Abstract
Three global regulators are known to control antibiotic production by Pseudomonas fluorescens. A two-component regulatory system comprised of the sensor kinase GacS (previously called ApdA or LemA) and GacA, a member of the FixJ family of response regulators, is required for antibiotic production. A mutation in rpoS, which encodes the stationary-phase sigma factor ςS, differentially affects antibiotic production and reduces the capacity of stationary-phase cells of P. fluorescens to survive exposure to oxidative stress. The gacA gene of P. fluorescens Pf-5 was isolated, and the influence of gacS and gacA on rpoS transcription, ςS levels, and oxidative stress response of Pf-5 was determined. We selected a gacA mutant of Pf-5 that contained a single nucleotide substitution within a predicted α-helical region, which is highly conserved among the FixJ family of response regulators. At the entrance to stationary phase, ςS content in gacS and gacA mutants of Pf-5 was less than 20% of the wild-type level. Transcription of rpoS, assessed with an rpoS-lacZ transcriptional fusion, was positively influenced by GacS and GacA, an effect that was most evident at the transition between exponential growth and stationary phase. Mutations in gacS and gacA compromised the capacity of stationary-phase cells of Pf-5 to survive exposure to oxidative stress. The results of this study provide evidence for the predominant roles of GacS and GacA in the regulatory cascade controlling stress response and antifungal metabolite production in P. fluorescens.
Certain strains of fluorescent pseudomonads inhabit root and seed surfaces, where they suppress plant diseases caused by soilborne plant pathogens. Antifungal metabolites produced by Pseudomonas spp. in situ contribute to the suppression of plant disease (47). Pseudomonas fluorescens Pf-5 suppresses plant diseases caused by the fungal pathogens Pythium ultimum (20) and Rhizoctonia solani (19) and produces at least four antifungal secondary metabolites: pyoluteorin (20), pyrrolnitrin (19), 2,4-diacetylphloroglucinol (37), and hydrogen cyanide (HCN) (26). Secondary metabolite production by Pseudomonas spp. does not occur uniformly in all environments but is subject to regulation by genes responding to unknown environmental or physiological signals. Mutations in regulatory genes that alter antifungal metabolite production can improve or diminish biological control by P. fluorescens (5, 12, 30, 44). Therefore, elucidation of molecular mechanisms regulating antifungal metabolite production of P. fluorescens is likely to provide opportunities for enhancement of biological control.
In P. fluorescens, antifungal metabolite production and biological control are controlled by a two-component regulatory system comprised of GacS and GacA, which are highly conserved among Pseudomonas spp. (25, 41). gacA (12, 30) encodes a response regulator in the FixJ family, and gacS (also called apdA, lemA, repA, or pheN) (6, 25) encodes the cognate sensor kinase. GacS (for global activator sensor kinase) was renamed recently to reflect the high degree of deduced amino acid sequence similarity and functional conservation among homologues present in various species of Pseudomonas (25). gacS and gacA are required for production of pyrrolnitrin, pyoluteorin, 2,4-diacetylphloroglucinol, HCN, extracellular protease(s), and tryptophan side chain oxidase (TSO) by strains of P. fluorescens. gacS and gacA mutants produce none of these secondary metabolites or exoenzymes (6, 12, 30) and are less effective than wild-type strains in suppressing disease (5, 12, 30, 39). Mutants with nucleotide substitutions in gacA accumulate in late-stationary-phase cultures of P. fluorescens (7, 10). The functional gacA allele gacA(Y49), which specifies a tyrosine residue at position 49 (30), apparently was isolated from such a mutant of P. fluorescens CHA0. The wild-type gacA gene from strain CHA0, termed gacA(D49), encodes an aspartate residue at position 49 (3, 40).
The stationary-phase sigma factor ςS is a third regulator of antibiotic production in P. fluorescens. In Escherichia coli, ςS directs the transcription of many genes expressed upon entry into stationary phase (31) and in response to starvation (34, 38) or osmotic stress (17). Some genes transcribed by the ςS-RNA polymerase holoenzyme confer stress tolerance on stationary-phase cells of E. coli (34, 42). In P. fluorescens Pf-5, an rpoS mutation is pleiotropic, reducing the bacterium’s capacity to survive oxidative stress and altering the spectrum of secondary metabolite production (44). An rpoS mutant of Pf-5 overproduces pyoluteorin and 2,4-diacetylphloroglucinol but produces no pyrrolnitrin (39, 44). Characterization of ςS-regulated phenotypes of Pf-5 provided the first evidence that a single regulatory gene can control both antibiotic production and stress response in P. fluorescens (44).
The research described here was undertaken to determine if the GacS-GacA two-component regulatory system and ςS interact or operate through independent regulatory circuits in Pf-5. In this study, we describe the nucleotide sequence of the gacA gene of Pf-5 and demonstrate that GacS and GacA influence ςS accumulation and rpoS transcription in Pf-5. We also demonstrate that gacS and gacA, like rpoS, are required for optimal survival of stationary-phase cells of Pf-5 when exposed to oxidative stress.
(Portions of this work were published earlier as abstracts [7, 48].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. P. fluorescens was grown at 27°C, with shaking at 200 rpm, in King’s medium B broth (KMB) (24) for routine culturing; in KMB broth amended with glycine (4.4 g/liter) for HCN assays; in Luria-Bertani (LB) medium (43) for transcriptional fusion studies and Western analysis; in nutrient broth (Difco Laboratories, Detroit, Mich.) supplemented with 2% (wt/vol) glucose or 1% (wt/vol) glycerol for antibiotic extractions; in nutrient broth supplemented with 1% (wt/vol) glycerol for TSO assays; or in M9 minimal medium (M9) supplemented with 0.4% glucose (43) for Western analysis and oxidative stress tests. Cells of P. fluorescens were enumerated by spreading serial dilutions of bacterial suspensions on KMB. Cultures of E. coli were routinely grown in LB medium at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| P. fluorescens | ||
| Pf-5 | Rhizosphere isolate | 19 |
| JL3985 | Derivative of Pf-5, rpoS::Tn5 | 44 |
| JL4135 | Derivative of Pf-5, gacS::Tn5 | 6 |
| JL4477 | Derivative of Pf-5, gacA(V203) | This study |
| JL4489 | Derivative of Pf-5, rpoS::lacZ | This study |
| JL4491 | Derivative of JL4135, gacS::Tn5 rpoS::lacZ | This study |
| JL4492 | Derivative of JL4477, gacA(V203) rpoS::lacZ | This study |
| E. coli DH5α | F−endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 λ− | 43 |
| Plasmids | ||
| pUC19 | ColE1 replicon, Ampr | 43 |
| pRK2013 | Mobilizing plasmid, Tra+ Kmr | 11 |
| pRK415 | IncP1 replicon, polylinker of pUC19, Mob+ Tcr | 23 |
| pME3066 | 1.65-kb BamHI-BglII fragment containing gacA(Y49) from P. fluorescens CHA0 cloned in pLAFR3, Mob+ Tcr | 30 |
| pMini-Tn5lacZ1 | Mini-Tn5 containing promoterless lacZ on a 4.1-kb SmaI fragment cloned in pUT, Kmr Tcr | 8 |
| pJEL01 | Stably maintained in E. coli or Pseudomonas spp., replicons from pVSP1 and pACYC184, Mob+ Tcr | 44 |
| pJEL5649 | 2.9-kb EcoRI fragment containing rpoS from Pf-5 cloned in pJEL01, Mob+ Tcr | 44 |
| pJEL5926 | rpoS-lacZ transcriptional fusion cloned in pRK415, Mob+ Tcr | This study |
| pJEL5937 | 1.65-kb BamHI-BglII fragment containing gacA from Pf-5 cloned in pUC19, Ampr | This study |
Abbreviations: Ampr, Kmr, and Tcr, resistance to ampicillin, kanamycin, and tetracycline, respectively.
Recombinant DNA techniques.
Methods for transformations, digestions with restriction enzymes, and gel electrophoresis were standard (43). Blunt-end ligation was performed by the thermal cycling method (33). Enzymes were from Gibco BRL Life Technologies (Gaithersburg, Md.). Plasmids were purified by an alkaline lysis procedure (43). Plasmids were mobilized from E. coli DH5α donors into Pf-5 in triparental matings with helper plasmid pRK2013 (11). Transconjugants were selected on KMB containing 200 μg of tetracycline per ml.
Derivation of a gacA mutant of Pf-5.
Strain JL4477, a derivative of Pf-5 containing a point mutation in gacA, was selected by the method described by Duffy and Défago (10). Pf-5 was grown in nutrient broth amended with 0.5% yeast extract at 27°C. After 6 days, dilutions of cultures were spread onto LB agar. Colonies that appeared orange in comparison to the wild-type strain after several days incubation at 27°C (a characteristic of gacA mutants [10]) were screened for loss of extracellular protease activity on Bacto Litmus milk agar (Difco). Protease-deficient mutants were evaluated for antibiotic production by reverse-phase thin-layer chromatography as described previously (26).
Cloning of gacA from Pf-5.
An extant genomic library of Pf-5 (39) was screened by colony hybridization (14) to identify cosmids that hybridized to gacA(Y49) of P. fluorescens CHA0 (30). The gacA(Y49) probe, a 1.65-kb BamHI-BglII fragment of pME3066, was labeled with [32P]dCTP or biotinylated dATP by using a nick translation kit (Gibco BRL Life Technologies) and purified over a D50 column (International Biotechnologies Inc., New Haven, Conn.). Southern analysis identified restriction fragments in cosmids that hybridized to the gacA(Y49) probe. A 1.65-kb BamHI-BglII fragment that hybridized to the probe was cloned into pUC19 to construct pJEL5937.
Sequence analysis of gacA alleles.
DNA sequencing and oligonucleotide syntheses were done at the Center for Gene Research and Biotechnology at Oregon State University, Corvallis. Sequencing of double-stranded templates was done on an ABI model 373A automated DNA sequencer using a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) according to the manufacturer’s protocol. Oligonucleotide primers were synthesized on an ABI model 380B DNA synthesizer using phosphoramidite chemistry (1). Sequencing of the gacA gene of Pf-5 was done with primers complementary to pUC19 DNA on either side of the polylinker and by oligonucleotide primers complementary to regions within the 1.65-kb fragment of pJEL5937 containing gacA. Sequencing of an allele of gacA with a point mutation [termed gacA (V203)] was performed directly on the PCR product amplified from the genome of JL4477 with primers designed from the sequence of the gacA gene cloned in pJEL5937. Analyses of DNA and deduced protein sequences and comparisons with sequences in the GenBank database were accomplished with software from the Genetics Computer Group, Inc., Madison, Wis. (9). Theoretical secondary structures of proteins encoded by alleles of gacA were predicted by PepPlot and PlotStructure programs (Genetics Computer Group).
Antibiotic quantification.
Antibiotics were extracted from cells and spent media of cultures grown in triplicate as described previously (37). Pyoluteorin and pyrrolnitrin concentrations were quantified from cultures grown for 2 days at 20°C in 5 ml of nutrient broth containing 1% glycerol, a medium that favors their production. The concentration of 2,4-diacetylphloroglucinol was quantified from cultures grown for 4 days in 5 ml of nutrient broth containing 2% glucose, a medium that favors its production. Restoration of antibiotic production in JL4477 harboring plasmid pME3066 was assessed in the absence of tetracycline, which decreased the growth rate of the strain. Culture supernatants were extracted twice with ethyl acetate, and excess water was removed with anhydrous MgSO4. The bacterial pellet was extracted with acetone. Extracts dissolved in methanol were analyzed by C18 reverse-phase high-performance liquid chromatography (0.8- by 10-cm Waters Nova-Pak radial compression cartridge; 45% water–30% acetonitrile–25% methanol [vol/vol]; 1.5 ml/min). Antibiotics were detected with a UV photodiode array detector at 225 (pyrrolnitrin), 310 (pyoluteorin), and 278 (2,4-diacetylphloroglucinol) nm and quantified against authentic standards. Quantification was done twice, with similar results.
Exoenzyme production.
Extracellular protease was assessed visually as a cleared zone around bacterial colonies on Bacto Litmus milk agar (Difco) following incubation at 27°C for 48 h.
TSO production was quantified from duplicate cultures of P. fluorescens grown at 27°C for 48 h with shaking. Cells from 1 ml of culture were harvested, washed, and suspended in 100 μl of ice-cold 50 mM potassium phosphate, pH 6.0. Cells were lysed by two sequential cycles of rapid freezing in liquid nitrogen followed by thawing at 45°C. Cell debris was harvested at 4°C, and the supernatant was incubated at room temperature in 1.0 mM acetyl-l-tryptophanamide–50 mM potassium phosphate, pH 6.0. The production of N-acetyl-α,β-didehydrotryptophanamide was monitored spectrophotometrically at 333 nm (ɛmM of 19.8 cm−1) at 5-min intervals for 30 min and at 60, 120, and 180 min (36). The rate of production (amount of N-acetyl-α,β-didehydrotryptophanamide produced per minute) was determined from the linear portion of a curve relating absorbance at 333 nm to time. TSO production was normalized to CFU and reported as enzymatic units per 1010 CFU. One unit of TSO is defined as the amount of enzyme that catalyzed the formation of 1 μmol of N-acetyl-α,β-didehydrotryptophanamide/min (36). Quantification was done twice, with similar results.
HCN production.
To quantify HCN production (15, 16), duplicate cultures were grown at 27°C for 48 h with shaking. A sample from each culture was incubated in the presence of 0.1 N NaOH at room temperature for 3 h in a chamber sealed with paraffin. The NaOH fraction was diluted with 0.1 N NaOH to a concentration within the linear range of a standard curve relating the concentration of an NaCN standard to absorbance at 578 nm; 0.04 ml of the diluted NaOH fraction was added to 1.0 ml of a solution comprised of 2 parts 0.2 M 4-nitrobenzaldehyde in ethylene glycol monomethyl ether, 2 parts 0.1 M o-dinitrobenzene in ethylene glycol monomethyl ether, and 1 part 0.088 N NaOH. After 25 min of incubation in the dark at room temperature, absorbance at 578 nm was measured. HCN was quantified against an NaCN standard curve and normalized to CFU. Quantification was done twice, with similar results.
Western analysis of ςS and GacS.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transblotting for Western analysis were performed as specified by the manufacturer (Bio-Rad Laboratories, Hercules, Calif.). Exponential-phase cells were obtained from cultures grown to an optical density at 600 nm (OD600) of 0.2 to 0.4 (for reference, see Fig. 3A, t = 0 to 1 h). Early-stationary-phase cells were obtained from cultures grown until the optical density stopped increasing exponentially (see Fig. 3A, t = 2 to 3 h), and an additional stationary-phase sample was obtained from cultures 4 h later (see Fig. 3A, t = 6 to 7 h). Optical densities were used to estimate the volume of each culture that would provide an equivalent number of bacterial cells. Cells from that volume were harvested, immediately frozen in an ethanol–dry-ice bath, and extracted by boiling in protein sample buffer (Bio-Rad) containing 5% 2-mercaptoethanol. Proteins were separated by SDS-PAGE on a 12% gel and transferred onto a nitrocellulose membrane (Bio-Rad) for Western analysis. Blots were incubated with polyclonal antibodies to E. coli ςS, generously supplied by K. Tanaka (46), and the antibodies were detected by enhanced chemiluminescence as specified by the manufacturer (Amersham Life Science Inc., Arlington Heights, Ill.). Blots were stripped at 65°C for 30 min in a solution of 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl, pH 6.7. Blots were then incubated with polyclonal antibodies to GacS (previously LemA) from Pseudomonas syringae, generously supplied by T. Kitten and D. K. Willis (41), and the antibodies were detected by enhanced chemiluminescence. ςS and GacS were quantified by using a Molecular Dynamics model SI personal densitometer and ImageQuant software, version 4.1 (Molecular Dynamics, Sunnyvale, Calif.), and the results were normalized based on Bradford assays for total protein (Bio-Rad). In the absence of a purified standard for quantification, the linear range of ςS detection was determined by using a dilution series of Pf-5 protein extracts. Samples quantified were within the linear range of detection. Cell content of ςS and GacS is reported relative to the amount in stationary-phase Pf-5 cells. Each experiment was done twice, with similar results.
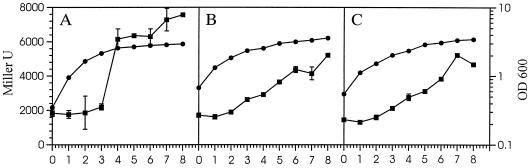
FIG. 3.
Growth and β-galactosidase activity of Pf-5 derivatives containing chromosomal rpoS-lacZ transcriptional fusions. OD600 (•) and β-galactosidase activity expressed in Miller units (■) were determined at 1-h intervals from duplicate cultures grown in LB medium. Bacterial strains were JL4489 (rpoS::lacZ) (A), JL4491 (gacS::Tn5 rpoS::lacZ) (B), and JL4492 [gacA(V203) rpoS::lacZ] (C). Error bars representing standard deviations may be obscured by symbols.
Transcription of rpoS.
A transcriptional fusion of lacZ to rpoS of Pf-5 was constructed by inserting a 4.1-kb blunt-ended SmaI fragment from pMini-Tn5lacZ1 (8) into a blunted XhoI site, located 36 nucleotides from the 3′ end of rpoS. The rpoS::lacZ transcriptional fusion cloned in pJEL5926 was exchanged with the genomic copy of rpoS in Pf-5 to derive JL4489, in JL4135 to derive JL4491, and in JL4477 to derive JL4492 by marker exchange mutagenesis as described previously (27). The rpoS::lacZ mutation in each strain was complemented with pJEL5649, a multicopy plasmid carrying the wild-type rpoS gene. From duplicate cultures of each strain grown in LB medium, β-galactosidase activity was determined at 1-h intervals for 8 h as described by Miller (35). Cells were made permeable with SDS and CHCl3 and then incubated for 10 min at 28°C, after which o-nitrophenyl-β-d-galactopyranoside was added to a final concentration of 0.66 mg/ml. β-Galactosidase was expressed as Miller units (35), and numbers of CFU were determined to verify that optical density was an accurate representation of cell density in all strains. The experiments were done twice, with similar results.
Stress response.
Survival of P. fluorescens following exposure to H2O2 was determined as previously described (44), with slight modifications. Cultures were grown in M9 with 0.4% glucose, and stationary-phase cells were harvested at 4 and 8 h after the optical density of cultures stopped increasing. Harvested cells were washed once and suspended in 5 ml of M9 without glucose to obtain an OD600 of 0.8. Suspended cells were exposed to 15 mM H2O2 and incubated with shaking at 27°C for 1 h; then CFU were enumerated at 20-min intervals. Three replicate cultures were evaluated for each treatment. The experiment was done twice, with similar results.
Nucleotide sequence accession number.
The GenBank accession number for the DNA sequence of the gacA gene of P. fluorescens Pf-5 is AF065156.
RESULTS
Sequence analysis of gacA alleles.
A 1.65-kb BamHI-BglII fragment that hybridized to gacA from P. fluorescens CHA0 (30) was identified from a genomic library of Pf-5. The deduced amino acid sequence of a 639-bp open reading frame present on the fragment is identical to that of the wild-type GacA(D49) of P. fluorescens CHA0 (40), and the open reading frame was therefore identified as gacA of Pf-5. JL4477, a spontaneous mutant of Pf-5 exhibiting the colony morphology described for gacA mutants of CHA0 (10), had an allele of gacA with a T rather than a C at nucleotide 607. Consequently, the deduced amino acid sequence of the mutant allele, heretofore called gacA(V203), has a valine rather than an alanine at position 203. The 314 nucleotides immediately upstream of gacA in JL4477 were identical to those upstream of gacA in Pf-5.
Phenotypic analysis of gacS::Tn5, gacA(V203), rpoS::Tn5, and rpoS::lacZ derivatives of Pf-5.
Pf-5 produced pyrrolnitrin, 2,4-diacetylphloroglucinol, pyoluteorin, extracellular protease(s), HCN, and TSO (Table 2). JL4135 (gacS::Tn5) produced no detectable antibiotics, extracellular protease(s), HCN, or TSO. JL4477 [gacA(V203)] produced no detectable antibiotics or extracellular protease(s), and it produced less HCN and TSO than Pf-5 produced. Production of pyoluteorin, pyrrolnitrin, 2,4-diacetylphloroglucinol, HCN, TSO, and extracellular protease(s) by JL4477 [gacA(V203)] was restored with plasmid pME3066, containing a functional gacA(Y49) allele from CHA0.
TABLE 2.
Secondary metabolite and exoenzyme production by P. fluorescens Pf-5 and derivatives
| Strain | Characteristics | Concn (mean ± SD)
|
Extracellular protease(s)d | ||||
|---|---|---|---|---|---|---|---|
| Pyrrolnitrin (μg/ml)a | 2,4-Diacetylphloro-glucinol (μg/ml)a | Pyoluteorin (μg/ml)a | HCN (μmol/ 1010 CFU)b | TSO (U/1010 CFU)c | |||
| Pf-5 | Wild type | 3.6 ± 0.4 | 26.2 ± 0.5 | 9.4 ± 1.5 | 30 ± 1 | 2,290 ± 290 | + |
| JL4135 | gacS::Tn5 | − | − | − | − | − | − |
| JL4477 | gacA(V203) | − | − | − | 9 ± 1 | 4 ± 3 | − |
| JL4477(pME3066) | gacA(V203) gacA(Y49) | 1.1 ± 0.4 | 32.3 ± 1.4 | 28.4 ± 1.1 | 38 ± 13 | 390 ± 3 | + |
| JL3985 | rpoS::Tn5 | − | 60.5 ± 15.6 | 30.9 ± 0.1 | 88 ± 15 | − | + |
| JL4489 | rpoS::lacZ | 0.5 ± 0.1 | 29.0 ± 1.1 | 27.8 ± 0.1 | 110 ± 34 | 59 ± 18 | + |
Mean from three replicate cultures; −, below the detection limit of 0.1 μg/ml.
Mean from two replicate cultures; −, below the detection limit of 1 μM.
Mean from two replicate cultures; −, below the detection limit of 0.1 U. One unit is defined as the amount of enzyme that catalyzed the formation of 1 μmol of N-acetyl-α,β-didehydrotryptophanamide/min (36).
+, detected; −, not detected.
Two derivatives of Pf-5 containing insertions in rpoS produced more pyoluteorin and HCN and less pyrrolnitrin than was produced by the wild-type strain (Table 2). JL3985 (rpoS::Tn5) overproduced 2,4-diacetylphloroglucinol, whereas JL4489 (rpoS::lacZ) produced wild-type levels of this antibiotic. Both JL3985 and JL4489 produced an extracellular protease(s). JL3985 produced no detectable TSO, but JL4489 produced trace levels of TSO. In JL4489, the lacZ insertion is located 36 nucleotides from the 3′ terminus of rpoS, corresponding to domain 4.2 of ςS, which is involved in recognition of the −35 region of target promoters (32). Like JL4489, mutants of E. coli that produce ςS proteins with altered lengths, due to insertions or deletions in the C-terminal region adjacent to domain 4.2, confer phenotypes that differ quantitatively from the wild-type phenotype (21, 49).
ςS accumulation.
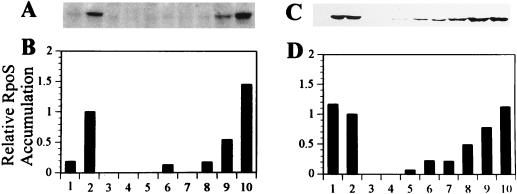
In LB medium, ςS was detected at a low level in exponentially growing cells of Pf-5. Upon entry into stationary phase, the cellular content of ςS increased by 500% (Fig. 1A and B, lanes 1 and 2). ςS was not detected in exponentially growing cells of JL4135 (gacS::Tn5) or JL4477 [gacA(V203)] (Fig. 1A and B, lanes 5 and 7). At the entrance to stationary phase, ςS content in JL4135 and JL4477 was less than 20% of the wild-type level (Fig. 1A and B, lanes 2, 6, and 8). Four hours after exponential growth ceased, the ςS content in JL4135 and JL4477 was 50% of that observed in Pf-5 (data not shown). Multiple copies of rpoS enhanced levels of ςS in derivatives of Pf-5 grown in LB medium (Fig. 1A and B, lanes 1, 2, 9, and 10).
FIG. 1.
Relative ςS accumulation. ςS was visualized with antibodies to E. coli ςS (46) from Western blots of protein extracted from cultures grown in LB medium (A and B) or M9 containing 0.4% glucose (C and D). ςS content in each lane was estimated by scanning Western blots with a densitometer. For each sample, ςS content was normalized to total protein content, determined from Bradford assays. The normalized ςS content (ςS content divided by total protein content) for each sample is reported relative to the normalized ςS content of stationary-phase cells of Pf-5 grown in the corresponding medium (B and D, lanes 2). Sample numbers correspond to extracts from cells growing exponentially (first of each pair; lanes 1, 3, 5, 7, and 9) or in early stationary phase (second of each pair; lanes 2, 4, 6, 8, and 10): 1 and 2, Pf-5; 3 and 4, JL3985 (rpoS::Tn5); 5 and 6, JL4135 (gacS::Tn5); 7 and 8, JL4477 [gacA(V203)]; 9 and 10, Pf-5 harboring pJEL5649, a multiple-copy plasmid containing rpoS cloned from Pf-5. Scanned images were reproduced for publication by using Adobe Photoshop version 4.0 (Adobe Systems Incorporated, San Jose, Calif.).
ςS was detected both in exponentially growing cells and in early-stationary-phase cells of Pf-5 in M9 containing 0.4% glucose (Fig. 1C and D, lanes 1 and 2). In contrast, the content of ςS in exponentially growing cells of JL4135 (gacS::Tn5) and JL4477 [gacA(V203)] was less than 20% of the wild-type level (Fig. 1C and D, lanes 1, 5, and 7). The contents of ςS in early-stationary-phase cells of JL4135 and JL4477 were 20 and 50%, respectively, of the wild-type level (Fig. 1C and D, lanes 2, 6, and 8). Four hours after exponential growth ceased, the ςS content increased in JL4135 and JL4477 to 40 and 50%, respectively, of that observed in Pf-5 (data not shown).
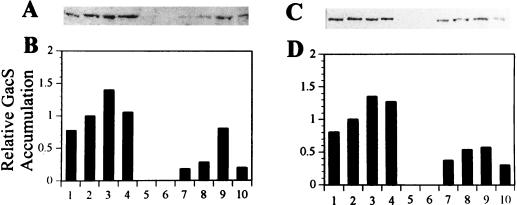
GacS accumulation.
The level of GacS increased slightly (by 20%) in Pf-5 during the transition from exponential growth to stationary phase (Fig. 2, lanes 1 and 2). The cellular GacS content was less in JL4477 [gacA(V203)] than in Pf-5, for both growth phases and culture media (Fig. 2, lanes 1, 2, 7, and 8). In exponentially growing cells, the GacS content in JL3985 (rpoS::Tn5) was greater than that in Pf-5 in one experiment (Fig. 2, lanes 1 and 3), but no difference was observed in a second experiment (data not shown). Multiple plasmid-borne copies of rpoS decreased GacS levels in stationary-phase cells of Pf-5 (Fig. 2, lanes 2 and 10) in both experiments.
FIG. 2.
Relative GacS accumulation. GacS was visualized with antibodies to GacS from P. syringae (41) on the Western blots used for quantification of ςS (Fig. 1). Protein was extracted from cultures grown in LB medium (A and B) or M9 containing 0.4% glucose (C and D). GacS content in each lane was estimated and reported as normalized values (B and D, lanes 2). Sample numbers correspond to extracts from cells growing exponentially (first of each pair; lanes 1, 3, 5, 7, and 9) or in early stationary phase (second of each pair; lanes 2, 4, 6, 8, and 10): 1 and 2, Pf-5; 3 and 4, JL3985 (rpoS::Tn5); 5 and 6, JL4135 (gacS::Tn5); 7 and 8, JL4477 [gacA(V203)]; 9 and 10, Pf-5 harboring pJEL5649, a multiple-copy plasmid containing rpoS cloned from Pf-5. The truncated form of GacS was not quantified in JL4135 (gacS::Tn5). Scanned images were reproduced for publication by using Adobe Photoshop version 4.0 (Adobe Systems Incorporated).
Transcription of rpoS assessed with a lacZ fusion.
β-Galactosidase activity conferred by a chromosomal rpoS-lacZ transcriptional fusion increased by 300% within a 1-h period when strain JL4489 (rpoS::lacZ) began the transition from exponential to stationary phase (Fig. 3A), reflecting an increase in rpoS transcription. In JL4491 (gacS::Tn5 rpoS::lacZ) (Fig. 3B) and JL4492 [gacA(V203) rpoS::lacZ] (Fig. 3C), induction of rpoS transcription occurred more gradually and to a smaller magnitude than in strains with functional GacS and GacA proteins (Fig. 3A). Multiple plasmid-borne copies of rpoS decreased β-galactosidase activity of stationary-phase cells of JL4489 by 50% (data not shown).
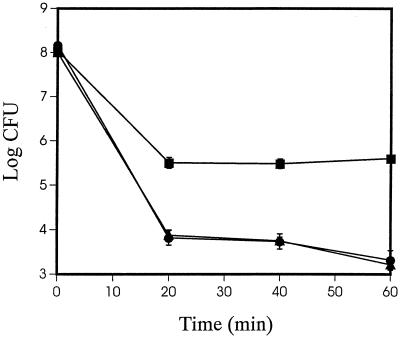
Survival of Pf-5 and gacS::Tn5 and gacA(V203) derivatives when exposed to oxidative stress.
In addition to influencing antibiotic and exoenzyme production, ςS influences the capacity of Pf-5 to survive oxidative stress (44). Stationary-phase cells of JL3985 (rpoS::Tn5) are more sensitive than stationary-phase cells of Pf-5 to hydrogen peroxide (44). Similarly, JL4135 (gacS::Tn5) and JL4477 [gacA(V203)] harvested from cultures 4 h (Fig. 4) or 8 h (data not shown) after cell density stopped increasing were more sensitive than Pf-5 to exposure to oxidative stress.
FIG. 4.
Survival of oxidative stress. Stationary-phase cells of P. fluorescens Pf-5 (■), JL4135 (gacS::Tn5) (•), and JL4477 [gacA(V203)] (▴) were exposed to 15 mM H2O2, and the numbers of culturable cells were estimated over time. Presented values are means of three replicate cultures; error bars representing standard deviations may be obscured by symbols.
DISCUSSION
This report provides the first evidence that the global regulators GacS and GacA influence the ςS accumulation and stress response of stationary-phase cells of Pseudomonas spp. Mutations in gacS and gacA reduced ςS accumulation in P. fluorescens Pf-5 and compromised the bacterium’s capacity to survive exposure to oxidative stress. These data are consistent with the hypothesis that the two-component regulatory system comprised of GacS and GacA can regulate gene expression by influencing ςS levels. Nevertheless, GacS and GacA are required for the expression of certain phenotypes (such as pyoluteorin and 2,4-diacetylphloroglucinol production) that are not positively regulated by rpoS, indicating that the two-component regulatory system also must function through a mechanism other than the control of ςS.
Transcription of rpoS, assessed with a lacZ fusion, was positively influenced by GacS and GacA in Pf-5, which is consistent with the pattern of rpoS transcription in Pseudomonas aeruginosa (29, 40). In that species, GacA positively controls the production of the autoinducer N-butyryl-homoserine lactone (40), which, through its interaction with the response regulator RhlR, positively influences the expression of rpoS (29). An autoinducer involved in quorum sensing has not been found in Pf-5, but it could be among the unknown components of regulatory circuits controlling antibiotic production and stress response in P. fluorescens.
GacS accumulation was diminished in the gacA(V203) mutant of Pf-5 and in the presence of multiple plasmid-borne copies of rpoS. Positive regulation of GacS content by GacA may be one mechanism by which the relative concentration of the two proteins is controlled. The proper stoichiometric balance of other response regulators within the FixJ family and their cognate sensor kinases is required for normal function of these two-component regulatory systems (18). In P. fluorescens CHA0 (40), multiple copies of gacA(D49) are not tolerated, and in Pf-5, multiple copies of gacA can partially compensate for gacS mutations (7), indicating that the system is sensitive to relative GacS and GacA contents. The regulatory mechanisms through which GacA and ςS influence GacS accumulation are not known, but the findings of this study highlight the complexity of interactions among the three global regulators.
Based on sequence similarities to better-characterized response regulators within the FixJ family, GacA contains two functional domains, an amino-terminal phosphorylation-induced activator domain and a carboxy-terminal output domain characterized by a helix-turn-helix DNA-binding motif (22). Amino acid substitutions within these functional domains typically destroy GacA function, manifested in the loss of multiple phenotypes controlled by the GacS-GacA two-component regulatory system (2). The gacA(V203) mutant evaluated in this study differed from those described previously because it lost only a subset of phenotypes controlled by the two-component regulatory system. Analysis of the theoretical secondary structures of GacA(V203) and GacA indicated that the valine substitution may interrupt an α-helical region, which is downstream of the helix-turn-helix motif and highly conserved within the FixJ family (22). This possibility is consistent with valine’s assignment as a strong β-sheet-forming residue, whereas the replaced alanine residue is a strong α-helix-forming residue (4, 13). Stibitz (45) demonstrated that mutations within this α-helical region in the response regulator BvgA eliminate expression of two genes but have little effect on the expression of a third gene under the control of the BvgS-BvgA two-component system. Thus, the differential effect of the gacA(V203) mutation on phenotypes regulated by GacS and GacA in Pf-5 is not unprecedented and may reflect the importance of the α-helical region as a specificity determinant for recognition of various promoters by GacA. The binding site(s) of GacA has not been described, however, and further exploration of this possibility would be facilitated by the identification of such target sequences.
The effects of gacS and gacA mutations on rpoS transcription and ςS accumulation were greater during the transition from exponential growth than later in stationary phase. Nevertheless, the ςS-mediated stress response of gacS and gacA(V203) mutants was diminished well into stationary phase. Induction of rpoS transcription at the transition between exponential and stationary phases may be critical to the process through which cells develop resistance to environmental stress. Consequently, the stress-resistant state could fail to develop fully if the level of ςS increases gradually or later in stationary phase. Although the present study focused on rpoS transcription and ςS accumulation, GacS and GacA also could influence rpoS translation or ςS stability. Indeed, posttranscriptional regulation plays a prominent role in controlling levels of ςS in E. coli (28, 50). In the event that GacS and GacA affect rpoS translation or ςS stability, the influence of the two-component regulatory system would likely persist beyond the transition period when rpoS transcription was most notably influenced.
ACKNOWLEDGMENTS
We thank T. Kitten and D. K. Willis for providing anti-GacS antiserum, K. Tanaka for providing anti-RpoS antiserum, D. Gentry and N. Thompson for supplying additional antisera evaluated for use in the study, J. LaVille and D. Haas for providing strains, F. Sidaner for evaluating antisera, and B. Nowak-Thompson for assisting in antibiotic quantification. We also thank D. K. Willis, C. T. Bull, and K. M. Culligan for reviewing the manuscript.
This work was supported in part by grant 93-38420-8753 from the U.S. Department of Agriculture, Food and Agricultural Sciences National Needs Graduate Fellowships Program (N.A.C. and C.A.W.), and by grant U-915213-01 from the U.S. Environmental Protection Agency, Science to Achieve Results Fellowships Program (C.A.W.).
REFERENCES
- 1.Alvarado-Urbina G, Chiarello R, Roberts E, Vilain G, Jurik F, Christensen L, Carmona C, Fang L, Watterson M, Crea R. Rapid automated synthesis via diisopropyl phosporamidite in situ activation. Chemical synthesis and cloning of a calmodulin gene. Biochem Cell Biol. 1986;64:548–555. doi: 10.1139/o86-077. [DOI] [PubMed] [Google Scholar]
- 2.Black T, Lam S, Gaffney T. Abstract book, 8th International Congress of Molecular Plant-Microbe Interactions, Knoxville, Tenn. 1996. Transcriptional analysis of LemA, GacA, RpoS and GacA-regulated genes in Pseudomonas fluorescens BL915. [Google Scholar]
- 3.Bull, C. T. 1998. Personal communication.
- 4.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–147. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Corbell, N. A. Unpublished data.
- 6.Corbell N A, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbell N A, Reifenrath C, Loper J E. Abstracts of the Sixth International Congress on Pseudomonas, Madrid, Spain. 1997. Global regulation of secondary metabolites in Pseudomonas fluorescens Pf-5: effects of point mutation and copy number of gacA; p. 30. [Google Scholar]
- 8.de Lorenzo V, Herrero M, Jacubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy B K, Défago G. Influence of cultural conditions on spontaneous mutations in Pseudomonas fluorescens CHA0. Phytopathology. 1995;85:1146. [Google Scholar]
- 11.Figurski K H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaffney T D, Lam S T, Ligon J, Gates K, Razelle A, Di Maio J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, Kempf H J, Becker J O. Global regulation of expression of anti-fungal factors by a Pseudomonas fluorescens biological control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 13.Garnier J, Osguthorpe D J, Robson B. Analysis and accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 14.Gergen J P, Stern R H, Wensink P C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979;7:2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gewitz H, Pistorius E K, Voss H, Vennesland B. Cyanide formation in preparations from Chlorella vulgaris Beijerinck: effect of sonication and amygdalin addition. Planta. 1976;131:145–148. doi: 10.1007/BF00389986. [DOI] [PubMed] [Google Scholar]
- 16.Guilbault G G, Kramer D N. Ultra sensitive, specific method for cyanide using p-nitrobenzaldehyde and o-dinitrobenzene. Anal Chem. 1966;38:834–836. [Google Scholar]
- 17.Hengge-Aronis R. Back to log phase: ςS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 18.Hertig C, Li R Y, Louarn A-M, Garnerone A M, David M, Batut J, Kahn D, Boistard P. Rhizobium meliloti regulatory gene fixJ activates transcription of R. meliloti nifA and fixK genes in Escherichia coli. J Bacteriol. 1989;171:1736–1738. doi: 10.1128/jb.171.3.1736-1738.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell C R, Stipanovic R D. Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology. 1979;69:480–482. [Google Scholar]
- 20.Howell C R, Stipanovic R D. Suppression of Pythium ultimum induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic pyoluteorin. Phytopathology. 1980;70:712–715. [Google Scholar]
- 21.Ivanova A, Renshaw M, Guntaka R V, Eisenstark A. DNA base sequence variability in katF (putative sigma factor) gene of Escherichia coli. Nucleic Acids Res. 1992;20:5479–5480. doi: 10.1093/nar/20.20.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahn D, Ditta G. Modular structure of FixJ: homology of the transcriptional activator domain with the −35 binding domain of sigma factors. Mol Microbiol. 1991;5:987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 23.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad host range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 24.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 25.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 26.Kraus J, Loper J E. Lack of evidence for a role of antifungal metabolite production by Pseudomonas fluorescens Pf-5 in biological control of Pythium damping-off of cucumber. Phytopathology. 1992;82:264–271. [Google Scholar]
- 27.Kraus J, Loper J E. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl Environ Microbiol. 1995;61:849–854. doi: 10.1128/aem.61.3.849-854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange R, Hengge-Aronis R. The cellular concentration of the ςS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 29.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 30.Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewen P C, Hengge-Aronis R. The role of the sigma factor RpoS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 32.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lund A H, Duch M, Pedersen F S. Increased cloning efficiency by temperature-cycle ligation. Nucleic Acids Res. 1996;24:800–801. doi: 10.1093/nar/24.4.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 36.Narumiya S, Takai K, Tokuyama T, Noda Y, Ushiro H, Hayaishi O. A new metabolic pathway of tryptophan initiated by tryptophan side chain oxidase. J Biol Chem. 1979;254:7007–7015. [PubMed] [Google Scholar]
- 37.Nowak-Thompson B, Gould S J, Kraus J, Loper J E. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can J Microbiol. 1994;40:1064–1066. [Google Scholar]
- 38.O’Neal C R, Gabriel W M, Turk A K, Libby S J, Fang F C, Spector M P. RpoS is necessary for both the positive and negative regulation of starvation survival genes during phosphate, carbon, and nitrogen starvation in Salmonella typhimurium. J Bacteriol. 1994;176:4610–4616. doi: 10.1128/jb.176.15.4610-4616.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfender W F, Kraus J, Loper J E. A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici-repentis in wheat straw. Phytopathology. 1994;83:1223–1228. [Google Scholar]
- 40.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 41.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sak B D, Eisenstark A, Touati D. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci USA. 1989;86:3271–3275. doi: 10.1073/pnas.86.9.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sarniguet A, Kraus J, Henkels M D, Muehlchen A M, Loper J E. The sigma factor ςS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stibitz S. Mutations in the bvgA gene of Bordetella pertussis that differentially affect regulation of virulence determinants. J Bacteriol. 1994;176:5615–5621. doi: 10.1128/jb.176.18.5615-5621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka K, Takahashi H. Cloning, analysis, and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 47.Thomashow L S, Weller D M. Current concepts in the use of introduced bacteria for biological control: mechanisms and antifungal metabolites. In: Stacey G, Keen N, editors. Plant microbe interactions. New York, N.Y: Chapman and Hall; 1995. pp. 187–235. [Google Scholar]
- 48.Whistler C A, Sarniguet A, Stockwell V O, Loper J E. Abstracts of the Sixth International Congress on Pseudomonas, Madrid, Spain. 1997. Four global regulators influence antibiotic production and stress response in Pseudomonas fluorescens Pf-5; p. 196. [Google Scholar]
- 49.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]