Abstract
We cloned several genes encoding an Na+/H+ antiporter of Staphylococcus aureus from chromosomal DNA by using an Escherichia coli mutant, lacking all of the major Na+/H+ antiporters, as the host. E. coli cells harboring plasmids for the cloned genes were able to grow in medium containing 0.2 M NaCl (or 10 mM LiCl). Host cells without the plasmids were unable to grow under the same conditions. Na+/H+ antiport activity was detected in membrane vesicles prepared from transformants. We determined the nucleotide sequence of the cloned 7-kbp region. We found that seven open reading frames (ORFs) were necessary for antiporter function. A promoter-like sequence was found in the upstream region from the first ORF. One inverted repeat followed by a T-cluster, which may function as a terminator, was found in the downstream region from the seventh ORF. Neither terminator-like nor promoter-like sequences were found between the ORFs. Thus, it seems that the seven ORFs comprise an operon and that the Na+/H+ antiporter consists of seven kinds of subunits, suggesting that this is a novel type of multisubunit Na+/H+ antiporter. Hydropathy analysis of the deduced amino acid sequences of the seven ORFs suggested that all of the proteins are hydrophobic. As a result of a homology search, we found that components of the respiratory chain showed sequence similarity with putative subunits of the Na+/H+ antiporter. We observed a large Na+ extrusion activity, driven by respiration in E. coli cells harboring the plasmid carrying the genes. The Na+ extrusion was sensitive to an H+ conductor, supporting the idea that the system is not a respiratory Na+ pump but an Na+/H+ antiporter. Introduction of the plasmid into E. coli mutant cells, which were unable to grow under alkaline conditions, enabled the cells to grow under such conditions.
The Na+/H+ antiporter is widely distributed in cell membranes from bacteria to animals. The antiporter plays important roles in the Na+ cycle across the cytoplasmic membrane of all living cells (22, 34, 54). In bacteria, the antiporter extrudes Na+ or Li+ in exchange for H+. The driving force for this process is an electrochemical potential of H+ across the membrane, which is established either by the respiratory chain or the H+-translocating ATPase (22). However, in animals, an H+ is extruded from cells in exchange for Na+ via the antiporter (called the exchanger in animal cells). The driving force is an electrochemical potential of Na+ which is established by the Na+,K+-ATPase.
The Na+/H+ antiporter has several roles in bacterial cells: (i) establishment of an electrochemical potential of Na+ across the cytoplasmic membrane, which is the driving force for Na+-coupled processes such as Na+/solute symport (4, 11, 18, 46, 47) and Na+-driven flagellar rotation (13); (ii) extrusion of Na+ and Li+, which are toxic if accumulated at high concentrations in cells (14, 31, 33, 37); (iii) intracellular pH regulation under alkaline conditions (22, 34); and (iv) cell volume regulation (10, 34). Mutants of Escherichia coli which lack the Na+/H+ antiporter activity have been isolated (9, 31). Such mutants facilitated cloning of the gene(s) encoding the Na+/H+ antiporter. So far, genes for three distinct Na+/H+ antiporters of E. coli, nhaA (19), nhaB (36), and chaA (17), have been cloned and sequenced. The NhaA and NhaB antiporters have been purified and biochemically characterized (38, 42). Furthermore, homologs of nhaA and nhaB have been found in several other bacteria. These homologous genes have been cloned and sequenced. They include nhaA in Salmonella enteritidis (35), Vibrio parahaemolyticus (24), and Vibrio alginolyticus (29) and nhaB in V. parahaemolyticus (31) and V. alginolyticus (30). Furthermore, it has become clear that homologs of the nhaA and nhaB genes are present in Haemophilus influenzae, of which the entire chromosomal DNA sequence has been determined (8). Genes for other Na+/H+ antiporters have also been cloned and sequenced and include nhaC in Bacillus firmus (16), napA in Enterococcus hirae (52), nhaP in Pseudomonas aeruginosa (51), and nhaD in V. parahaemolyticus (32). Only one gene, and therefore one protein, is involved in Na+/H+ antiport in all of these Na+/H+ antiporters. Recently, a unique antiporter, called TetA(L), has been reported in Bacillus subtilis. TetA(L) is both an Na+/H+ antiporter and a tetracycline/H+ antiporter (5, 6). It should be stressed that only one protein [TetA(L)] mediates these two distinct antiport activities. For the two types of Na+/H+ antiporters, some similarities and diversities in primary structure and functional properties do exist. Thus, a comparison of the primary structures and the properties of many Na+/H+ antiporters would help to gain insight into their structure-function relationships.
Staphylococcus aureus is a halotolerant bacterium (20). This microorganism can survive even in the presence of 3 M NaCl or 1 M LiCl. We detected Na+/serine symport activity in S. aureus (1). S. aureus cells are able to grow under alkaline conditions, up to pH 9.5. Therefore, it seems that S. aureus possesses a strong Na+/H+ antiport activity. Indeed, in everted membrane vesicles prepared from cells of S. aureus, we detected Na+/H+ antiport activity (21). Here we report on a putative multisubunit Na+/H+ antiporter of S. aureus and its characteristics.
MATERIALS AND METHODS
Organisms, media, and growth.
S. aureus 209P was grown in nutrient broth (0.5% beef extract, 1.5% polypepton, 0.5% K2HPO4, 85 mM NaCl). The E. coli strains TG1 and KNabc, which lacks three major Na+/H+ antiporters (NhaA, NhaB, and ChaA) (31), were grown in modified L medium (2) [NaCl in the original medium was replaced with KCl; hereafter called L(K) medium]. NaCl or LiCl was added at the indicated concentration to the medium when necessary. To test the effect of pH on growth, E. coli HITΔAB− (44) or HITΔAB−/pNAS20 was grown in a minimal medium (100 mM Tris-HCl [at indicated pHs], 20 mM (NH4)2SO4, 50 mM KCl, 1 mM K2HPO4, 0.3 mM MgSO4, 0.01 mM CaCl2) supplemented with 40 mM glycerol (15). Cells were grown under aerobic conditions at 37°C. Cell growth was monitored turbidimetrically at 650 nm.
Preparation of membrane vesicles.
E. coli cells were grown in L(K) medium supplemented with 20 mM potassium lactate. Everted membrane vesicles were prepared by passing cells through a French press as described previously (24).
Na+/H+ antiport assay.
Activity of the Na+/H+ antiporter was measured by the quinacrine fluorescence quenching method with everted membrane vesicles as described previously (24) by use of a Hitachi F2000 fluorescence spectrophotometer.
Na+ extrusion assay.
E. coli cells grown in L(K) medium were washed twice with a buffer containing 0.2 M 3-[N-morpholino]propanesulfonic acid (MOPS) and 5 mM MgSO4 (pH adjusted to 7.5 with tetramethylammonium hydroxide) and then suspended in a buffer containing 0.2 M N-tris[(hydroxymethyl)methyl]glycine (Tricine) and 5 mM MgSO4 (pH adjusted to 8.0 with tetramethylammonium hydroxide). Movement of Na+ out of cells was measured with an Na+-selective electrode as described previously (47).
Cloning of genes.
E. coli KNabc is a mutant lacking all three major Na+/H+ antiporters and a restriction system (hsdΔ5) (31). Therefore, this mutant is useful for the cloning of an Na+/H+ antiporter gene(s) from another organism (31, 32, 51). S. aureus 209P cells were grown in nutrient broth. Ampicillin (final concentration, 100 μg/ml) was added to the culture medium at the exponential phase of growth to weaken its peptidoglycan layer. Chromosomal DNA was prepared from S. aureus cells by the method of Ausubel et al. (3). The DNA was partially digested with Sau3A1. Restriction fragments between 4 to 10 kbp were separated by sucrose density gradient centrifugation. The DNA fragments were ligated into either pUC19 or pBR322 (which had been digested with BamHI and dephosphorylated with bacterial alkaline phosphatase) by using T4 DNA ligase. Competent cells of E. coli KNabc were transformed with the ligated recombinant plasmids and spread onto 1.5% agar plates containing L(K) medium, 0.2 M NaCl (or 10 mM LiCl), and 100 μg of ampicillin per ml. The plates were incubated at 37°C for 1 day. The clones formed were picked. Plasmid DNA was prepared from the transformants. Competent cells of E. coli KNabc were retransformed and spread onto the same plates again. The plates were incubated at 37°C for 1 day. Many colonies appeared on the plates. Plasmids that were harbored by the retransformants were prepared. We obtained 25 candidate plasmids (13 candidates from the NaCl plate and 12 candidates from the LiCl plate; 20 candidates from the pUC19 vector and 5 candidates from the pBR322 vector). All of the plasmids possessed common restriction fragments in addition to several different ones. We therefore concluded that all of the plasmids carry a common portion of chromosomal DNA of S. aureus. We selected one of the plasmids, pNAS20, which carried the shortest DNA insert, for further analysis.
Southern analysis.
Southern hybridization analysis was performed with the Enhanced Chemiluminescence Detection System of Amersham Corp., as suggested by the manufacturer. The probe used was an EcoRV-MluI fragment (1.0 kbp) derived from the mnhD gene of S. aureus. Chromosomal DNA prepared from S. aureus or E. coli was digested with EcoRV and MluI, separated by electrophoresis in a 1% agarose gel, and blotted onto a nitrocellulose membrane. The probe was hybridized with the resulting Southern blot.
Sequencing.
A series of deletion plasmids for sequencing were constructed by using exonuclease III and mung bean nuclease from pNAS20. The nucleotide sequence was determined by the dideoxy chain termination method (40) using a DNA sequencer (ALF Express; Pharmacia Co.). Sequencing of both the sense and antisense strands was completed.
Computer analysis of sequence data.
Sequence data were analyzed with the GENETYX sequence analysis software (Software Development Co.). The SwissProt database was screened for sequence similarities.
Other assays and materials.
Protein content was determined by the method of Lowry et al. with bovine serum albumin as a standard (28). Reagents for DNA manipulation, sequencing, bacteriological media, and other chemicals were obtained from the usual commercial sources.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to the DDBJ, GenBank, and EMBL databases under accession no. AB015981.
RESULTS
Cloning of genes.
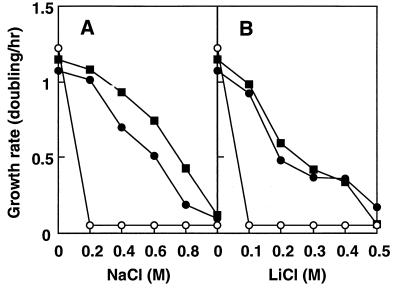
E. coli KNabc cells were not able to grow in the presence of concentrations equal to or higher than 0.2 M NaCl or 10 mM LiCl (Fig. 1). The parental cells from strain TG1, grew well under such conditions. We tried to clone a gene(s) for an Na+/H+ antiporter(s) or an Na+-extruding system(s) of S. aureus. By employing the shotgun method of cloning, we obtained 25 candidate plasmids carrying fragments of S. aureus chromosomal DNA that enabled the host cells, from strain KNabc, to grow in the presence of 0.2 M NaCl or 10 mM LiCl. However, restriction mapping suggested that all 25 candidate plasmids carried a common region of the S. aureus chromosome. KNabc cells harboring each one of the candidate plasmids showed similar growth capabilities in the presence of 0.2 M NaCl (data not shown). Thus, we chose one of them, pNAS20, which carried the shortest DNA insert. KNabc cells containing pNAS20 restored growth in the presence of 0.2 M NaCl or 0.1 M LiCl (Fig. 1). The transformed cells were able to grow even in the presence of 0.8 M NaCl or 0.4 M LiCl. Thus, it is likely that plasmid pNAS20 carries a gene(s) for Na+/H+ antiporter activity in S. aureus. In addition, it is likely that the putative antiporter gene(s) of S. aureus has been expressed in E. coli cells and that the gene product(s) has been synthesized in a functional form. It is unlikely that the gene product(s) activated the inactive E. coli Na+/H+ antiporters NhaA, NhaB, and ChaA, because KNabc is a deletion mutant of nhaA, nhaB, and chaA (31).
FIG. 1.
Effect of Na+ and Li+ on the growth of cells. E. coli TG1 (■), KNabc (○), and KNabc/pNAS20 (•) were grown in L(K) medium containing various concentrations of NaCl (A) or LiCl (B) under aerobic conditions at 37°C.
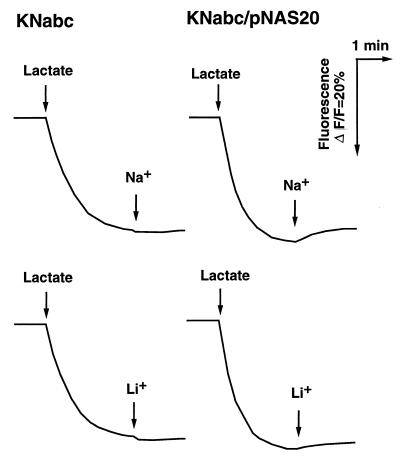
To test whether the pNAS20 really carries the Na+/H+ antiporter gene(s), we measured Na+/H+ antiport activity with everted membrane vesicles from KNabc/pNAS20 cells. Indeed, we detected Na+/H+ antiport activity in membrane vesicles of KNabc/pNAS20 but not in those of KNabc (Fig. 2). A weak Li+/H+ antiport activity was also detected in KNabc/pNAS20 (Fig. 2). The antiport activities were observed at pH 7.0. This suggests that the Na+/H+ antiporter of S. aureus is not of the NhaA type, the principal Na+/H+ antiporter of E. coli or V. parahaemolyticus, the activity of which is not measurable at pH 7.0 but very high at pH 8.5 (24, 33).
FIG. 2.
Na+/H+ antiport and Li+/H+ antiport activities in membrane vesicles. Everted membrane vesicles were prepared from E. coli KNabc and KNabc/pNAS20 cells by the French press method. Antiport activities were measured by the quinacrine fluorescence quenching method. At the time point indicated by the first arrow on the left, potassium lactate (final concentration, 5 mM) was added to the assay mixture to initiate respiration. At time point indicated by the second arrow from the left, NaCl (final concentration, 5 mM) or LiCl (final concentration, 5 mM) was added.
Therefore, we tested the effect of pH on the Na+/H+ antiporter activity in membrane vesicles of KNabc/pNAS20. The activity was at its maximum at pH 7.0 to 7.5 (data not shown). Thus, the pH profile for the Na+/H+ antiporter activity derived from S. aureus is obviously different from that of NhaA of E. coli (33) or V. parahaemolyticus (24). In addition, the pH profile is not similar to that of the NhaB Na+/H+ antiporter of E. coli (14, 33) or V. parahaemolyticus (31). Activity of the NhaB system is measurable at pH 7.0 and higher at alkaline pHs such as pH 8.0 or 8.5. The ChaA system of E. coli utilizes Ca2+ as an efficient substrate (17). We detected no significant difference in Ca2+/H+ antiport activity between membrane vesicles prepared from cells of KNabc/pNAS20 and those from cells of KNabc (data not shown). Thus, the properties of the Na+/H+ antiporter derived from pNAS20 are different from those of NhaA, NhaB, and ChaA.
Sequences.
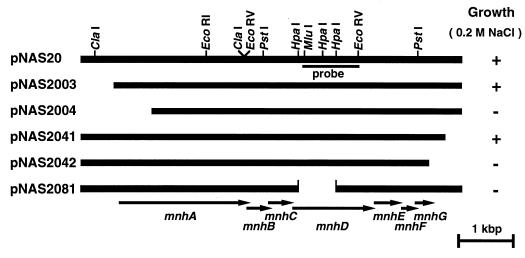
We constructed many deletion derivatives from pNAS20 in order to localize the region of the DNA insert where the antiporter gene(s) is located (Fig. 3). The length of the insert derived from chromosomal DNA of S. aureus in the pNAS20 was about 7 kbp. This is too long for an Na+/H+ antiporter possessing about 12 transmembrane domains. It has been shown that the length of the gene for NhaA (19), NhaB (35), or ChaA (17) (these antiporters possess about 12 transmembrane domains) is about 1.5 kbp. Deletion plasmids were constructed and introduced into KNabc cells. The growth capability in the presence of 0.2 M NaCl was tested. Surprisingly, we found that most (about 6 kbp) of the DNA insert carried on the pNAS20 plasmid was necessary for growth in the presence of 0.2 M NaCl (Fig. 3). One deletion plasmid, pNAS2081, which lacks an internal portion of the DNA insert, did not enable KNabc cells to grow in the presence of 0.2 M NaCl (Fig. 3). These results suggest that either the Na+/H+ antiporter of S. aureus consists of multiple subunits, multiple components are involved in regulation of the antiporter, or the antiporter protein is a giant protein with roughly 2,000 amino acid residues.
FIG. 3.
Plasmids and restriction map. Physical maps of the DNA insert derived from the S. aureus chromosome in each plasmid are shown. The DNA inserts are aligned. Restriction sites are present at the same horizontal position in each insert. Locations and directions of each ORF (mnhA to mnhG) which were revealed by sequencing are shown at the bottom. The probe used for the Southern analysis is also shown. The growth capability of E. coli KNabc harboring each plasmid in L(K) medium supplemented with 0.2 M NaCl is shown on the right. Symbols: +, cell growth occurred; −, no cell growth occurred.
We determined the sequence of 6,995 nucleotides of the DNA insert of pNAS20 (DDBJ/EMBL/GenBank accession no. AB015981). We found eight open reading frames (ORFs) and a part of another ORF in the sequenced region. All of the ORFs were preceded by Shine-Dalgarno sequences (41). Amino acid sequences were deduced from these ORFs. Analysis of a series of deletion plasmids revealed that the first ORF and the incomplete ORF found in the last portion of the DNA insert were not necessary for the Na+/H+ antiporter function (Fig. 3). KNabc cells harboring plasmid pNAS2003, carrying the seven ORFs except the first ORF (or the last incomplete ORF), grew in the presence of 0.2 M NaCl (Fig. 3). Furthermore, membrane vesicles prepared from such cells showed Na+/H+ antiport activity (data not shown). Thus, we conclude that the seven ORFs are necessary and sufficient for Na+/H+ antiporter function. A promoter-like sequence (−35 region and −10 region) was found in the upstream region from the seven ORFs (data not shown). An inverted repeat followed by T-cluster was found in the downstream region from the seven ORFs (data not shown). Neither a terminator-like nor a promoter-like sequence was found between the seven ORFs. Therefore, it seems that the seven ORFs comprise an operon. We designated the operon mnh, and the genes included in this operon were designated mnhA to mnhG (from upstream to downstream).
As a result of a Southern hybridization analysis using a DNA fragment derived from mnhD as a probe (Fig. 3), we detected a dense band in a sample of S. aureus chromosomal DNA but not in a sample of E. coli chromosomal DNA (data not shown).
Characteristics of the primary structure.
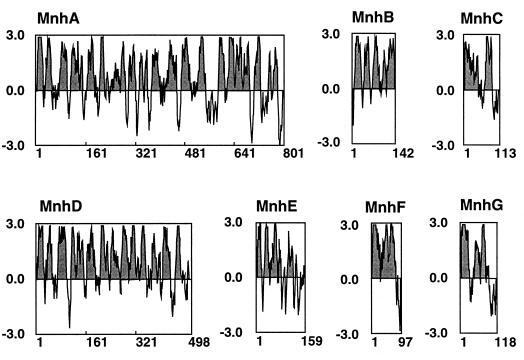
Judging from the deduced amino acid sequences, each putative protein (MnhA to MnhG) consists of 801, 142, 113, 498, 159, 97, and 118 amino acid residues, respectively. Calculated molecular weights for MnhA to MnhG are 89,385, 15,682, 12,260, 54,755, 18,319, 10,616, and 12,818, respectively. These proteins are all of low polarity (58 to 71% hydrophobic residues).
Hydropathy profiles calculated by the method of Kyte and Doolittle (26) and plotted along the amino acid sequences suggested that all subunits have hydrophobic domains which seem to be membrane-spanning regions (Fig. 4). In particular, MnhA and MnhD possess many hydrophobic domains. It seems that all of the Mnh proteins are intrinsic membrane proteins.
FIG. 4.
Hydropathy patterns of deduced Mnh proteins. Hydropathy values were calculated by the method of Kyte and Doolittle (28) for the deduced amino acid sequences of MnhA to MnhG. The values were plotted from the NH2 terminus to the COOH terminus. The portions above and below the midpoint line indicate hydrophobic and hydrophilic regions, respectively. The hydrophobic regions are shaded.
We searched for homology between the deduced amino acid sequences of MnhA to MnhG and reported sequences in a protein sequence database (SwissProt) (Table 1). We found putative homologs of MnhA to MnhG in B. subtilis, for which the complete nucleotide sequence of the genome has been determined (23). A comparison of sequences of the primary structures of MnhA and YufT, MnhB and YufU, MnhC and YufV, MnhD and YufD, MnhF and YufC, and MnhG and YufB revealed 39 to 54% identities and 82 to 89% similarities. Although an ORF corresponding to MnhE has not been identified in the sequence of B. subtilis (23), a homolog of MnhE emerges if we remove one nucleotide from the reported sequence of the corresponding region of the B. subtilis DNA. The amino acid sequence of the resulting hypothetical homolog showed 38% identity and 79% similarity with the MnhE sequence. Comparison of amino acid residues between MnhA and YufT revealed that the MnhA has 27 additional residues at the NH2 terminus. We checked whether there is an initiation codon preceded by a Shine-Dalgarno sequence in the corresponding upstream region from the proposed yufT gene (EMBL/GenBank/DDBJ accession no. Z93937). In fact, we found a TTG codon preceded by a Shine-Dalgarno sequence in the same frame as the original yufT. If this TTG is the initiation codon for the yufT gene, the product YufT consists of 801 amino acid residues, exactly the same number of residues that MnhA has. The newly added 27 residues in the YufT sequence had a high sequence similarity with the corresponding region of MnhA. The numbers of amino acid residues in all of the corresponding homologs were similar (data not shown). Thus, the Yuf proteins may form a complex and function as an Na+/H+ antiporter in B. subtilis. MnhA, MnhB, and MnhC showed sequence similarities (38 to 48% identities and 78 to 87% similarities) with putative genes (ORF1, ORF2, and ORF3) responsible for an Na+/H+ antiporter of alkaliphilic Bacillus sp. strain C-125 (12) (Table 1). Also, we found homologs of MnhA to MnhG in Rhizobium meliloti (EMBL/GenBank/DDBJ database accession no. X93358) (39). They are designated PhaA to PhaG. Sequence similarities between MnhA and PhaA, MnhB and PhaB, MnhC and PhaC, MnhD and PhaD, MnhE and PhaE, MnhF and PhaF, and MnhG and PhaG were demonstrated (21 to 41% identities and 58 to 73% similarities) (Table 1). Interestingly, the sequence of the N-terminal portion (80 residues) of PhaB showed significant similarity (43% identity and 83% similarity) with the C-terminal sequence of MnhA, in addition to similarity between the C-terminal portion (125 residues) of PhaB and the entire region of MnhB (26% identity and 73% similarity). The molecular weights of PhaA to PhaG, except PhaB, roughly correspond to those of MnhA to MnhG, except MnhB.
TABLE 1.
Identities and similarities of amino acid sequence between Mnhs and homologs
| S. aureus Na+/H+ antiporter | % Identity/% similarity (homolog)
|
||||
|---|---|---|---|---|---|
| B. subtilis hypothetical homolog | Bacillus sp. strain C-125 Na+/H+ antiportera | R. meliloti Pha | E. coli NDH-1 | Bovine complex I | |
| MnhA | 54/86 (YufT) | 49/84 (ORF1) | 36/76 (PhaA) | 31/71 (NuoL) | 24/67 (ND5) |
| MnhB | 45/82 (YufU) | 39/81 (ORF2) | 43/83 (PhaB) | NAb | NA |
| MnhC | 52/89 (YufV) | 47/88 (ORF3) | 42/75 (PhaC) | NA | NA |
| MnhD | 49/86 (YufD) | NA | 32/71 (PhaD) | 22/67 (NuoN) | 18/64 (ND2) |
| MnhE | 38/79 (YufX)c | NA | 24/73 (PhaE) | NA | NA |
| MnhF | 51/88 (YufC) | NA | 25/74 (PhaF) | NA | NA |
| MnhG | 39/82 (YufB) | NA | 28/73 (PhaG) | NA | NA |
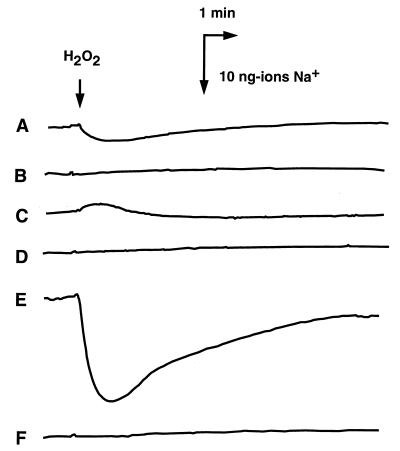
We also found significant sequence similarities between MnhA and NuoL (53) of E. coli (20% identity and 46% similarity), MnhA and ND5 (1a) of bovine mitochondria (19% identity and 53% similarity), MnhD and NuoN (53) of E. coli (18% identity and 55% similarity), and MnhD and ND2 (1) of bovine mitochondria (13% identity and 46% similarity). NuoL, NuoN, ND5, and ND2 are components of the respiratory chain (1a, 53). Thus, it seemed possible that the protein products MnhA to MnhG are components of the respiratory chain of S. aureus and that Na+ extrusion via the Mnh complex is directly driven by respiration, similar to the case of the respiratory Na+ pump reported in V. alginolyticus (45) and V. parahaemolyticus (49). We tested these possibilities. Previously, we reported Na+ extrusion from E. coli that is driven by an electrochemical potential of H+ across the membranes by the Na+/H+ antiport mechanism (48) and from V. parahaemolyticus that is directly driven by the respiratory chain, the respiratory Na+ pump (49). A key experiment to distinguish between these two mechanisms is to test the effect of an H+ conductor on the respiration-driven Na+ extrusion from cells. If the Na+ extrusion is mediated by an Na+/H+ antiporter, an H+ conductor should inhibit the Na+ extrusion (48). On the other hand, if Na+ extrusion is mediated by the respiratory complex, an H+ conductor should not inhibit the Na+ extrusion (49). In control experiments, we observed Na+ extrusion from cells of wild-type E. coli (Fig. 5A), which was completely inhibited by an H+ conductor, carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Fig. 5B). We detected no Na+ extrusion in KNabc cells, which lack all of the major Na+/H+ antiporters. Thus, the Na+ extrusion observed with wild-type E. coli is mediated by Na+/H+ antiporters. We detected some Na+ uptake driven by respiration in KNabc cells (Fig. 5C). Perhaps this Na+ uptake is due to the membrane potential established by respiration. We observed a large Na+ extrusion from cells of KNabc/pNAS20 which was elicited by respiration (Fig. 5E). This large Na+ extrusion was completely inhibited by CCCP (Fig. 5F). Thus, it seems that the Mnh complex is not a respiratory Na+ pump but an Na+/H+ antiporter.
FIG. 5.
Na+ extrusion from cells elicited by respiration. Extrusion of Na+ from cells of E. coli TG1 (A and B), KNabc (C and D), and KNabc/pNAS20 (E and F) was measured with an Na+ electrode. At the time point indicated by the arrow labeled H2O2, a small amount (5 μl of a 0.5% solution) of H2O2 was added to the cell suspension (2.5 ml) to supply O2 and to initiate respiration. In three cases (B, D, and F), CCCP was present at 100 μM in the assay medium. A downward deflection represents Na+ extrusion from the cells.
pH resistance in growth.
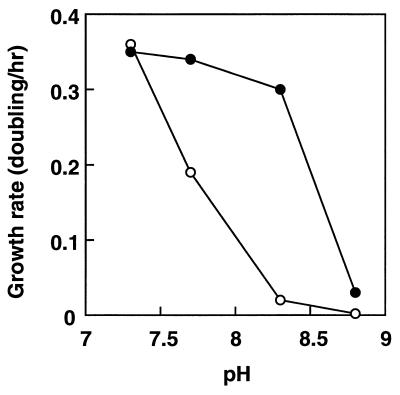
Previously we reported the isolation of a mutant of E. coli HIT-1 which showed a defect in the Na+/H+ antiporter and a defect in growth under alkaline conditions (15). It is not clear yet, however, whether another mutation is present in the mutant. We constructed a series of Na+/H+ antiporter mutants (44). The mutant strain HITΔAB− is one of the mutants lacking both NhaA and NhaB antiporter activities (44). We tested whether the Na+/H+ antiporter genes from S. aureus confer upon E. coli HITΔAB− the ability to grow under alkaline conditions, in addition to exhibiting Na+/H+ antiporter activity. The wild-type E. coli grew well at pHs up to 8.5 (15). As shown in Fig. 6, the growth rate of HITΔAB− was greatly reduced under alkaline conditions (especially at pHs above 8.0) compared with that at neutral pH. Introduction of the plasmid pNAS20 into HITΔAB− cells restored growth at pHs between 7.6 and 8.3 (Fig. 6). It has been reported that pha genes of R. meliloti are involved in pH adaptation (39).
FIG. 6.
Effects of pH on cell growth of E. coli HITΔAB− and HITΔAB−/pNAS20. E. coli HITΔAB− (○) and HITΔAB−/pNAS20 (•) were grown in minimal medium supplemented with glycerol at the indicated pHs at 37°C under aerobic conditions.
DISCUSSION
We found putative Na+/H+ antiporter genes in S. aureus. Our data suggest that the putative Na+/H+ antiporter consists of seven kinds of subunits. All of the Na+/H+ antiporters or Na+/H+ exchangers so far identified in microbial or animal cells are of a single subunit, although we do not yet know whether they function as monomers or homo-oligomers. Several examples of Na+ extrusion systems which consist of multiple components present in cell membranes are known; these include the respiratory Na+ pump of halophilic bacteria (50), the F0F1-type Na+-translocating ATPase of Propionigenium modestum (27), the Na+-translocating ATPase of Enterococcus hirae (43), and a two-gene ATP-binding cassette (ABC)-type Na+ extrusion system of B. subtilis (7). No similarity was found in the primary structure of components of the S. aureus Na+/H+ antiporter and components of these Na+ extrusion systems. No ABC motif was found in any of the subunits of the Na+/H+ antiporter of S. aureus (data not shown).
We have found homologs of the mnh genes in B. subtilis, namely, the yuf genes. Recently, we cloned a DNA fragment containing the yuf region from the B. subtilis genome. E. coli KNabc cells harboring a plasmid carrying this DNA region restored Na+/H+ antiport activity (8a). It is also very likely that a similar Na+/H+ antiporter exists in the alkaliphilic Bacillus sp. strain C-125 (12). Hamamoto et al. reported a mutant of the alkaliphilic Bacillus in which the activity of the putative Na+/H+ antiporter was lost (12).
The overall GC content of the cloned gene was 33%, which is close to that of the genomic DNA of S. aureus (34%) (20). On the other hand, R. meliloti is a GC-rich bacterium in which the GC content in the pha operon (EMBL/GenBank/DDBJ accession no. X93358) is 63%. R. meliloti is a gram-negative and rod-shaped bacterium. S. aureus is a gram-positive coccus. It is interesting that genes of the same type are present both in S. aureus and in a quite different bacterium, R. meliloti.
Growth of KNabc/pNAS20 was observed in the presence of 0.8 M NaCl (or 0.4 M LiCl) (Fig. 1). This suggested that S. aureus Na+/H+ antiporter activity produced in E. coli is fairly high. In fact, we observed very high Na+ extrusion activity in cells due to the Na+/H+ antiporter in KNabc/pNAS20 (Fig. 5). However, Na+/H+ antiporter activity was very low, as measured by the fluorescence quenching method, in everted membrane vesicles prepared from KNabc/pNAS20 (Fig. 2). At present, we do not know the reason for this discrepancy. Although cells of S. aureus can grow in the presence of a high concentration of NaCl or at alkaline pHs, which suggests the presence of a strong Na+/H+ antiporter in this microorganism, we detected weak (or moderate) Na+/H+ antiporter activity in membrane vesicles (21). Thus, some factor(s) present in the cytoplasm may be necessary for the full activity of the Mnh Na+/H+ antiporter. Addition of ATP to the mixture of the fluorescence quenching assay showed no effect (data not shown). Another possibility is that some component(s) of the Mnh system is unstable in membrane vesicles.
We obtained 25 candidate plasmids that enabled E. coli KNabc transformants to grow in the presence of 0.2 M NaCl or 10 mM LiCl. All of them seemed to carry a common DNA region of the S. aureus chromosome. This suggests that the Mnh Na+/H+ antiporter is the sole major Na+ (and Li+) extrusion system in S. aureus. Another possibility is that another Na+/H+ antiporter(s) of S. aureus is not expressed in E. coli.
We observed some Na+ uptake driven by respiration in E. coli KNabc which lacks all of the major Na+/H+ antiporters, although Na+ was extruded from parental cells. This suggests the presence of a membrane potential-driven Na+ influx system in E. coli, perhaps a Na+ channel. Recently, we developed a patch clamp method which was applicable to the measurement of ion flux through bacterial ion channels or ion pumps (25). This method could be utilized to analyze not only ion channels of bacterial cells but also the putative multisubunit Na+/H+ antiporter, Mnh, if the antiport process is electrogenic. In fact, it has been reported that the Na+/H+ antiporter probably consisting of ORF1 to ORF3 (and perhaps some others) of Bacillus sp. strain C-125 functions as an electrogenic transporter (12).
ACKNOWLEDGMENTS
We thank Manuel Varela of Eastern New Mexico University for critically reading the manuscript.
This study was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan. T.K. was a research fellow of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Akamatsu, Y., and T. Tsuchiya. Unpublished results.
- 1a.Anderson S, de Bruijn M H, Coulson A R, Eperon I C, Sanger F, Young I G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. p. 1.1.2. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. p. 2.4.3. [Google Scholar]
- 4.Chen C C, Tsuchiya T, Yamane Y, Wood J M, Wilson T H. Na+(Li+)-proline cotransport in Escherichia coli. J Membr Biol. 1985;84:157–164. doi: 10.1007/BF01872213. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Guffanti A A, Krulwich T A. The chromosomal tetracycline resistance locus of Bacillus subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated pH. J Biol Chem. 1994;269:27365–27371. [PubMed] [Google Scholar]
- 6.Cheng J, Baldwin K, Guffanti A A, Krulwich T A. Na+/H+ antiport activity conferred by Bacillus subtilis tetA(L), a 5′ truncation product of tetA(L), and related plasmid genes upon Escherichia coli. Antimicrob Agents Chemother. 1996;40:852–857. doi: 10.1128/aac.40.4.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J, Guffanti A A, Krulwich T A. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8a.Gohda, T., and T. Tsuchiya. Unpublished results.
- 9.Goldberg E B, Arbel T, Chen J, Karpel R, Mackie G A, Schuldiner S, Padan E. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grinstein S, editor. In Na+/H+ exchange. Boca Raton, Fla: CRC Press Inc.; 1988. [Google Scholar]
- 11.Hama H, Shimamoto T, Tsuda M, Tsuchiya T. Properties of a Na+-coupled serine-threonine transport system in Escherichia coli. Biochim Biophys Acta. 1987;905:231–239. doi: 10.1016/0005-2736(87)90451-2. [DOI] [PubMed] [Google Scholar]
- 12.Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 13.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1988;21:705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 14.Inaba K, Kuroda T, Shimamoto T, Kayahara T, Tsuda M, Tsuchiya T. Lithium toxicity and Na+(Li+)/H+ antiporter in Escherichia coli. Biol Pharm Bull. 1994;17:395–398. doi: 10.1248/bpb.17.395. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa T, Hama H, Tsuda M, Tsuchiya T. Isolation and properties of a mutant of Escherichia coli possessing defective Na+/H+ antiporter. J Biol Chem. 1987;262:7443–7446. [PubMed] [Google Scholar]
- 16.Ivey D M, Guffanti A A, Bossewitch J S, Padan E, Krulwich T A. Molecular cloning and sequencing of a gene from alkaliphilic Bacillus firmus OF4 that functionally complements an Escherichia coli strain carrying a deletion in the nhaA Na+/H+ antiporter gene. J Biol Chem. 1991;266:23483–23489. [PubMed] [Google Scholar]
- 17.Ivey D M, Guffanti A A, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich T A. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J Biol Chem. 1993;268:11296–11303. [PubMed] [Google Scholar]
- 18.Jackowski S, Alix J-H. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J Bacteriol. 1990;172:3842–3848. doi: 10.1128/jb.172.7.3842-3848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpel R, Olami Y, Taglicht D, Schuldiner S, Padan E. Sequencing of the gene ant which affects the Na+/H+ antiporter activity in Escherichia coli. J Biol Chem. 1988;263:10408–10414. [PubMed] [Google Scholar]
- 20.Kloos W E, Schleifer K H. Genus IV. Staphylococcus Rosenbach 1884, 18AL. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1013–1035. [Google Scholar]
- 21.Kodama K, Hashimoto A, Morita Y, Tomochika K, Tsuchiya T. Preparation and characterization of everted membrane vesicles from cells of Staphylococcus aureus. Biol Pharm Bull. 1998;21:5–9. doi: 10.1248/bpb.21.5. [DOI] [PubMed] [Google Scholar]
- 22.Krulwich T A. Na+/H+ antiporters. Biochim Biophys Acta. 1983;726:245–264. doi: 10.1016/0304-4173(83)90011-3. [DOI] [PubMed] [Google Scholar]
- 23.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda T, Shimamoto T, Inaba K, Tsuda M, Tsuchiya T. Properties and sequence of the NhaA Na+/H+ antiporter of Vibrio parahaemolyticus. J Biochem. 1994;116:1030–1038. doi: 10.1093/oxfordjournals.jbchem.a124624. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda T, Okuda N, Saitoh N, Hiyama T, Terasaki Y, Anazawa H, Hirata A, Mogi T, Kusaka I, Tsuchiya T, Yabe I. Patch clamp studies on ion pumps of the cytoplasmic membrane of Escherichia coli: formation, preparation, and utilization of giant vacuole-like structures consisting of everted cytoplasmic membrane. J Biol Chem. 1998;273:16897–16904. doi: 10.1074/jbc.273.27.16897. [DOI] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Laubinger W, Dimroth P. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0 type. Eur J Biochem. 1987;168:475–480. doi: 10.1111/j.1432-1033.1987.tb13441.x. [DOI] [PubMed] [Google Scholar]
- 28.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 29.Nakamura T, Komano Y, Itaya E, Tsukamoto K, Tsuchiya T, Unemoto T. Cloning and sequencing of an Na+/H+ antiporter gene from the marine bacterium Vibrio alginolyticus. Biochim Biophys Acta. 1994;1190:465–468. doi: 10.1016/0005-2736(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T, Enomoto H, Unemoto T. Cloning and sequencing of the nhaB gene encoding an Na+/H+ antiporter from Vibrio alginolyticus. Biochim Biophys Acta. 1996;1275:157–160. doi: 10.1016/0005-2728(96)00034-5. [DOI] [PubMed] [Google Scholar]
- 31.Nozaki K, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem Biophys Res Commun. 1996;222:774–779. doi: 10.1006/bbrc.1996.0820. [DOI] [PubMed] [Google Scholar]
- 32.Nozaki K, Kuroda T, Mizushima T, Tsuchiya T. A new Na+/H+ antiporter, NhaD, of Vibrio parahaemolyticus. Biochim Biophys Acta. 1998;1369:213–220. doi: 10.1016/s0005-2736(97)00223-x. [DOI] [PubMed] [Google Scholar]
- 33.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s) J Biol Chem. 1989;264:20297–20302. [PubMed] [Google Scholar]
- 34.Padan E, Schuldiner S. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim Biophys Acta. 1994;1185:129–151. doi: 10.1016/0005-2728(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 35.Pinner E, Carmel O, Bercovier H, Sela S, Padan E, Schuldiner S. Cloning, sequencing and expression of the nhaA and nhaR genes from Salmonella enteritidis. Arch Microbiol. 1992;157:323–328. doi: 10.1007/BF00248676. [DOI] [PubMed] [Google Scholar]
- 36.Pinner E, Padan E, Schuldiner S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1992;267:11064–11068. [PubMed] [Google Scholar]
- 37.Pinner E, Kotler Y, Padan E, Schuldiner S. Physiological role of NhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1993;268:1729–1734. [PubMed] [Google Scholar]
- 38.Pinner E, Padan E, Schuldiner S. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J Biol Chem. 1994;269:26274–26279. [PubMed] [Google Scholar]
- 39.Putnoky P, Kereszt A, Nakamura T, Endre G, Grosskopf E, Kiss P, Kondorosi A. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type K+ efflux system. Mol Microbiol. 1998;28:1091–1101. doi: 10.1046/j.1365-2958.1998.00868.x. [DOI] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 43.Takase K, Kakinuma S, Yamato I, Konishi K, Igarashi K, Kakinuma Y. Sequencing and characterization of the ntp gene cluster for vacuolar-type Na+-translocating ATPase of Enterococcus hirae. J Biol Chem. 1994;269:11037–11044. [PubMed] [Google Scholar]
- 44.Thelen P, Tsuchiya T, Goldberg E B. Characterization and mapping of a major Na+/H+ antiporter gene of Escherichia coli. J Bacteriol. 1991;173:6553–6557. doi: 10.1128/jb.173.20.6553-6557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokuda H, Unemoto T. A respiration-dependent primary sodium extrusion system functioning at alkaline pH in the marine bacterium Vibrio alginolyticus. Biochem Biophys Res Commun. 1981;102:265–271. doi: 10.1016/0006-291x(81)91516-3. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchiya T, Hasan S M, Raven J. Glutamate transport driven by an electrochemical gradient of sodium ions in Escherichia coli. J Bacteriol. 1977;162:794–798. doi: 10.1128/jb.131.3.848-853.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchiya T, Wilson T H. Cation-sugar cotransport in the melibiose transport system of Escherichia coli. Membr Biochem. 1978;2:63–79. doi: 10.3109/09687687809063858. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchiya T, Takeda K. Extrusion of sodium ions energized by respiration and glycolysis in Escherichia coli. J Biochem. 1979;86:225–230. [PubMed] [Google Scholar]
- 49.Tsuchiya T, Shinoda S. Respiration-driven Na+ pump and Na+ circulation in Vibrio parahaemolyticus. J Bacteriol. 1985;162:794–798. doi: 10.1128/jb.162.2.794-798.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unemoto T, Hayashi M. Na+-translocating NADH-quinone reductase of marine and halophilic bacteria. J Bioenerg Biomembr. 1993;25:385–391. doi: 10.1007/BF00762464. [DOI] [PubMed] [Google Scholar]
- 51.Utsugi J, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim Biophys Acta. 1998;1398:330–334. doi: 10.1016/s0167-4781(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 52.Waser M, Hess-B D, Davies K, Solioz M. Cloning and disruption of a putative Na+/H+ antiporter gene of Enterococcus hirae. J Biol Chem. 1992;267:5396–5400. [PubMed] [Google Scholar]
- 53.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH: ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 54.Yun C H, Tse C M, Nath S K, Levine S A, Brant S R, Donowitz M. Mammalian Na+/H+ exchanger gene family: structure and function studies. Am J Physiol. 1995;269:G1–G11. doi: 10.1152/ajpgi.1995.269.1.G1. [DOI] [PubMed] [Google Scholar]