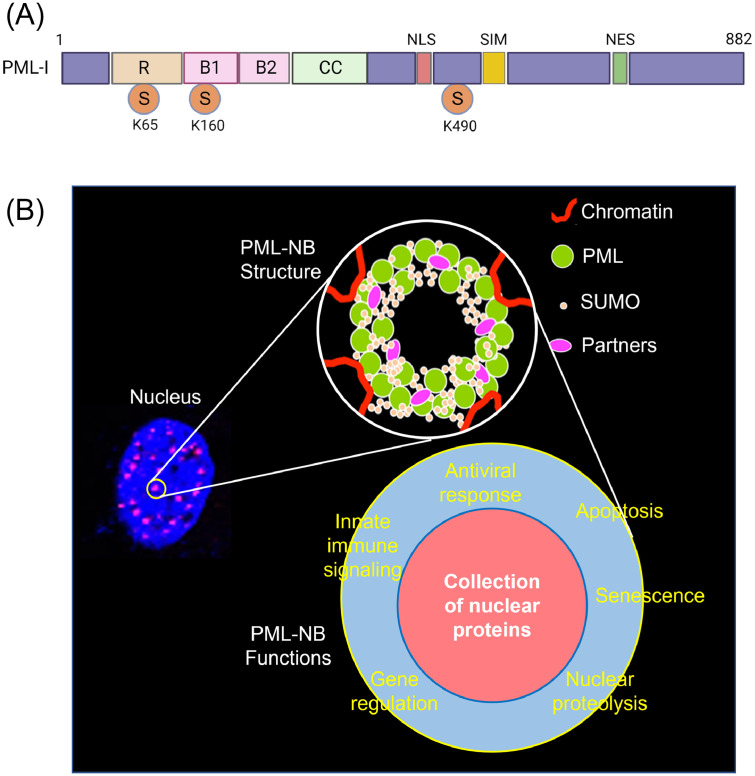
TRIM19/promyelocytic leukemia protein (PML) and PML nuclear bodies (NBs). Of more than 80 members of TRIM proteins in humans, TRIM19 is of particular interest. TRIM19 is known as the promyelocytic leukemia (PML) protein, and by alternative splicing in the 3′ half of the gene, seven different isoforms of PML protein are generated, PML-I through PML-VII. All isoforms share the N-terminal RBCC domains of TRIM and contain the nuclear localization signal (NLS) (Fig 1A), except the smallest isoform PML-VII, and thus show a predominantly nuclear pattern. PML protein undergoes extensive PTMs, mainly SUMOylation. The main SUMOylation sites are K65, K160, and K490, and occasionally, K616 can also be SUMOylated. Besides SUMO sites, PML contains a SUMO interacting motif (SIM) at amino acid positions of 556–562, enabling it to interact with other SUMOylated proteins (Fig. 1A) (Bernardi and Pandolfi, 2007).
Fig. 1.
Structure and function of promyelocytic leukemia nuclear bodies (PML NBs). (A), Primary structure of the isotype I of PML. PML has a conserved structural arrangement of three domains (RBCC): RING (R) domain, two B-box (B1 and B2) domains, coiled-coil (CC) domain. The RING (really interesting new gene) domain possesses an E3-ubiquitin ligase activity which can conjugate ubiquitin to their targets as well as ubiquitin-like proteins such as ISG15 (interferon-stimulated gene 15) and SUMO (small ubiquitin-like modifier). Two B-box domains are thought to modulate protein-protein interactions. The CC domain mediates the homomeric or heteromeric assembly of PML proteins. PML is SUMOylated at three lysine residues of K65, K160, and K490. NLS, nuclear localization signal; SIM, SUMO-interacting motif; NES, nuclear export signal; S, SUMOylation sites (Adapted from Bernardi and Pandolfi, 2007). (B), PML NBs are dynamic membrane-less protein aggregates in the nucleus of the cell. PML NBs appear as dot-shaped spherical structures in the interchromatin nuclear space. PML is a key component for the structural integrity of PML NBs. PML NBs are formed initially by disulfide linkages of PML monomers as well as non-covalent interactions to drive the assembly of PML oligomerization. PML is SUMOylated and recruits other partner proteins to the inner core, and mature PML NBs are formed. PML NBs participate in a wide range of cellular processes such as cell cycle regulation, apoptosis, senescence, DNA repair, gene regulations, and antiviral response. Of these processes, antiviral response is the first line of intracellular defense against invading pathogens, and PML NBs can confer intrinsic immunity (Adapted from Hoischen et al., 2018).
PML is a key component for the structural integrity of PML nuclear bodies (NBs; also called nuclear domain 10). PML NBs are dynamic membrane-less protein aggregates. PML NBs are formed initially by intermolecular disulfide linkages of PML monomers, as well as non-covalent interactions between RBCC domains to drive the assembly of PML oligomerization. Then, UBC9, which is the only known SUMO E2-conjugating enzyme, is recruited to mediate SUMOylation of PML, triggering PML-PML dimerization and subsequently oligomerization via SUMO-SIM interactions. SUMOylated PML oligomers then recruit other client proteins to the inner core through their SIM, and mature PML NBs are assembled (Fig. 1B) (Hoischen et al., 2018).
PML NBs appear as spherical structures in the interchromatin nuclear space, and more than 170 nuclear proteins reside within the nuclear bodies either constitutively or transiently (Paulus et al., 2020). Among the client proteins residing in the PML NBs, Daxx (death domain-associated protein), Sp100 (speckled 100 kDa nuclear antigen), ATRX (alpha-thalassemia X-linked), and SUMO members SUMO1, SUMO2, SUMO3, and SUMO5 are found along with the nuclear isoforms of PML, except PML-VII which lacks NLS. PML NBs are hubs of SUMOylation, and client proteins associated with these organelles undergo PTMs by SUMO paralogs (McManus et al., 2018; Paulus et al., 2020). Depending on the cell type and physiological state, 5 to 30 PML NBs are typically present in the nucleus.
PML NBs and antiviral function. PML NBs participate in a wide range of cellular processes such as cell cycle regulation, apoptosis, senescence, DNA repair, and antiviral response (Lallemand-Breitenbach and de Thé, 2010, 2018; van Gent et al., 2018). Of those processes, the antiviral response is the first line of intracellular defense against invading pathogens, and PML NBs can confer intrinsic immunity by entrapping viral genomes or capsids (Catez et al., 2012; Reichelt et al., 2011). Sp100, Daxx, PML, and others associated with PML NBs can act individually as restriction factors for viruses (Paulus et al., 2020; Scherer and Stamminger, 2016). Sp100 and Daxx are transcriptional repressors and restrict gene expression for DNA viruses. PML has also been identified as a key regulator of cytokine responses and innate immunity. Certain PML isoforms are positive regulators of IFN induction and promote ISG expressions (Ashley et al., 2017; Chen et al., 2015; Scherer and Stamminger, 2016). Interestingly, PML proteins themself are ISGs (Ashley et al., 2017). PML proteins can also be associated with IRF3 and STAT1, both of which are key regulators for IFN induction and ISG expressions, and contribute to establishing an IFN-induced antiviral state (Chee et al., 2003; Chen et al., 2015; Regad et al., 2001; Ulbricht et al., 2012). These findings demonstrate the essential role of PML in antiviral restriction and cytokine-induced inflammatory states. Many viruses have evolved to equip mechanisms to inactivate PML-associated antiviral responses. Targeting PML can lead to changes in the structural integrity of the organelles, which is one common mechanism by which viruses antagonize the intrinsic and innate immune responses (Lallemand-Breitenbach and de Thé, 2010; Scherer et al., 2017).
Porcine arterivirus PRRSV and viral IFN antagonism. Porcine reproductive and respiratory syndrome virus (PRRSV) is a single-stranded positive-sense RNA virus that belongs to the family Arteriviridae in the order Nidovirales together with the Coronaviridae family. The Arteriviridae family consists of 6 subfamilies (Equarterivirinae, Simarterivirinae, Variarterivirinae, Zealarterivirinae, Heroarterivirinae, and Crocarterivirinae), and within the family, 23 species are listed according to the recent version of the International Code of Virus Classification and Nomenclature 2022 (https://talk.ictvonline.org/taxonomy). The most studied and frequently sampled arteriviruses are represented by PRRSV, equine arteritis virus (EAV), lactate dehydrogenase elevating virus (LDV) of mice, and simian hemorrhagic fever virus (SHFV). EAV is the prototype virus of the family, but a wealth of information has become available for PRRSV. The PRRSV genome varies from 14.9 to 15.5 kb in length and consists of the 5′ untranslated region (UTR), 11 open reading frames (ORFs) followed by the 3′ UTR and polyadenylated tail (Kappes and Faaberg, 2015; Wootton et al., 2000), regardless of the two distinct species of PRRSV-1 (Betaarterivirus suid 1) and PRRSV-2 (Betaarterivirus suid 2).
A notable immunological characteristic has been observed in PRRSV-infected cells and pigs. The induction of type I IFNs are unusually poor upon infection, and eight viral proteins (nsp1α, nsp1β, nsp2, nsp2TF, nsp2N, nsp4, nsp11, and N) have been identified to participate in the blocking of IFN production and signaling (Beura et al., 2010; Chen et al., 2014; Han and Yoo, 2014a; Lunney et al., 2016; Patel et al., 2010; Sagong and Lee, 2011; Subramaniam et al., 2010; Wang et al., 2013b; Yoo et al., 2010). Among these proteins, nsp1 is cleaved into the nsp1α and nsp1β subunits and exhibits the potent IFN antagonistic property. Interestingly, the nsp1α, nsp1β, and N proteins are localized in the nucleus in addition to their normal cytoplasmic distribution (Pei et al., 2008; Rowland and Yoo, 2003). Their roles in the nucleus are not fully understood, but according to the available data, they seem to contribute to efficient viral replication (Han et al., 2014; Su et al., 2023a; Yoo et al., 2003). The nsp1α subunit inhibits the production of type I IFNs and impedes the promoter activity when stimulated with the dsRNA analog (Chen et al., 2010). It also downregulates IFN induction by degrading the CREB (cyclic-AMP-responsive element binding)-binding protein (CBP), which is likely through the proteasome-ubiquitin pathway (Kim et al., 2010). Furthermore, PRRSV nsp1α suppresses NF-κB activation in a RIG-I-dependent manner, leading to the suppression of type I IFN production (Song et al., 2010; Subramaniam et al., 2010). The nsp1β subunit has also been identified as an effective IFN antagonist (Li et al., 2013). PRRSV nsp1β inhibits the STAT1 phosphorylation and disrupts the nuclear translocation of ISGF3, thereby suppressing the JAK-STAT signaling pathway and inhibiting the expression of ISGs (Chen et al., 2010; Patel et al., 2010). In addition, nsp1β degrades karyopherin-α1 which is a nuclear transporter required for ISGF3 nuclear translocation (Wang et al., 2013a). A recent study shows that nsp1β binds to nucleoporin 62 (Nup62) and disrupts the nuclear pore complex. The disruption of the nuclear pore complex results in the blocking of nucleocytoplasmic export of host mRNAs and the reduction of host protein synthesis including type I IFNs, ISGs, and IRF3 (Ke et al., 2019). The SAP motif [SAF-A/B (scaffold attachment factors A and B), Acinus, and PIAS (protein inhibitor of activated STAT)] has been identified in nsp1β and is the critical domain for host mRNA nuclear retention and IFN suppression (Han et al., 2017). The combined activities of nsp1α and nsp1β targeting both IFN production and signaling pathways make the nsp1 protein the most potent type I IFN antagonist for PRRSV.
In the present study, we studied the modulation of PML functions by PRRSV. We found that PML suppressed the viral replication, and PRRSV nsp1β degraded and suppressed the PML expression, resulting in productive viral replication. The nsp1 orthologs in other arteriviruses also possessed the ability of PML NB downregulation. This is a novel strategy for immune evasion and enhanced viral replication in arteriviruses.
2. Results and discussions
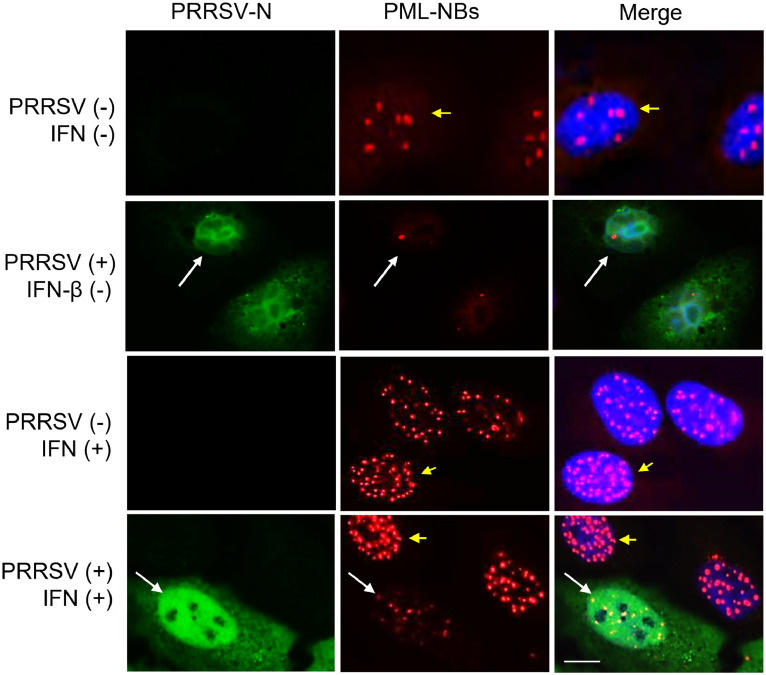
Downregulation of PML NBs by PRRSV. The PML protein is one of ISGs and has emerged as a significant regulator for viral replication. Conversely, viruses have evolved to counteract the antiviral function of PML. To determine whether PRRSV possessed the ability to counteract the PML function, MARC-145 which is the only established cell line permissive for PRRSV was infected with PRRSV for 24 h and stimulated with IFN, given that PML is an IFN-inducible gene product. The potential of PRRSV for PML regulation was then examined by co-staining with anti-PML antibody and anti-PRRSV N antibody. In uninfected cells, the numbers of PML-NBs in the nucleus were increased significantly after IFN stimulation (Fig. 2, short yellow arrows in third panel from top), while their numbers without stimulation remained minimal (short yellow arrows in top panel). In virus-infected cells, however, the numbers of PML-NBs were decreased significantly even after IFN stimulation (long white arrows in bottom panel,) compared to those in unstimulated cells (long white arrows in second panel from top).
Fig. 2.
Reduction of PML-NBs in PRRSV-infected cells. MARC-145 cells were infected with the PA8 strain of PRRSV at 1 multiplicity of infection for 24 h, followed by incubation with 1000 units/ml of human IFN-β for 6 h. Cells were stained with anti-PRRSV-N-specific monoclonal Ab (MR40) (mouse) (green) and anti-PML polyclonal Ab (rabbit) (red) as described in Materials and Methods. Nuclei were stained with DAPI (blue). Long white arrows indicate PRRSV-infected cells, and short yellow arrows indicate uninfected cells. In virus-infected cells (long white arrows), the numbers of PML-NBs were decreased compared to the numbers in uninfected cells (short yellow arrows). Images were taken by confocal microscopy. Scale bar represents 10 µm.
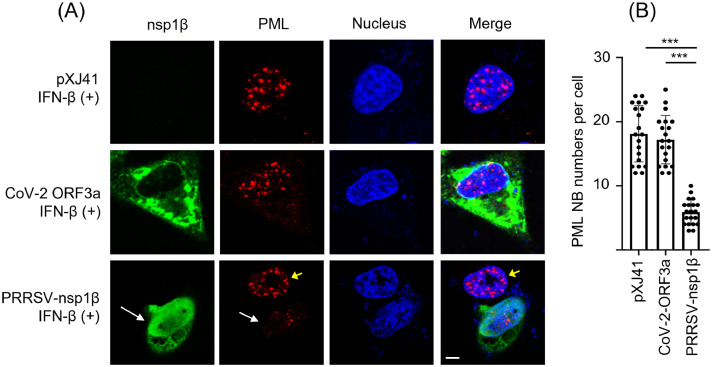
PRRSV nsp1β protein as the PML suppressor. The nsp1β protein is the potent IFN antagonist of PRRSV. The nsp1β protein downregulates the nucleocytoplasmic trafficking of host mRNAs and inhibits the type I IFNs and ISG transcriptions (Han and Yoo, 2014b; Ke et al., 2019, 2018). Since PML functions as an ISG, it was postulated that the nsp1β protein might play a role in the reduction of PML in PRRSV-infected cells. We thus examined PML-NBs in cells expressing nsp1β by gene transfection, followed by staining with PML antibody. The ORF3a protein of SARS-CoV-2 was previously shown to be irrelevant to IFN response (Su et al., 2023b) and thus was included as a negative control. After stimulation with IFN, the numbers of PML-NBs decreased significantly in nsp1β-expressing cells (Fig. 3A, bottom panels, white arrows) compared to empty vector pXJ41 (upper panels) or SARS-CoV-2 ORF3a-expressing control cells (middle panels). The decrease of PML-NBs was quantified by counting the numbers of PML-NB puncta per cell for 20 cells (Fig. 3B). A considerable decrease in the average number of PML-NBs per nucleus was observed in nsp1β gene-expressing cells compared to pXJ41 or CoV-2-ORF3a-expressing cells. The decrease was pronounced with the average number of 18.5 PML-NBs in control compared to the average number of 5.8 in nsp1β-expressing cells. Our data demonstrates that PML downregulation was mediated by the nsp1β protein and suggests a potential association between the IFN antagonism mediated by nsp1β and the reduction of PML NBs.
Fig. 3.
Downregulation of PML NBs by the nsp1β protein of PRRSV. (A), Numbers of PML-NBs were decreased in PRRSV nsp1β-expressing cells. HeLa cells were transfected with SARS-CoV-2 ORF3a, PRRSV-nsp1β, or empty vector pXJ41 for 24 h and stimulated by incubation with 1000 units/ml of IFN-β for 6 h. The cells were fixed and stained with anti-FLAG MAb (green) and anti-PML PAb (red). Nuclei were stained with DAPI (blue) as described in Materials and Methods. Long white arrows indicate nsp1β-expressing cells and short yellow arrows indicate uninfected cells. The images were taken by confocal microscopy (Nikon A1R). Scale bar represents 5 µm. (B), Quantification of the reduction of PML-NBs in nsp1β-expressing cells. Twenty cells expressing the nsp1β protein were randomly chosen, and numbers of PML-NB puncta per nucleus per cell were counted. The reduction of PML was quantified as described in Materials and Methods. Statistical significance is indicated as follows: ***P<0.01.
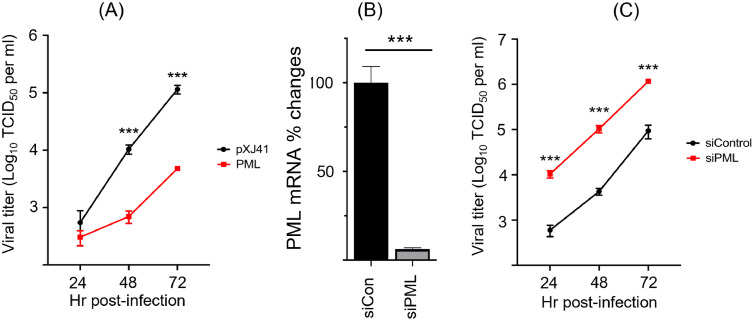
Suppression of PRRSV replication by PML. PML has been shown to possess antiviral activity against a number of viruses (Gongora et al., 2000; Grötzinger et al., 1996; Neerukonda, 2021; Wang et al., 2018). To determine if the antiviral role of PML was also functional for PRRSV, PML isotypes I through VI were overexpressed in MARC-145 cells for 24 h, and the PML-overexpressing cells were infected with PRRSV for 3 days, followed by viral titration. In PML over-expressing cells, the viral titers were decreased at 48 h and 72 h post-infection compared to those of control (Fig. 4A), demonstrating that PML possessed the anti-PRRSV activity and inhibited PRRSV replication.
Fig. 4.
Restriction of PRRSV replication by PML. (A), Suppression of PRRSV replication in PML-overexpressing cells. Plasmids containing the genes for six individual isoforms of PML were equally mixed and transfected to MARC-145 cells for overexpression. At 48 h of PML expression, cells were infected with PRRSV at 1 multiplicity of infection (MOI) for indicated times. Culture supernatants were collected at 24, 48, and 72 h post-infection, and viral titers were determined by TCID50. (B), siRNA-mediated gene silencing of PML as indicated as PML mRNA% changes (Y axis). MARC- 145 cells were grown to 50 % confluence in 6-well plates, and 100 pmol of siRNA per well was transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At 48 h of siRNA treatment, total cellular RNA was extracted and subjected to RT-qPCR using PML-specific primers as described in Materials and Methods. (C), Enhanced replication of PRRSV in PML-knockdown cells. Endogenous PML gene expression was knockdown using siRNAs for 48 h as described above (B), and PML-knockdown cells were infected with PRRSV at 1 MOI. Culture supernatants were collected at 24, 48, and 72 h, and viral titers in the culture supernatants were determined by TCID50. pXJ41, empty vector; Red lines indicate PRRSV titers in PML-overexpressing cells (A) or PML-siRNA (siPML)-treated cells (C). Black lines indicate PRRSV titers in cells treated with control siRNA. Error bars represent means ± standard deviation. The experiments were repeated three times (n = 3). ***, P<0.001.
To confirm the suppression of viral replication by PML, PML expression was knocked down by siRNA-mediated gene silencing, and PML-knockdown cells were infected with PRRSV for 3 days followed by TCID50 titration. The efficacy of PML gene-silencing was first examined (Fig. 4B). Specific small interfering RNA (designated siPML) that targets all six isoforms of PML was transfected to MARC-145 cells, and total cellular RNA was extracted for RT-qPCR for PML mRNAs. As shown in Fig. 4B, siPML effectively downregulated PML-specific mRNA by approximately 95 % compared to control at 24 h post-treatment (Fig. 4B). Then, siPML-transfected cells were infected with PRRSV, and viral titers were determined by TCID50 for viral titration. The viral titers were significantly higher in PML-knockdown cells by more than 1 log as early as 24 h post-infection, and the titer continued to increase until 72 h compared to control cells (Fig. 4C). Together, these data confirm the inhibitory role of PML on PRRSV replication during infection. It is plausible to conclude that the PRRSV protein nsp1β antagonizes the PML-mediated antiviral response and as a result, viral replication is enhanced.
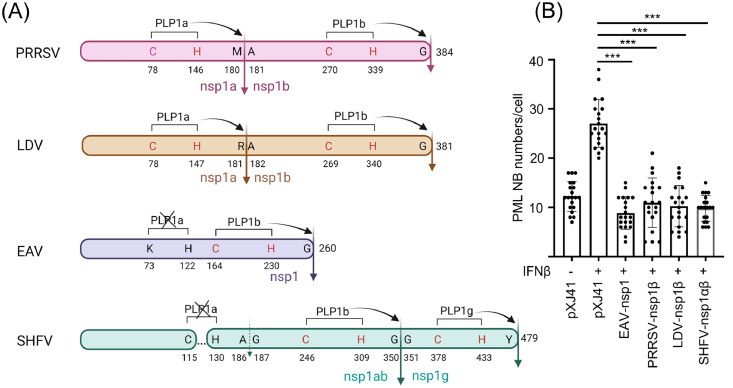
PML downregulation by nsp1 is common in other arteriviruses. PRRSV is a member of the family Arteriviridae, and thus, we expanded our findings to other arteriviruses to examine whether the role of nsp1 against PML was conserved in the family. Besides PRRSV, EAV, LDV of mice, and SHFV are the most studied members in the family, and their processing and biogenesis of nsp1 protein is diverse (Fig. 5A). For PRRSV, cysteine (C) at position 73 and histidine (H) at position 146 are crucial for PLP1α to cleave M180-A181 and release nsp1α subunit. For LDV, nsp1 contains both PLP1α and PLP1β activities, and their catalytic residues C78 as well as H147 are conserved. Thus, nsp1 is cleaved into nsp1α and nsp1β subunits. For EAV, however, C73 is mutated to lysine (K), and this mutation makes the PLP1α inactive, thus leaving nsp1 uncleaved. The processing of SHFV nsp1 is also complicated. C115 and H140 are conserved and mimic the catalytic residues for PLP1α, and A186-G187 is predicted to be the cleavage site. However, a small deletion of 15 residues is found in the region between C115 and H140, which renders PLP1α inactive. Thus, nsp1α subunit is predicted to be uncleaved. Indeed, the inability of SHFV PLP1α to cleave off nsp1α was demonstrated by Han et al. (2014). In contrast, the PLP1β and PLP1γ activities, utilizing C246 and H309 and C378 and H433, respectively, remain active, and thus, cleave at G350-G351 and after Y479, respectively. Thus, nsp1 is cleaved into two subunits nsp1αβ and nsp1γ, instead of 3 subunits. For the present study, therefore, PRRSV nsp1β, LDV nsp1β, EAV nsp1, and SHFV nsp1αβ were selected. Each gene was individually transfected to HeLa cells, and at 48 h post-transfection, PML NBs were examined by co-staining for each subunit and PML. The numbers of PML NBs per cell were then counted and quantified (Fig. 5B). The decrease in the numbers of PML-NBs was significant in cells expressing each of PRRSV nsp1β, LDV nsp1β, EAV nsp1, and SHFV nsp1αβ, compared to the vector control after IFN stimulation, demonstrating that the inhibition of PML NBs was common in arteriviruses.
Fig. 5.
Reduction of PML NBs by nsp1 of different arteriviruses. (A), Biogenesis, structures, and cleavages of nsp1 proteins of arteriviruses. The full-length uncleaved nsp1 proteins for PRRSV, LDV, EAV, and SHFV are 384, 381, 260, and 479 amino acids (aa), respectively. The catalytic amino acid residues for PLPs are indicated in red. Solid vertical arrows represent the PLP-mediated cleavage sites. The dotted vertical arrow for SHFV indicates predicted cleavage site which is in fact uncleaved because PLP1α is inactive due to the deletion between 115 and 130 (dotted horizontal line). Crossing-outs indicate non-functional PLPs. Numbers indicate aa positions with respect to the full length of nsp1. The space between aa positions 115 and 130 in SHFV-nsp1 indicates a deletion in comparison with nsp1 of other arteriviruses (Adapted from Han et al., 2014). (B), Quantification of the reduction of PML-NBs in nsp1β-expressing cells. HeLa cells were transfected with indicated FLAG-tagged arterivirus nsp1 subunit genes for 24 h and stimulated with 1000 units/ml of IFN-β by incubation for 6 h. The cells were fixed and stained with anti-FLAG Ab (green) and anti-PML Ab (red) as described in Materials and Methods. Nuclei were stained with DAPI (blue). The images were taken by confocal microscopy (Nikon A1R). Twenty cells expressing nsp1 were randomly chosen, and numbers of PML-NB puncta per nucleus per cell were counted. The average numbers of PML NBs were calculated for each gene and used to construct the bar graphs. PLP, papain-like proteinase; PRRSV, porcine reproductive and respiratory syndrome virus; LDV, lactate dehydrogenase elevating virus of mice; EAV, equine arteritis virus; SHFV, simian hemorrhagic fever virus. ***, P<0.001. The figure was created with BioRender.com.
Type I IFNs play an important role in the innate immune response of a host. PML is one of the ISGs expressed in response to viral infection and thus is IFN inducible. Since PML NBs are nuclear organelles, nucleus-replicating viruses have mainly been studied for their action on PML to establish a productive infection (Hsu and Kao, 2018). Recent studies, however, indicate that cytoplasm-replicating viruses can also counteract PML functions (Chen et al., 2018; Conde et al., 2020; Dubuisson et al., 2018; El McHichi et al., 2010; Giovannoni et al., 2019). In the present study, we have shown that PRRSV nsp1β protein suppresses PML expression. PRRSV nsp1β is a nuclear protein and functions as an IFN antagonist. It also blocks host mRNA nuclear export and downregulates host cell gene expression (Han et al., 2017; Ke et al., 2019). Nevertheless, a preliminary study shows that PML transcription was not affected in nsp1β-expressing cells, and PML downregulation was a post-translational event (data not shown). Indeed, one of the viral mechanisms for PML downregulation is viral protease-dependent degradation as shown for encephalomyocarditis virus and enterovirus 71 (Chen et al., 2018; El McHichi et al., 2010). For these viruses, 3Cpro cleaves the isoforms of PML and inhibits their expressions. Similarly, PRRSV nsp1β is a viral proteinase, implicating that PLP1β activity of nsp1β may participate in PML degradation. The co-localization of nsp1β with the PML protein supports this possibility, and it will be interesting to determine if PML degradation is mediated by PLPβ. Besides, PML NBs contain UBC9, so it is possible that nsp1β becomes SUMOylated once recruited to NBs. In summary, we have shown that nsp1 of four arteriviruses suppressed the PML expression. This finding implies that arteriviruses may have evolved to establish a common mechanism for PML regulation. Studies are in progress to reveal the mechanism for PML downregulation and immune evasion in arteriviruses.
3. Materials and methods
Cells and viruses. HeLa and MARC-145 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Corning Inc., Corning, NY) supplemented with 10 % heat-inactivated fetal bovine serum (FBS; Gibco, Grand Island, NY), in a humidified incubator with 5 % CO2 at 37 °C. North American genotype PRRSV strain PA8 was propagated in MARC-145 cells and used for this study. For infection, MARC-145 cells were grown to approximately 70 % confluence and infected at a multiplicity of infection of 1. Viral titers were determined by a 50 % tissue culture infectious dose (TCID50) assays in MARC-145 cells.
Antibodies and chemicals. Antibodies and chemicals used in the present study are listed as follows. Anti (α)-PRRSV-N protein MAb (MR40; mouse monoclonal antibody) was obtained from E. Nelson (South Dakota State University, Brookings, SD). Anti-PML PAb (rabbit) (sc-5621) and anti-β-actin MAb (mouse) (C4; sc-47778) were purchased from Santa Cruz Biotechnologies Inc. (Santa Cruz, CA). Anti-FLAG PAb (rabbit) was purchased from Rockland Inc. (Gilbertsville, PA). Anti-FLAG MAb (rat) (M2) was purchased from Agilent (Santa Clara, CA). Alexa-Fluor 488-conjugated, and Alexa-Fluor 568-conjugated secondary antibodies were purchased from ThermoFisher (Rockford, IL). DAPI (4′, 6′-diamidino-2-phenylindol) was obtained from Sigma (St. Louis, MO). Human interferon-β recombinant protein was purchased from Millipore Sigma (St. Louis, MO). For stimulation of HeLa or MARC-145 cells, 1000 units/ml was added to cells for 6 h (Han et al., 2017).
Genes and plasmids. The nsp1β gene was cloned from PRRSV-2 strain VR2332 and inserted as a fusion with the FLAG tag into the pXJ41 expression plasmid as described previously (Han et al., 2017). FLAG-nsp1β of LDV, FLAG-nsp1 of EAV, FLAG-nsp1αβ of SHFV, and FLAG-ORF3a of SARS-CoV-2 were cloned into pXJ41 vectors as described elsewhere (Han et al., 2017; Ke et al., 2018; Su et al., 2021). The HA-PML-I, HA-PML-II, HA-PML-III, HA-PML-IV, HA-PML-V, and HAPML-VI genes were subcloned from individual plasmids phNGX-PML I through VI into pXJ41 using the Hind III and Xho I recognition sequences. E. coli strain DH5α was used for transformation and plasmid DNA preparation.
DNA transfection and immunofluorescence assay. DNA transfection was performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Cells were grown on microscope coverslips and fixed at indicated times with 4 % paraformaldehyde in phosphate-buffered saline (PBS) for 1 h at room temperature (RT), followed by three washes with PBS. The cells were permeabilized with 0.1 % Triton X-100 for 15 min at RT, followed by three washes with PBS. After incubation with 1 % bovine serum albumin (BSA) in PBS for 1 h at RT, cells were incubated with a primary antibody (1:200 dilution) in blocking buffer for 2 h, followed by three washes with PBS and incubation with a secondary antibody (1:200 dilution) for 1 h. Cells were stained with DAPI (1:5000 dilution) for 5 min, and after a final wash with PBS, the coverslips were mounted on microscope slides using Fluoromount-G mounting medium (Southern Biotech, Birmingham, AL). The cells were examined using a Nikon A1R confocal microscope. The reduction of PML was quantified using the following formula: (number of PML NBs per nucleus for 20 cells expressing nsp1) per (20 cells expressing nsp1).
Reverse transcription‐quantitative PCR (RT‐qPCR). Total cellular RNA was extracted using the TRIzol reagent according to the manufacturer's instructions (Invitrogen). RT-qPCR was performed in the ABI Sequence Detector System (ABI Prism 7000 and accompanying software, Applied Biosystems) using a final volume of 25 μl containing 2 μl of cDNA from reverse-transcription reaction, a primer mix (2.5 pM each of sense and antisense primers), 12.5 μl of SYBR Green Master Mix (Applied Biosystems), and 8 μl of distilled water. The primer sequences were listed as follows: for PML, forward 5′-CATCACCCAGGGGAAAGATG-3′, reverse 5′-GGTCAACGTCAATAGGGTCC-3′; for GAPDH, forward 5′-CGGAGTCAACGGATTTGGTCGTA-3′, reverse 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The amplification parameters were 40 cycles of two steps each cycle comprised of heating to 95 °C and 60 °C. The mRNA levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) and normalized using GAPDH.
Statistical analysis. Statistical significance was determined by two-tailed Student's t-test. Data analyses were performed using GraphPad Prism version 9.00 (San Diego California USA).
Declaration of generative AI and AI-assisted technologies in the writing process
The authors declare that they did not use generative artificial intelligence (AI) and AI-assisted technologies in the writing process of this manuscript.
CRediT authorship contribution statement
Chia-Ming Su: Writing – original draft, Methodology, Investigation, Formal analysis, Conceptualization. Yu Fan Hung: Methodology, Investigation, Formal analysis. Junyu Tang: Methodology, Investigation, Formal analysis. Mingyuan Han: Methodology, Investigation, Formal analysis, Conceptualization. Roger Everett: Resources, Methodology. Dongwan Yoo: Writing – review & editing, Validation, Supervision, Project administration, Methodology, Funding acquisition, Data curation, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project was supported by Agriculture and Food Research Initiative (AFRI) Competitive Grants nos. 2018–67015–28287 and 2023–67015–39710 from the U.S. Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA) awarded to DY.
Data availability
Data will be made available on request.
References
- Ashley C.L., Glass M.S., Abendroth A., McSharry B.P., Slobedman B. Nuclear domain 10 components upregulated via interferon during human cytomegalovirus infection potently regulate viral infection. J Gen Virol. 2017;98(7):1795–1805. doi: 10.1099/jgv.0.000858. [DOI] [PubMed] [Google Scholar]
- Bernardi R., Pandolfi P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol. 2010;84(3):1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F., Picard C., Held K., Gross S., Rousseau A., Theil D., Sawtell N., Labetoulle M., Lomonte P. HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons. PLoS Pathog. 2012;8(8) doi: 10.1371/journal.ppat.1002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee A.V., Lopez P., Pandolfi P.P., Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77(12):7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Feng C., Tian X., Zheng N., Wu Z. Promyelocytic leukemia restricts Enterovirus 71 replication by inhibiting autophagy. Front Immunol. 2018;9:1268. doi: 10.3389/fimmu.2018.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wright J., Meng X., Leppard K.N. Promyelocytic leukemia protein isoform ii promotes transcription factor recruitment to activate interferon beta and interferon-responsive gene expression. Mol Cell Biol. 2015;35(10):1660–1672. doi: 10.1128/MCB.01478-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Lawson S., Sun Z., Zhou X., Guan X., Christopher-Hennings J., Nelson E.A., Fang Y. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398(1):87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Li M., He Q., Du J., Zhou L., Ge X., Guo X., Yang H. The amino acid at residue 155 in nonstructural protein 4 of porcine reproductive and respiratory syndrome virus contributes to its inhibitory effect for interferon-β transcription in vitro. Virus Res. 2014;189:226–234. doi: 10.1016/j.virusres.2014.05.027. [DOI] [PubMed] [Google Scholar]
- Conde J.N., Schutt W.R., Mladinich M., Sohn S.Y., Hearing P., Mackow E.R. NS5 sumoylation directs nuclear responses that permit zika virus to persistently infect human brain microvascular endothelial cells. J Virol. 2020;94(19) doi: 10.1128/JVI.01086-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.E., Gack M.U. Ubiquitination in the antiviral immune response. Virology. 2015;479-480:52–65. doi: 10.1016/j.virol.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson L., Lormières F., Fochi S., Turpin J., Pasquier A., Douceron E., Oliva A., Bazarbachi A., Lallemand-Breitenbach V., De Thé H., Journo C., Mahieux R. Stability of HTLV-2 antisense protein is controlled by PML nuclear bodies in a SUMO-dependent manner. Oncogene. 2018;37(21):2806–2816. doi: 10.1038/s41388-018-0163-x. [DOI] [PubMed] [Google Scholar]
- El McHichi B., Regad T., Maroui M.A., Rodriguez M.S., Aminev A., Gerbaud S., Escriou N., Dianoux L., Chelbi-Alix M.K. SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J Virol. 2010;84(22):11634–11645. doi: 10.1128/JVI.01321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni F., Ladelfa M.F., Monte M., Jans D.A., Hemmerich P., García C. Dengue non-structural protein 5 polymerase complexes with promyelocytic leukemia protein (PML) isoforms III and IV to disrupt PML-nuclear bodies in infected cells. Front Cell Infect Microbiol. 2019;9:284. doi: 10.3389/fcimb.2019.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongora C., Degols G., Espert L., Hua T.D., Mechti N. A unique ISRE, in the TATA-less human Isg20 promoter, confers IRF-1-mediated responsiveness to both interferon type I and type II. Nucleic Acids Res. 2000;28(12):2333–2341. doi: 10.1093/nar/28.12.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grötzinger T., Jensen K., Will H. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J Biol Chem. 1996;271(41):25253–25260. doi: 10.1074/jbc.271.41.25253. [DOI] [PubMed] [Google Scholar]
- Han M., Ke H., Zhang Q., Yoo D. Nuclear imprisonment of host cellular mRNA by nsp1β protein of porcine reproductive and respiratory syndrome virus. Virology. 2017;505:42–55. doi: 10.1016/j.virol.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim C.Y., Rowland R.R., Fang Y., Kim D., Yoo D. Biogenesis of non-structural protein 1 (nsp1) and nsp1-mediated type I interferon modulation in arteriviruses. Virology. 2014;458-459:136–150. doi: 10.1016/j.virol.2014.04.028. [DOI] [PubMed] [Google Scholar]
- Han M., Yoo D. Engineering the PRRS virus genome: updates and perspectives. Vet Microbiol. 2014;174(3–4):279–295. doi: 10.1016/j.vetmic.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Yoo D. Modulation of innate immune signaling by nonstructural protein 1 (nsp1) in the family Arteriviridae. Virus Res. 2014;194:100–109. doi: 10.1016/j.virusres.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen C., Monajembashi S., Weisshart K., Hemmerich P. Multimodal light microscopy approaches to reveal structural and functional properties of promyelocytic leukemia nuclear bodies. Front Oncol. 2018;8:125. doi: 10.3389/fonc.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.S., Kao H.Y. PML: regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018;8:5. doi: 10.1186/s13578-018-0204-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes M.A., Faaberg K.S. PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology. 2015;479-480:475–486. doi: 10.1016/j.virol.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H., Han M., Kim J., Gustin K.E., Yoo D. Porcine reproductive and respiratory syndrome virus nonstructural protein 1 beta interacts with nucleoporin 62 To promote viral replication and immune evasion. J Virol. 2019;93(14) doi: 10.1128/JVI.00469-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H., Han M., Zhang Q., Rowland R., Kerrigan M., Yoo D. Type I interferon suppression-negative and host mRNA nuclear retention-negative mutation in nsp1β confers attenuation of porcine reproductive and respiratory syndrome virus in pigs. Virology. 2018;517:177–187. doi: 10.1016/j.virol.2018.01.016. [DOI] [PubMed] [Google Scholar]
- Kim O., Sun Y., Lai F.W., Song C., Yoo D. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402(2):315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5) doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., de Thé H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154–161. doi: 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhu L., Lawson S.R., Fang Y. Targeted mutations in a highly conserved motif of the nsp1β protein impair the interferon antagonizing activity of porcine reproductive and respiratory syndrome virus. J Gen Virol. 2013;94(Pt 9):1972–1983. doi: 10.1099/vir.0.051748-0. [DOI] [PubMed] [Google Scholar]
- Lunney J.K., Fang Y., Ladinig A., Chen N., Li Y., Rowland B., Renukaradhya G.J. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. 2016;4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- McManus F.P., Bourdeau V., Acevedo M., Lopes-Paciencia S., Mignacca L., Lamoliatte F., Rojas Pino J.W., Ferbeyre G., Thibault P. Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9. Sci Rep. 2018;8(1):7754. doi: 10.1038/s41598-018-25150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G. Genomics and evolution of the TRIM gene family. Adv Exp Med Biol. 2012;770:1–9. doi: 10.1007/978-1-4614-5398-7_1. [DOI] [PubMed] [Google Scholar]
- Neerukonda S.N. Interplay between RNA viruses and promyelocytic leukemia nuclear bodies. Vet Sci. 2021;8(4) doi: 10.3390/vetsci8040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Nan Y., Shen M., Ritthipichai K., Zhu X., Zhang Y.J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol. 2010;84(21):11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus C., Harwardt T., Walter B., Marxreiter A., Zenger M., Reuschel E., Nevels M.M. Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1. PLoS Pathog. 2020;16(5) doi: 10.1371/journal.ppat.1008537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Hodgins D.C., Lee C., Calvert J.G., Welch S.K., Jolie R., Keith M., Yoo D. Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res. 2008;135(1):107–114. doi: 10.1016/j.virusres.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Regad T., Saib A., Lallemand-Breitenbach V., Pandolfi P.P., de Thé H., Chelbi-Alix M.K. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. Embo j. 2001;20(13):3495–3505. doi: 10.1093/emboj/20.13.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt M., Wang L., Sommer M., Perrino J., Nour A.M., Sen N., Baiker A., Zerboni L., Arvin A.M. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003;95(1–2):23–33. doi: 10.1016/S0168-1702(03)00161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagong M., Lee C. Porcine reproductive and respiratory syndrome virus nucleocapsid protein modulates interferon-β production by inhibiting IRF3 activation in immortalized porcine alveolar macrophages. Arch Virol. 2011;156(12):2187–2195. doi: 10.1007/s00705-011-1116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M., Schilling E.M., Stamminger T. The human CMV IE1 protein: an offender of PML nuclear bodies. Adv Anat Embryol Cell Biol. 2017;223:77–94. doi: 10.1007/978-3-319-53168-7_4. [DOI] [PubMed] [Google Scholar]
- Scherer M., Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. 2016;90(13):5850–5854. doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Krell P., Yoo D. Nonstructural protein 1α subunit-based inhibition of NF-κB activation and suppression of interferon-β production by porcine reproductive and respiratory syndrome virus. Virology. 2010;407(2):268–280. doi: 10.1016/j.virol.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Su C.M., Du Y., Rowland R.R.R., Wang Q., Yoo D. Reprogramming viral immune evasion for a rational design of next-generation vaccines for RNA viruses. Front Immunol. 2023;14 doi: 10.3389/fimmu.2023.1172000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Shen S., Hu Y., Chen S., Cheng L., Cai Y., Wei W., Wang Y., Rui Y., Yu X.F. SARS-CoV-2 ORF3a inhibits cGAS-STING-mediated autophagy flux and antiviral function. J Med Virol. 2023;95(1):e28175. doi: 10.1002/jmv.28175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Kwon B., Beura L.K., Kuszynski C.A., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus non-structural protein 1 suppresses tumor necrosis factor-alpha promoter activation by inhibiting NF-κB and Sp1. Virology. 2010;406(2):270–279. doi: 10.1016/j.virol.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Ulbricht T., Alzrigat M., Horch A., Reuter N., von Mikecz A., Steimle V., Schmitt E., Krämer O.H., Stamminger T., Hemmerich P. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J Cell Biol. 2012;199(1):49–63. doi: 10.1083/jcb.201112015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent M., Sparrer K.M.J., Gack M.U. TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol. 2018;5(1):385–405. doi: 10.1146/annurev-virology-092917-043323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Nan Y., Yu Y., Zhang Y.J. Porcine reproductive and respiratory syndrome virus Nsp1β inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-α1 degradation. J Virol. 2013;87(9):5219–5228. doi: 10.1128/JVI.02643-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xiao Y., Opriessnig T., Ding Y., Yu Y., Nan Y., Ma Z., Halbur P.G., Zhang Y.J. Enhancing neutralizing antibody production by an interferon-inducing porcine reproductive and respiratory syndrome virus strain. Vaccine. 2013;31(47):5537–5543. doi: 10.1016/j.vaccine.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Wang S., Wang W., Hao C., Yunjia Y., Qin L., He M., Mao W. Antiviral activity against enterovirus 71 of sulfated rhamnan isolated from the green alga Monostroma latissimum. Carbohydr Polym. 2018;200:43–53. doi: 10.1016/j.carbpol.2018.07.067. [DOI] [PubMed] [Google Scholar]
- Wootton S., Yoo D., Rogan D. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch Virol. 2000;145(11):2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Song C., Sun Y., Du Y., Kim O., Liu H.C. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res. 2010;154(1–2):48–60. doi: 10.1016/j.virusres.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Wootton S.K., Li G., Song C., Rowland R.R. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J Virol. 2003;77(22):12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.