Abstract
The humpback grouper (Cromileptes altivelis), a medium-sized coral reef teleost, is a naturally rare species distributed in the tropical waters of the Indian and Pacific Oceans. It has high market value, but artificial reproduction and breeding remain limited and need to be improved. Here, we assembled the genome with 1.08 Gb, with a contig N50 of 43.78 Mb. A total of 96.59% of the assembly anchored to 24 pseudochromosomes using Hi-C technology. It contained 24,442 protein-coding sequences, of which 99.3% were functionally annotated. The completeness of the assembly was estimated to be 97.3% using BUSCO. The phylogenomic analysis suggested that humpback grouper should be classified into the genus Epinephelus rather than Cromileptes. The comparative genomic analysis revealed that the gene families related to circadian entrainment were significantly expanded. The high-quality reference genome provides useful genomic tools for exploiting the genomic resource of humpback grouper and supports the functional genomic study of this species in the future.
Subject terms: Genetics, Evolution
Background & Summary
Groupers, as a series of important commercial and ecological reef fish, are distributed in tropical and subtropical waters worldwide. On present understanding, groupers consist of 165 species in 16 genera and vary considerably in terms of lifestyle, growth rate, and body appearance1. The humpback grouper is a naturally rare species that is widely distributed in the tropical waters of the Indian and Pacific Oceans2. The term “humpback grouper” is because its body is relatively higher than its head, which gives a humpback aspect. The humpback grouper is a medium-sized fish, which grows up to 70 cm. As a protogynous hermaphroditic species, all humpback grouper individuals are born female and can transform into male when they grow up and experience 2–5 spawning seasons. This fish has high market value and is exceedingly favored by consumers due to their high nutritional value, tasty flesh, and beautiful appearance. In recent years, overfishing has led to a sharp decrease in the wild humpback grouper population, whereas the market demand has increased rapidly. Its relatively slow growth rate, unique sex-change strategy, and susceptibility to various pathogenic diseases during cultivation severely restrict the development of artificial culture. Previous studies of humpback grouper focused on immunology, the establishment of cell lines, classification, and feed supplement3–6. The decoding of a high-quality reference genome could support more information on molecular biology, genetics, breeding, and conservation biology.
Recently, several types of grouper genomes have been assembled, such as giant grouper (Epinephelus lanceolatus), leopard coral grouper (Plectropomus leopardus), and red-spotted grouper (Epinephelus akaara)7–9. Traditionally, grouper identification was primarily dependent on the surface profile and phenotype. Actually, it could cause errors and challenges in taxonomy. The groupers had a close relationship in evolution. To better understand the evolutionary relationship and taxonomy, it was necessary to acquire a specific solution by molecular biology. Besides, a high-quality reference genome resource could also provide an effective tool for genetic improvement and germplasm conservation. At present, the long-read and short-read sequencing technologies have been applied to the assembled genome. It was able to obtain highly integrated genome assemblies, especially circular consensus sequencing (CCS) improved the accuracy of PacBio SMRT sequencing. The HiFi sequence updated the genome assembly between read length and base quality significantly.
In 2021, a humpback grouper genome was constructed with the assembly of 1.013 Gb (contig N50 of 18.09 Mb)10. In this study, we represent a chromosome-scale genome assembly and annotation of humpback grouper with the PacBio HiFi and Hi-C sequencing technologies. Approximately 1.08 Gb genome was assembled with the contig N50 43.78 Mb. BUSCO analysis showed that 97.3% of the final assembly was complete BUSCOs. Overall, this high-quality reference genome provides a valuable basis for further genetic improvement and understanding the functional genes and molecular mechanisms in humpback grouper
Methods
DNA sample collection, library construction, and sequencing
A female humpback grouper was collected from Hainan Chenhai Aquatic Co., Ltd. The muscle tissue was collected for DNA extraction and library construction. Genomic DNA was extracted by the QIAamp DNA purification kit (Qiagen, USA). The short fragment library was generated using the Truseq Nano DNA HT Sample Preparation Kit (Illumina, USA) with an insert size of 350 bp and the Illumina NovaSeq 6000 platform. For the HiFi read generation, DNA fragment > 30 kb was selected using BluePippin Systerm (Sage Science, USA). The library was generated using the SMRTbell Template PrepKit 2.0 (PacBio, USA), and the library was sequenced in CCS on the PacBio Sequel II platform. The Hi-C library was constructed following the standard protocol described previously with certain modifications11, and it was sequenced using the Illumina NovaSeq 6000 platform. A total of 53.1 Gb of Illumina data, 21.5 Gb PacBio of PacBio data, and 96 Gb of Hi-C data after trimming the low-quality reads and adaptor sequences from the raw data.
RNA sample collection, library construction, and sequencing
The samples of eight embryonic development stages (one cell, morula, high blastula, low blastula, gastrula, somite, neurula, and before the hatching stage) were collected for RNA extraction using TRIzol reagent (Invitrogen, USA). RNA-seq libraries were constructed using Illumina TruSeq Stranded mRNA Library Prep Kit (Illumina, USA) and sequenced by the Illumina NovaSeq 6000 platform. Further, RNA extracted from embryonic samples was mixed for Iso-seq. The Iso-seq library was constructed and sequenced on the PacBio Sequel II platform. The clean data was obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from the raw data. Around 55.6 Gb of RNA-seq data and 69.1 Gb of Iso-seq data were generated for genome annotation.
Genome assembly and quality assessment
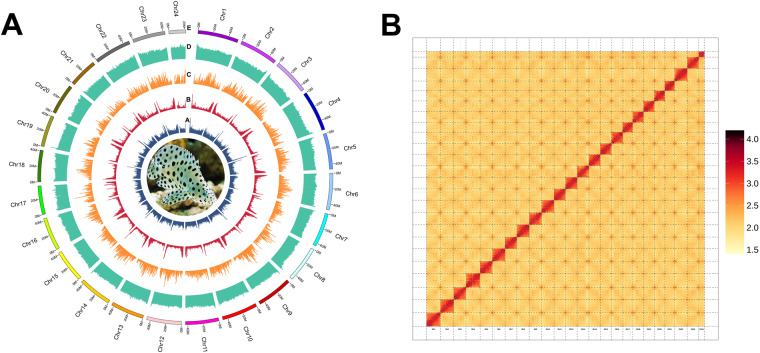
The characterization of the genome was estimated using the Illumina short-read data, and the 17 bp k-mer analysis was applied for estimation. The estimated genome size was 1,091.59 Mb, the heterozygosity rate was approximately 0.19%, and the repeated content was 45.81%. The genome was assembled using SOAPdenovo2 with k-mer set at 41 bp12. The gaps were filled with GapCloser. Then, the draft genome was corrected and re-assembled using HiFi long reads by Hifiasm 0.12-r304 with the parameters “-t 30 -D 10”13. The genome assembly was 1.08 Gb, with a contig N50 size of 43.78 Mb (Fig. 1A). To obtain the chromosome-level genome, we applied ALLHiC pipeline to link the mapped contigs to 24 pseudochromosomes14. Finally, 96.59% of scaffolds were mapped to 24 chromosomes (Fig. 1B).
Fig. 1.
Genome assembly of the humpback grouper. (A) Genomic features. From inner to outer tracks: A, distribution of DNA TEs across the genome; B, distribution of RNA TEs across the genome; C, gene density across the genome; D, GC content across the genome. E, humpback grouper chromosomes. (B) Hi-C contact map of the humpback grouper genome. The blocks represent the contacts between one location and another. The color illustrates the contact density from red (high) to low (orange).
To evaluate the assembled genome, BUSCO was applied to evaluate the completeness of genome assembly. A total of 3,345 BUSCO genes were identified, with 3,263 complete genes, 3,230 single-copy genes, 33 multi-copy genes, 47 fragmented genes, and 44 missing genes accounting for 97.3%, 96.3%, 1.0%, 1.4%, and 1.3% of the whole genome, respectively (Table 1).
Table 1.
BUSCO evaluation result of humpback grouper genome.
| Genome evaluation | Gene number | Percentage % |
|---|---|---|
| Complete BUSCOs | 3,263 | 97.3 |
| Complete and single-copy BUSCOs | 3,230 | 96.3 |
| Complete Duplicated BUSCOs | 33 | 1.0 |
| Fragmented BUSCOs | 47 | 1.4 |
| Missing BUSCOs | 44 | 1.3 |
| Total BUSCO groups searched | 3,345 | 100 |
Repeat and noncoding RNA annotation
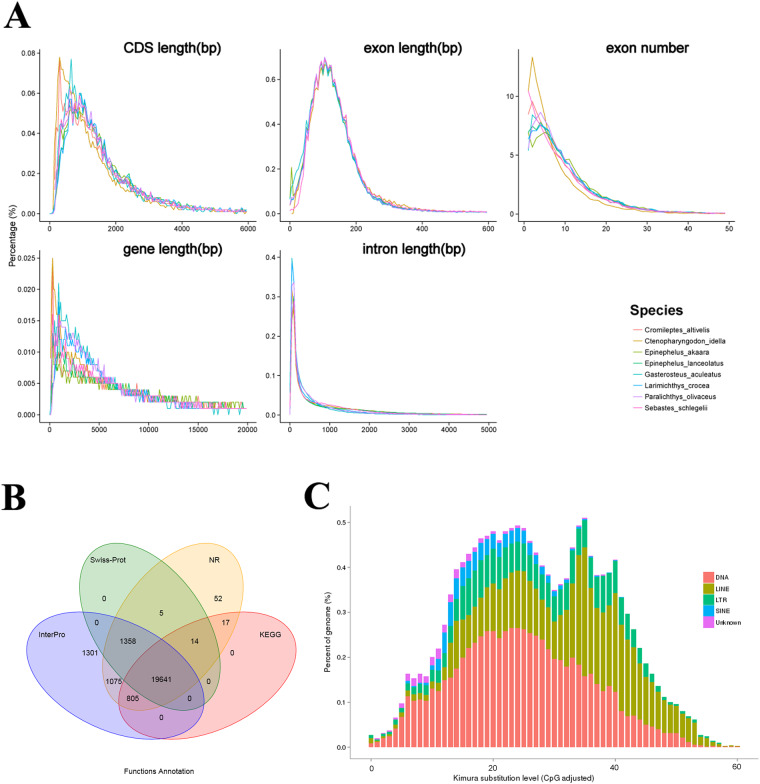
Repeat sequences of the humpback grouper genome were identified using a combination of homology-based and de novo approaches. For the ab initio method, the RepeatModeler (v2.0.1)15, RepeatScout (v1.0.5)16, and LTR_finder (v1.0.6)17 were used to build the humpback grouper custom repeat database. In the homology-based method, the Repbase database18 was used to identify repeats with the RepeatMasker and RepeatProteinMask. The total length of the repetitive elements accounted for 44.38% of the humpback grouper genome (Fig. 2C). DNA transposons represented the most abundant class of repeats (17.85% of the genome) followed by long interspersed elements (LINEs, 15.20%), long terminal repeats (LTRs, 5.38%), and short interspersed elements (SINEs, 1.11%) (Table 2).
Fig. 2.
The structural and functional annotation of humpback grouper. (A) Comparisons of the predicted gene models between the humpback grouper genome and other teleosts, including CDS length, exon length, exon number, gene length, and intron length. (B) The functional annotation of humpback grouper using different databases. (C) The percentage of different types of repetitive elements in the humpback grouper genome.
Table 2.
Statistic results of different types of annotated repeat content.
| Type | Denovo + Repbase | TE proteins | Combined TEs | |||
|---|---|---|---|---|---|---|
| Length (bp) | % in genome | Length (bp) | % in genome | Length (bp) | % in genome | |
| DNA | 185,549,534 | 17.13 | 17,809,686 | 1.64 | 193,277,492 | 17.85 |
| LINE | 164,630,235 | 15.20 | 45,118,481 | 4.17 | 184,303,613 | 17.02 |
| SINE | 12,052,908 | 1.11 | 0 | 0 | 12,052,908 | 1.11 |
| LTR | 83,542,399 | 7.71 | 7,526,000 | 0.69 | 86,240,213 | 7.96 |
| Other | 0 | 0 | 0 | 0 | 0 | 0 |
| Unknow | 22,465,884 | 2.07 | 0 | 0 | 22,465,884 | 2.07 |
| Total | 451,213,734 | 41.66 | 70,355,897 | 6.50 | 468,059,310 | 43.22 |
Noncoding RNAs, including rRNAs, snRNAs, miRNAs, and tRNAs, were identified by adopting INFERNAL (v1.1.2) through the Rfam database (release 13.0) for the humpback grouper genome using BLASTN (E-value ≤ 1e−5)19–21. Transfer RNA was predicted using tRNAscan (v1.3.1)22 with default parameters for eukaryotes. Ribosome RNAs and their subunits were predicted using the RNAmmer (v1.2)23. For non-coding RNA annotation, a total of 1,905 miRNA, 2,107 tRNA, 3,360 rRNA, and 1,637 snRNA were identified (Table 3).
Table 3.
Summary statistics of noncoding RNA.
| Type | Copy | Average length (bp) | Total length (bp) | % of genome |
|---|---|---|---|---|
| miRNA | 1,905 | 110.68 | 210,842 | 0.019468 |
| tRNA | 2,107 | 75.32 | 158,693 | 0.014653 |
| rRNA | 1,680 | 173.81 | 291,993 | 0.026960 |
| 18 S | 153 | 384.06 | 58,761 | 0.005426 |
| 28 S | 329 | 307 | 101,003 | 0.009326 |
| 5.8 S | 26 | 152.50 | 3,965 | 0.000366 |
| 5 S | 1,172 | 109.44 | 128,264 | 0.011843 |
| snRNA | 846 | 149.29 | 126,296 | 0.011661 |
| CD-box | 149 | 114.12 | 17,004 | 0.001570 |
| HACA-box | 88 | 151.48 | 13,330 | 0.001231 |
| splicing | 554 | 155.32 | 86,050 | 0.007945 |
Gene prediction and annotation
Firstly, three strategies were used for gene structure prediction, including de novo prediction, homology-based, and RNA-seq data-based prediction. Augustus (v2.5.5)24, Glimm erHMM (v3.01)25, SNAP26, Geneid27, and Genescan28, were used for de novo gene prediction with default settings. Protein sequences of giant grouper, black rockfish (Sebastes schlegelii), stickleback (Gasterosteus aculeatus), large yellow croaker (Larimichthys crocea), grass carp (Ctenopharyngodon idella), Japanese flounder (Paralichthys olivaceus), and red-spotted grouper were downloaded from Ensembl and NCBI databases. These sequences were aligned to the humpback grouper genome with TBLASTN (E-value ≤ 10−5), and homologous genome sequences were then aligned against matching proteins by GeneWise (v2.4.0)29 to generate a gene structure based on the alignment. Furthermore, the RNA-seq data from different embryonic development stages were assembled using Trinity (v2.1.1)30 and mapped to the humpback grouper genome by using the Cufflinks (v2.1.1)31. Gene prediction from the above methods was merged to a consensus gene set using the EVM (v1.1.1)32. The functional annotation of the predicted genes of humpback grouper was performed by alignment to the SwissProt33, NR34, KEGG35, Interpro36, GO37, and Pfam databases38. A total of 24,442 protein-coding genes were predicted (Table 4), of which 24, 268 (99.3%) genes were annotated (Fig. 2B). The lengths of average transcript and CDS were 19,080.10 and 1,607.91 bp, respectively (Fig. 2A).
Table 4.
Summary statistics of predicted protein-coding genes in the assembled genome.
| Gene set | Nmuber | Average transcript length (bp) | Average CDS length (bp) | Average exons per gene | Average exon length (bp) | Average intron length (bp) |
|---|---|---|---|---|---|---|
| De novo | ||||||
| Augustus | 30,800 | 13,113.62 | 1,312.94 | 7.34 | 178.81 | 1,860.59 |
| GlimmerHMM | 97,560 | 9,725.14 | 606.75 | 4.26 | 142.57 | 2,800.74 |
| SNAP | 40,866 | 34,778.53 | 1,097.41 | 7.83 | 140.20 | 4933.02 |
| Geneid | 31,659 | 21,781.54 | 1,369.00 | 6.80 | 201.27 | 3,518.41 |
| Genscan | 33,606 | 22,898.26 | 1,545.07 | 8.63 | 178.96 | 2,797.26 |
| Homolog | ||||||
| C. altivelis | 44,047 | 6,114.79 | 960.00 | 4.29 | 223.62 | 1,565.37 |
| E. akaara | 46,506 | 8,715.41 | 1,0811.7 | 5.35 | 202.13 | 1,755.44 |
| E. lanceolatus | 35,787 | 11,103.76 | 1,343.05 | 6.55 | 294.99 | 1,758.08 |
| G. aculeatus | 37,918 | 8,509.73 | 1,042.30 | 5.31 | 196.38 | 1,733.56 |
| L. crocea | 30,196 | 12,466.57 | 1,524.71 | 7.25 | 210.23 | 1,749.94 |
| P. olivaceus | 35,614 | 10,087.70 | 1,219.13 | 6.02 | 202.57 | 1,767.19 |
| S. schlegelii | 35,473 | 9,460.80 | 1,236.28 | 5.96 | 207.35 | 1,657.41 |
| RNA-seq | ||||||
| PASA | 76,981 | 15,673.39 | 1,357.35 | 8.31 | 163.26 | 1,957.31 |
| Cufflinks | 66,026 | 25,660.70 | 3,700.11 | 10.38 | 356.61 | 2,342.28 |
| EVM | 30,917 | 15,398.43 | 1,368.41 | 7.86 | 174.20 | 2,046.52 |
Data Records
The genome assembly and raw reads of the genome and transcriptome sequencing for humpback grouper were deposited under the Sequence Read Archive SRP32259439. The genome assembly was deposited at GenBank with the accession number GCA_019925165.140. Besides, the assembled genome, predicted peptide, CDS, and GO term files were available in the figshare database with the DOI number: 10.6084/m9.figshare.24145230.v241.
Technical Validation
Evaluation of the genome assembly and annotation
To evaluate the integrity and accuracy of the genome assembly, the completeness of the final genome assembly was assessed using BUSCO (v4.0)42 with the lineage database vertebrata_odb10 and CEGMA (v2.5)43. It was shown that the assembly contained 97.3% complete and 1.4% fragmented conserved single copy orthologue genes, and 94.35% of the 248 core eukaryotic genes. By aligning Illumina sequencing reads to the genome using BWA (v0.7.8)44, the reads mapping rate and the coverage rates were 99.68% and 99.91%, respectively. It was indicating high mapping efficiency and comprehensive coverage. Thus, all of the above results indicated that we obtained the high-quality genome of humpback grouper.
Acknowledgements
This work was supported by the Project of Sanya Yazhou Bay Science and Technology City (SCKJ-JYRC-2023-63, SCKJ-2023-01-004), and the Hainan Special PhD Scientific Research Foundation of Sanya Yazhou Bay Science and Technology City (HSPHDSRF-2022-02-009). This work was supported by the High-performance Computing Platform of YZBSTCACC. We appreciate the help from Novogene company for the sequencing.
Author contributions
J.X.L., H.B.S., Y.X.M., and Q.Q.Z. conceived the study. J.X.L. and Y.J.W. interpreted the data. L.T. and H.H. prepared the material. J.X.L., H.B.S., and Z.G.W. drafted the manuscript. J.X.L., H.H., and Q.Q.Z. contributed to the final manuscript editing.
Code availability
No specific code was used in this study. The data analyses used standard bioinformatic tools specified in the methods.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jinxiang Liu, Huibang Sun.
Contributor Information
Hai Huang, Email: huanghai74@126.com.
Quanqi Zhang, Email: qzhang@ouc.edu.cn.
References
- 1.Ma KY, Craig MT, Choat JH, van Herwerden L. The historical biogeography of groupers: Clade diversification patterns and processes. Molecular Phylogenetics and Evolution. 2016;100:21–30. doi: 10.1016/j.ympev.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Ortega-Recalde O, Goikoetxea A, Hore TA, Todd EV, Gemmell NJ. The Genetics and Epigenetics of Sex Change in Fish. Annual Review of Animal Biosciences. 2020;8:47–69. doi: 10.1146/annurev-animal-021419-083634. [DOI] [PubMed] [Google Scholar]
- 3.Ketut M, Zafran, Asami Y, Teruo M. Susceptibility of juvenile humpback grouper Cromileptes altivelis to grouper sleepy disease iridovirus (GSDIV) Diseases of Aquatic Organisms. 2004;59:1–9. doi: 10.3354/dao059001. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, et al. Establishment and characterization of a new cell line from the muscle of humpback grouper (Cromileptes altivelis) Fish Physiology and Biochemistry. 2020;46:1897–19075. doi: 10.1007/s10695-020-00841-5. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, Hu D, Yang W, Xiao J. Complete mitochondrial genome of the humpback grouper Cromileptes altivelis. Mitochondrial DNA. 2014;25:200–201. doi: 10.3109/19401736.2013.796463. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, et al. Evaluation of Lactococcus lactis HNL12 combined with Schizochytrium limacinum algal meal in diets for humpback grouper (Cromileptes altivelis) Fish & Shellfish Immunology. 2019;94:880–888. doi: 10.1016/j.fsi.2019.09.059. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, et al. A chromosome-level genome assembly of the giant grouper (Epinephelus lanceolatus) provides insights into its innate immunity and rapid growth. Molecular Ecology Resources. 2019;19:1322–1332. doi: 10.1111/1755-0998.13048. [DOI] [PubMed] [Google Scholar]
- 8.Wang, Y. et al. Chromosome Genome Assembly of the Leopard Coral Grouper (Plectropomus leopardus) With Nanopore and Hi-C Sequencing Data. Frontiers in Genetics11, (2020). [DOI] [PMC free article] [PubMed]
- 9.Ge H, et al. De novo assembly of a chromosome-level reference genome of red-spotted grouper (Epinephelus akaara) using nanopore sequencing and Hi-C. Molecular Ecology Resources. 2019;19:1461–1469. doi: 10.1111/1755-0998.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, Y. et al. Chromosome Genome Assembly of Cromileptes altivelis Reveals Loss of Genome Fragment in Cromileptes Compared with Epinephelus Species. Genes12 (2021). [DOI] [PMC free article] [PubMed]
- 11.Belton J-M, et al. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience1 (2012). [DOI] [PMC free article] [PubMed]
- 13.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zhang S, Zhao Q, Ming R, Tang H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nature Plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- 15.Tarailo-Graovac M, Chen N. Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Current Protocols in Bioinformatics. 2009;25:4.10.11–14.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 16.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 17.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Research. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics (Oxford, England) 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalvari I, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Research. 2018;46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Research. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 26.Leskovec J, Sosič R. SNAP: A General Purpose Network Analysis and Graph Mining Library. ACM Transactions on Intelligent Systems and Technology. 2016;8:1. doi: 10.1145/2898361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanco E, Parra G, Guigó R. Using geneid to Identify Genes. Current Protocols in Bioinformatics. 2007;18:4.3.1–4.3.28. doi: 10.1002/0471250953.bi0403s18. [DOI] [PubMed] [Google Scholar]
- 28.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA11Edited by F. E. Cohen. Journal of Molecular Biology. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 29.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Research. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S, Chan C-KK. Plant Bioinformatics: Methods and Protocols. New York: Springer; 2016. pp. 339–361. [Google Scholar]
- 32.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biololgy. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bairoch A, et al. The Universal Protein Resource (UniProt) Nucleic Acids Research. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn RD, et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Research. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Research. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Research. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.2020. NCBI Sequence Read Archive. SRP322594
- 40.2021. NCBI GenBank. Figshare. GCA_019925165.1
- 41.Liu J. 2023. The humpback grouper genome. Figshare. [DOI]
- 42.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 43.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2020. NCBI Sequence Read Archive. SRP322594
- 2021. NCBI GenBank. Figshare. GCA_019925165.1
- Liu J. 2023. The humpback grouper genome. Figshare. [DOI]
Data Availability Statement
No specific code was used in this study. The data analyses used standard bioinformatic tools specified in the methods.