Abstract
Background
Nuclear pleomorphism and tumor microenvironment (TME) play a critical role in cancer development and progression. Identifying most predictive nuclei and TME features of basal cell carcinoma (BCC) may provide insights into which characteristics pathologists can use to distinguish and stratify this entity.
Objectives
To develop an automated workflow based on nuclei and TME features from basaloid cell tumor regions to differentiate BCC from trichoepithelioma (TE) and stratify BCC into high‐risk (HR) and low‐risk (LR) subtypes, and to identify the nuclear and TME characteristics profile of different basaloid cell tumors.
Methods
The deep learning systems were trained on 161 H&E ‐stained sections which contained 51 sections of HR‐BCC, 50 sections of LR‐BCC and 60 sections of TE from one institution (D1), and externally and independently validated on D2 (46 sections) and D3 (76 sections), from 2015 to 2022. 60%, 20% and 20% of D1 data were randomly splitted for training, validation and testing, respectively. The framework comprised four stages: tumor regions identification by multi‐head self‐attention (MSA) U‐Net, nuclei segmentation by HoVer‐Net, quantitative feature by handcrafted extraction, and differentiation and risk stratification classifier construction. Pixel accuracy, precision, recall, dice score, intersection over union (IoU) and area under the curve (AUC) were used to evaluate the performance of tumor segmentation model and classifiers.
Results
MSA‐U‐Net model detected tumor regions with 0.910 precision, 0.869 recall, 0.889 dice score and 0.800 IoU. The differentiation classifier achieved 0.977 ± 0.0159, 0.955 ± 0.0181, 0.885 ± 0.0237 AUC in D1, D2 and D3, respectively. The most discriminative features between BCC and TE contained Homogeneity, Elongation, T‐T_meanEdgeLength, T‐T_Nsubgraph, S‐T_HarmonicCentrality, S‐S_Degrees. The risk stratification model can well predict HR‐BCC and LR‐BCC with 0.920 ± 0.0579, 0.839 ± 0.0176, 0.825 ± 0.0153 AUC in D1, D2 and D3, respectively. The most discriminative features between HR‐BCC and LR‐BCC comprised IntensityMin, Solidity, T‐T_minEdgeLength, T‐T_Coreness, T‐T_Degrees, T‐T_Betweenness, S‐T_Degrees.
Conclusions
This framework hold potential for future use as a second opinion helping inform diagnosis of BCC, and identify nuclei and TME features related with malignancy and tumor risk stratification.
Keywords: artificial intelligence, basal cell carcinoma vs trichoepithelioma, histopathology images, tumor microenvironment
1. INTRODUCTION
Basal cell carcinoma (BCC) is the most common skin malignancy worldwide, approximately 80% of skin cancer occurrences per year attributed to BCC. 1 The rising incidence of BCC is putting a heavy burden on healthcare systems and economics in terms of diagnosis, treatment and follow‐up. 2
Microscopic examination of pathology slides is the cornerstone of BCC diagnosis. During this process, the diagnosis between BCC and trichoblastoma or trichoepithelioma (TE) is one of the most common clinical practices for dermatopathologist. 3 If a definite diagnosis of BCC is made, pathological subtypes of BCC, which could be stratified as low‐risk BCCs (LR‐BCCs) such as nodular, superficial, pigmented, fibroepithelial, infundibulocystic variants, and high‐risk BCCs (HR‐BCCs), including micronodular, infiltrating, sclerosing/morphoeic, basosquamous and sarcomatoid subtypes should be stated, in order to provide a guidance for proper therapeutic choices. 4
TE and BCC share similar cytology in term of the basaloid epithelial component while the surrounding stromal reaction is different. To differentiate these two entities, a lot of efforts has been made including immunohistochemical stains with minimal help. 5 HR‐BCCs represent the population which are more likely to recurrence and metastasis and need more extensive care in contrast to LR one. They are featured by infiltrating architecture of the epithelial composition and sclerosing mesenchyme with different immune reaction, comparing with the LR counterpart in pathology. These differences including the neoplastic epithelial cells, the tumor microenvironment (TME) and their cross talk are thought to play a critical role in cancer development and its consequent prognosis. 6 Therefore, characterization of the tumor cytology such as nuclear pleomorphism and the surrounding TME may provide subtle evidence to recognize and even stratify BCCs more exactly. However, during the practice of diagnosing BCC, it is difficult for pathologists to quantify the minor difference of nuclei features of basaloid cell tumors and describe their TME signatures. As a result, TME, such as lymphocyte and fibroblast infiltration, is not routinely presented in the pathological diagnostic reports of BCC.
Recently, deep learning algorithms have shown promising results in computing the microscopic features for further quantitative characterization and classification of tissue images of cancers. Early in 1997, Herrera‐Espiñeira C et al. used planar graphs to discriminate benign and malignant breast lesions by measuring nuclear shape, orientation, and texture features from H&E ‐stained images. 7 In 2011, Kararizou et al. reported that the mean value of nuclear orientation was highly correlated to degree of malignancy of human gliomas. 8 In 2021, Mohsin bilal et al. applied HoVer‐Net (a nuclear segmentation model) to segment and classify cell nuclei on H&E ‐stained slides from colorectal cancer, and predicted the status of key molecular pathways and mutations based the nuclear features of tumor images. 9
Traditionally, the classification and segmentation of pathological primitives among different cancer types including BCC have been developed for years. The algorithms have shown excellent performance in recognizing the neoplasms. 10 , 11 , 12 Our team previously developed a cascaded framework based on smartphone‐captured microscopic ocular images to recognize BCC which had a classification model for identifying hard cases and a segmentation model for further in‐depth analysis of them. 13 However, less deep learning study has focused on demonstrating nuclear and TME signatures of basaloid cell tumors and clarified the association between these characteristics and corresponding entities.
Therefore, in this work, we aimed to develop two classifiers, based on tumor images output from (multi‐head self‐attention) MSA‐U‐Net and combined with the fine nuclear features which contain morphology, texture, spatial arrangement of tumor cell nucleus, interrelationships between tumor cells, fibroblasts and lymphocytes, to differentiate and stratify BCC. We also identified the nuclear and TME characteristics profile of different basaloid cell tumors and investigated their diagnosis significance in clinical setting.
2. MATERIALS AND METHODS
2.1. General dataset introduction
A total of 161 H&E slides representing typical cases of LR‐BCC (50), HR‐BCC (51), and TE (60) were obtained from Hospital for Skin Diseases and Institute of Dermatology, Chinese Academy of Medical Sciences & Peking Union Medical College, Nanjing (D1) from January 2015 to October 2022. Two external validation cohorts were tested, the first consisting of 46 slides from Department of Dermatology, School of Medicine, Zhong Da Hospital, Southeast University, Nanjing (D2), the second including 76 slides from Dermatology Hospital, Southern Medical University, Guangzhou (D3) (Table S1). Slides from D1 were manually labelled by three dermatopathologists in consultation with a pathologist. The D1 and 2 were digitized using a Hamamatsu NaNoZoomer S360 digital slide scanner, while D3 was scanned using a Hamamatsu NaNoZoomer‐S60, all at a 40× magnification with 0.23 um pixel‐size.
All methods and procedures were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all participants, and this study was approved by the Institutional Review Board of the Institute of Dermatology, Peking Union Medical College & Chinese Academy of Medical Sciences (2021‐KY‐042).
2.2. Tumor regions identification and nuclei segmentation
The ground‐truth annotations were created in QuPath by experienced pathologists over the full‐resolution images with four categories: normal epithelial tissue (containing epidermis and hair follicle), tumor region (containing BCC and TE), background, other normal tissue. The whole slide segmentation model consisted of an implementation of MSA‐U‐Net model that is trained using 512 × 512 patches sampled from 10× magnification (μm/pixel). To manage class imbalances and prevent overfitting during training, additional rotation, blurring, contrast and color augmentation were applied to each patch. Sixty percent patches were used for training, 20% were used for validation, and the remaining 20% comprised an unseen test set, which was used to test model performance. The segmentation model trained on D1 was able to outline normal epithelial tissue, tumor region, background, other normal tissue by predicting a pixel level classification in a given test image. We calculated pixel accuracy, precision, recall, intersection over union (IoU), and dice score to report the performance of the four classes segmentation model on the test set. Batch level predictions were aggregated to generate WSI level prediction for downstream feature analysis from each independent patch. Then, only tissue with evident tumor features (prediction probability, >average pixel accuracy rate) were included for training in nuclei segmentation model, HoVer‐Net which can segment different types of cell nuclei: non‐neoplastic epithelial cells, neoplastic epithelial cells, inflammatory cells, mesenchymal cells and necrotic cells. 9
2.3. Feature extraction
A total of 262 dimensions of nuclear features (variance in nuclear texture, morphological, spatial arrangement characteristic) and TME profiles (variance in the spatial relationship between tumor nuclei and connective nuclei/ tumor nuclei and immune nuclei/ immune nuclei and connective nuclei) were manually extracted from tumor areas. The features are related to nuclear texture, shape, nuclear spatial arrangement and TME features as described below.
Nuclear morphological/texture features (46 descriptors): this set of characteristics focused on quantifying nuclear texture and shape using different measurements including: area, areaBbox, cellEccentricities, circularity, elongation, and so on.
Nuclear spatial arrangement and TME features (216 descriptors): These features are intended to capture the difference of nuclear topology and spatial structural relationship between tumor nuclei in corresponding conditions. They contain Nsubgraph, degrees, coreness, closeness, and so on.
2.4. Feature selection, and classifier construction and evaluation
Three different feature selection methods, Random Forest, minimum redundancy maximum relevance (mRMR) and Chi‐square test were used to identify the best features that maximally distinguished two classes (BCC and TE; HR‐BCC and LR‐BCC). The most frequently selected features were identified and were quantitatively evaluated using violin plots to compare features expression between BCC and TE (HR‐BCC and LR‐BCC). We limited the number of features for inclusion in the classifier to 20 to avoid the curse of dimensionality problem and model overfitting.
Four different machine learning classifiers, logistic regression, decision tree, support vector machine (SVM), and Random Forest classifier were implemented in conjunction with the selected features. The top‐performing combination of feature selection scheme and machine learning classifier was identified with a five‐fold cross‐validation based on the AUC of patch level within the modeling set. To classify a whole slide image, we constructed an integration module that uses the average‐probability aggregation strategy to average the predicted probabilities for each and tumor type and assigned the type with the larger probability. AUCs was generated based the performance of classifiers on test dataset, and then we applied differentiation and risk stratification models to two external independent test sets (D2, D3) to assess their performance and generalizability. AUC, accuracy, recall, and specificity were used to evaluate to overall performance of the classifier. The workflow is illustrated in Figure 1.
FIGURE 1.
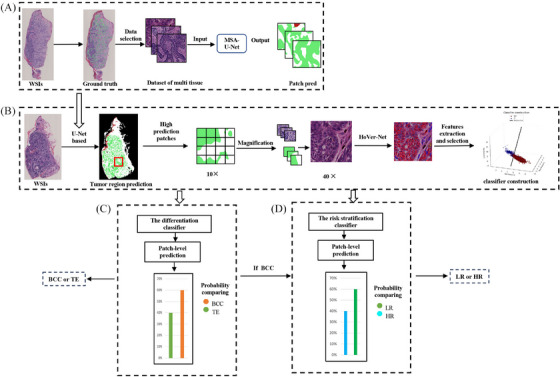
Experimental workflow. (A) Four classes MSA U‐Net based segmentation model (B) Tumor regions identification, nuclear features extraction and selection, and classifier construction. (C) The differentiation classifier generated patch‐level prediction, then compared the probability of BCC and TE, and finally assigned the type with the larger probability. (D) If differentiation classifier predicted the case as BCC, it would transfer to risk stratification classifier for further analysis. BCC, basal cell carcinoma; MSA, multi‐head self‐attention; TE, trichoepithelioma.
3. RESULTS
3.1. Patients and tumor characteristics of three datasets
In D1, regarding BCC, the mean age was 65.69 years (SD 16.15; rang 27–90 years); 38.61% were males. Ninety‐five percent lesions were located on the head. In LR‐BCC dataset, there consisted mainly four subtypes of BCC: superficial (7), nodular (32), pigmented (5), infundibulocystic (6). In HR‐BCC dataset, three histologic subtypes were included: micronodular (13), infiltrating and sclerosing (38). Since it is difficult to distinguish infiltrating type from sclerosing one, these two histologic patterns were regarded as one type. 14 In TE dataset, the mean age was 32.48 years (SD 16.07; rang 6–70); 16.66% were males. All the lesions were located on the head.
In D2, the mean age of BCCs was 72 years (SD 9.61; rang 55–93 years); 45.94% were males. 86.48% were located on the head. There were four subtypes of LR‐BCC, superficial (3), nodular (16), pigmented (2), infundibulocystic (3). Three subtypes of HR‐BCC contained micronodular (4), infiltrating and sclerosing (9). In TE dataset, the mean age was 47 years (SD 10.42; rang 36–70); 55.55% were males.
In D3, the mean age of BCCs was 61.08 years (SD 16.00; rang 11–87 years); 52.63% were males. 84.21% were located on the head. There were four subtypes of LR‐BCC, superficial (10), nodular (18), pigmented (3), infundibulocystic (3). Three subtypes of HR‐BCC contained micronodular (9), infiltrating and sclerosing (14). In TE dataset, the mean age was 32.73 years (SD 19.43; rang 8–82); 42.10% were males. Demographics of the patients, histologic subtypes and anatomic location from three datasets are available in Table 1.
TABLE 1.
Patients and tumor characteristics of three datasets.
| Variable (Dataset 1) | Variable (Dataset 2) | Variable (Dataset 3) | |||
|---|---|---|---|---|---|
| LR‐BCC | Total (50) | LR‐BCC | Total (24) | LR‐BCC | Total (34) |
| Mean age | 59.75 ± 11.91 | Mean age | 72.37 ± 9.31 | Mean age | 64.79 ± 13.82 |
| Sex, n (%) | Sex, n (%) | Sex, n (%) | |||
| Female | 27(54) | Female | 13(54.16) | Female | 12(35.29) |
| Male | 23(46) | Male | 11(45.83) | Male | 22(64.70) |
| Location | Location | Location | |||
| Head and neck | 48(96) | Head and neck | 19(79.16) | Head and neck | 27(79.41) |
| Extremities | 1(2) | Extremities | 1(4.16) | Extremities | 2(5.88) |
| Trunk | 1(2) | Trunk | 2(8.33) | Trunk | 4(11.76) |
| Other | 0 | Other | 2(8.33) | Other | 1(2.94) |
| Diagnosis, n (%) | Diagnosis, n (%) | Diagnosis, n (%) | |||
| Superficial | 7(14) | Superficial | 3(4.16) | Superficial | 10(29.41) |
| Nodular | 32(64) | Nodular | 16(66.66) | Nodular | 18(52.94) |
| Pigmented | 5(10) | Pigmented | 2(8.33) | Pigmented | 3(8.23) |
| Infundibulocystic | 6(12) | Infundibulocystic | 3(12.5) | Infundibulocystic | 3(8.23) |
| HR‐BCC | Total (51) | HR‐BCC | Total (13) | HR‐BCC | Total (23) |
| Mean age | 68.25 ± 16.07 | Mean age | 71.30 ± 10.09 | Mean age | 55.60 ± 17.37 |
| Sex, n (%) | Sex, n (%) | Sex, n (%) | |||
| Female | 35(68.62) | Female | 7(53.84) | Female | 15(56.21) |
| Male | 16(31.37) | Male | 6(46.15) | Male | 8(34.78) |
| Location | Location | Location | |||
| Head and neck | 50(98.03) | Head and neck | 13(100) | Head and neck | 21(91.30) |
| Extremities | 0 | Extremities | 0 | Extremities | 0 |
| Trunk | 0 | Trunk | 0 | Trunk | 2(8.69) |
| Other | 1(1.96) | Other | Other | ||
| Diagnosis, n (%) | Diagnosis, n (%) | Diagnosis, n (%) | |||
| Micronodular | 13(25.49) | Micronodular | 4(30.76) | Micronodular | 9(39.13) |
| Infiltrating and sclerosing | 38(74.50) | Infiltrating and sclerosing | 9(69.23) | Infiltrating and sclerosing | 14(60.86) |
| TE | Total (60) | TE | Total (9) | TE | Total (19) |
| Mean age | 32.48 ± 16.07 | Mean age | 47 ± 10.42 | Mean age | 32.73 ± 19.43 |
| Sex, n (%) | Sex, n (%) | Sex, n (%) | |||
| Female | 50(83.33) | Female | 4(44.44) | Female | 11(57.89) |
| Male | 10(16.66) | Male | 5(55.55) | Male | 8(42.10) |
| Location | Location | Location | |||
| Head and neck | 60(100) | Head and neck | 9(100) | Head and neck | 18(94.73) |
| Extremities | Extremities | Extremities | 1(5.26) |
Abbreviations: BCC, basal cell carcinoma; Infiltrating, infiltrating BCC; Infundibulocystic, Infundibulocystic BCC; Micronodular, Micronodular BCC; Nodular, Nodular BCC; Pigmented, Pigmented BCC; sclerosing, sclerosing BCC; Superficial, superficial BCC; TE, trichoepithelioma.
3.2. Tumor regions identification and the performance of two classifiers
The MSA‐U‐Net algorithm segmented tumor regions with 91.0% precision, 86.9% recall, 88.9% dice score and 80.0% IoU, and the network can achieve a per‐pixel accuracy and an average class accuracy of 93.2% and 88.9% respectively across four classes (background, normal epithelia tissue, tumor and other tissues (Table 2).
TABLE 2.
The performance of four‐class segmentation model.
| Class | Precision | Recall | Dice score | IoU |
|---|---|---|---|---|
| Background | 0.961 | 0.972 | 0.966 | 0.935 |
| Other tissue | 0.880 | 0.874 | 0.877 | 0.781 |
| Normal epithelial tissue | 0.808 | 0.770 | 0.789 | 0.652 |
| Tumor regions (BCC+TE) | 0.910 | 0.869 | 0.889 | 0.800 |
Note: Normal epithelial tissue containing epidermis and hair follicle.
Abbreviations: BCC, basal cell carcinoma; IoU, intersection over union; TE, trichoepithelioma.
Regarding to the differentiation classifier, the performance of the 12 combination of feature selection and classifier schemes in terms of classification performance for differentiation BCC from TE on patch level are summarized in Table 2S. The combination of Random Forest and SVM analysis achieved the best performance on WSI in distinguishing BCC and TE (AUC = 0.98 ± 0.02, accuracy = 0.91 ± 0.0006, recall = 0.97 ± 0.002, specificity = 0.92 ± 0.009); AUC in D2 was 0.95 ± 0.02 (accuracy = 0.91 ± 0.02, recall = 0.92 ± 0.0001, specificity = 0.87 ± 0.0001); AUC in D3 was 0.89 ± 0.02 (accuracy = 0.82 ± 0.001, recall = 0.92 ± 0.0001, specificity = 0.61 ± 0.002) (Figure 2).
FIGURE 2.
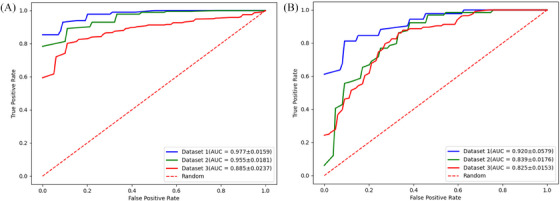
Performance of the differentiation and risk stratification classifier on three independent datasets. (A) The differentiation classifier achieved 0.977 ± 0.0159, 0.955 ± 0.0181, 0.885 ± 0.0237 AUC on D1 (blue), D2 (green), and D3 (red), respectively. The dashed line represents the random prediction 0.5. (B) The risk stratification classifier obtained 0.920 ± 0.0.0579, 0.839 ± 0.0579, 0.825 ± 0.0153 on D1(blue), D2 (green), and D3 (red), respectively.
With regard to risk grading classifier, the performance of the 12 combination of feature selection and classifier schemes in terms of classification performance for risk grading of BCC on patch level are summarized in Table 3S. The feature selection scheme‐Random Forest, combination of SVM classifier achieved the best performance on WSI in discriminating LR‐BCC from HR‐BCC (AUC = 0.92 ± 0.06, accuracy = 0.81 ± 0.02, recall = 0.75 ± 0.03, specificity = 0.87 ± 0.01); AUC in D2 was 0.84 ± 0.02 (accuracy = 0.75 ± 0.0001, recall = 0.60 ± 0.0004, specificity = 0.90 ± 0.001); AUC in D3 was 0.82 ± 0.02 (accuracy = 0.82 ± 0.02, recall = 0.69 ± 0.001, specificity = 0.82 ± 0.001) (Figure 2)
3.3. The discriminative features of differentiation classifier
In the modeling set, there were 20‐dimension discriminative features used to construct the differentiation classifier (11 dimensions related to nuclear texture features, 3 dimensions related to nuclear morphology features, 6 dimensions related to nuclear topology features) (Figure 3) (Table 4S). We ranked the features by the contribution of each feature to the classification target, which is obtained by the feature_importances function of the random forest. These top three discriminative features between BCC and TE were related to the nuclear texture and topological relationship between tumor cells: IntensityMin_mean, IntensityMean_mean, T‐T_meanEdgeLength_mean. The comparison of standardized feature values of corresponding nuclear features between BCC and TE is visualized by violin plots (Figure 3) and for more feature definition and explanation please refer to Table 4S.
FIGURE 3.
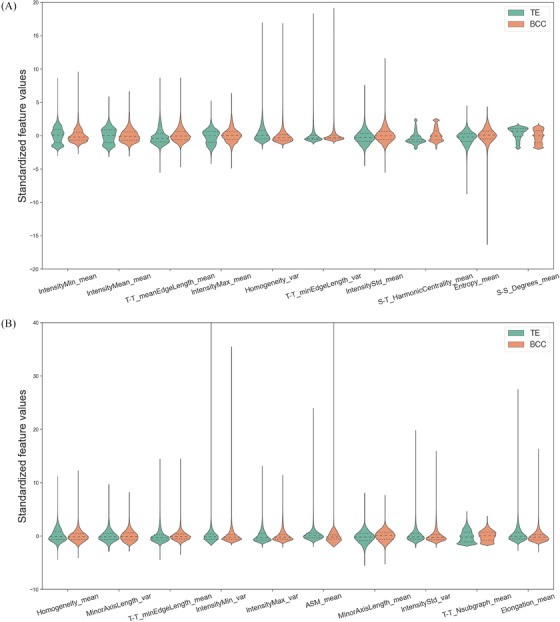
Discriminative features of the differentiation classifier. (A) Top 1 to top 10 nuclear features. (B) Top 11 to top 20 features. Violin plots are used to visualize and compare features expression between BCC and TE. These 20 dimensions nuclear features were statistically different between BCC (orange) and TE (green) (p < 0.05). BCC, basal cell carcinoma; TE, trichoepithelioma.
For nuclear texture features, the nuclear intensity of TE appeared to be more uniform which was supported by the features Texture_Homogeneity_var, Texture_Homogeneity_mean. By contrast, there was a greater variation in nuclear intensity of BCC, evidenced by the feature associated with minimum, mean, maximum or the standard deviation intensity value of nuclear image. The constituent tumor nuclei of TE had a higher ratio of major axis length over the minor axis length than that of BCC in terms of morphological features (indicated by the features Elongation_mean, MinorAxisLength_mean, MinorAxisLength_var). Regarding to topological features, the distance between two connected tumor nuclei in TE is shorter than that of BCC (supported by the features T‐T_meanEdgeLength_mean, T‐T_minEdgeLength_var, T‐T_minEdgeLength_mean). Moreover, BCC had more complex image compositions than TE (supported by T‐T_Nsubgraph_mean). With regard to TME features, the distance between a nucleus of stroma cell and tumor nucleus in TE is shorter than that of BCC (indicated by S‐T_HarmonicCentrality_mean). TE had tighter connectivity between the stromal cells, supported by S‐S_Degrees_mean.
3.4. The discriminative features of risk stratification classifier
There were 20‐dimension discriminative features used to construct the risk stratification classifier (12 dimensions related to nuclear texture features, 1 dimension related to nuclear morphology feature, 7 dimensions related to nuclear topology features) (Figure 4) (Table 5S). We also ranked the features by the contribution of each feature to the classification target, which is obtained by the feature_importances function of the random forest. These top three discriminative features between HR‐BCC and LR‐BCC were all related to the nuclear texture: IntensityMean_var, Homogeneity_var, ASM_mean. The comparison of standardized feature values of corresponding nuclear features between HR‐BCC and LR‐BCC is visualized by violin plot (Figure 4) and for more feature definition and explanation please refer to Table 5S.
FIGURE 4.
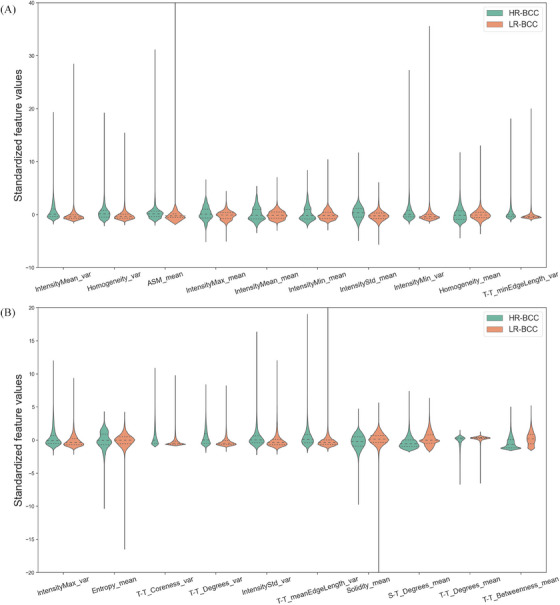
Discriminative features of the risk grading classifier. (A) Top 1 to top 10 nuclear features. (B) Top 11 to top 20 features. Violin plots are used to visualize and compare features expression between HR‐BCC and LR‐BCC. These 20 dimensions nuclear features were statistically different between HR‐BCC (green) and LR‐BCC (orange) (p < 0.05). BCC, basal cell carcinoma; HR, high‐risk; LR, low‐risk.
Regarding nuclear texture features, HR‐BCC had higher intensity value of nucleus than that of LR‐BCC (indicated by the IntensityMin_var, IntensityMin_mean, IntensityMean_var, Homogeneity_var and other nuclear texture feature). For nuclear morphologic features, LR‐BCC had a regular nuclear edge than that of HR‐BCC (supported by Solidity_mean). With regard to topological features, LR‐BCC had a shorter distance between tumor cells and a greater number of connections between tumor cells (supported by T‐T_minEdgeLength_var) than that of HR‐BCC. HR‐BCC had more dispersed tumor masses than LR‐BCC (indicated by T‐T_Coreness_var, T‐T_Degrees_var, T‐T_Degrees_mean, T‐T_Betweenness_mean). LR‐BCC had a greater number of tumor nucleus to stromal nucleus connections in terms of TME features (supported by S‐T_Degrees_mean) in tumor region compared to HR‐BCC.
4. DISCUSSION
In the daily pathological diagnostic workflow of BCC, distinguishing BCC from potential mimics and stating aggressive (HR‐BCC) or nonaggressive types (LR‐BCC) are essential skills for dermatopathologists. 15 Deep learning models have been applied separately for the differentiation and risk stratification of BCC, but not yet been used to construct a pathological framework to accomplish such clinical procedure for daily dermatopathological diagnosis. 16 , 17 In addition, the importance of nuclear and TME features which have been proved to be important factors among other cancer types, and have been used to predict tumor prognosis, benign and malignant judgment, and disease risk classification in non‐dermatopathological area, although these characteristics are often underestimated in dermatopathology. 18 , 19 , 20 , 21 Thus, we developed an automated deep learning system which integrated the nuclear and TME features of basaloid cell tumors to diagnose BCC in a clinical setting.
We constructed two end‐to‐end algorithms to recognize BCC and if a BCC was identified, it would be sent into the risk stratification classifier. Such a combination is similar to pathologist's diagnostic thinking and is expected to be applied in clinical practice. When dealing with common clinical problem (hesitation between BCC and TE), the differentiation classifier can discriminate BCC from TE in model setting with comparable performance to some of previous studies. 17 , 19 The performance of the this classifiers is above mean level (sensitivity = 89.2%, specificity = 81.1%) for machine learning in the diagnosis of non‐melanoma skin cancers, but under the top level to differentiate BCC from TE (recall = 100%). 17 , 19 However, the later model was trained only on 5 sections of TE and test on one slide to differentiate BCC from TE. 17 The small sample size of TE and single center study reduced its confidence of differentiation, while our differentiation classifier is robust to variable H&E stains from different institutions and AUCs were above 0.88 in three different datasets, demonstrating broad applicability and robustness of this algorithm.
Furthermore, in this work, we described the nuclear and TME features difference between BCC and TE, and revealed most representative indicators of them which may help differentiation in future clinicopathological diagnosis. For nuclear texture features, features related to nuclear intensity suggest that nuclear staining in TE is more uniform compared to that of BCC and the indicator of nuclear staining consistency is Homogeneity, followed by other indicators associated with maximum, minimum and mean of the intensity of nuclear texture. The discriminating nuclear texture features signify the uniformity or variance in nuclear texture may help distinguish between benign and malignant lesions. This is intuitive since in benign tumor, the constituted tumor cells are relatively well differentiated as a result of absence of nuclear atypia or nuclear pleomorphism. 22 Also, we checked the measurements related to nuclear morphological features and found the constituent tumor nuclei shared more spindle shape in comparison to BCC. The most suggestive indicators are Elongation and this nuclear morphological can be used as one of key differentiation points between BCC and TE in future clinicopathological diagnosis. Meanwhile, we have looked at features relating to nuclear topology and found increased distance within tumor nuclei in BCC compared with TE. This is in alignment with pathological manifestations which BCC tumor cells are arranged loosely for mucin presenting within or surrounding tumor nodules. 5 The most suggestive indicator of minimum distance between tumor nuclei is T‐T_meanEdgeLength. Furthermore, some other topological features also implied the histopathological images of BCC were relatively more complex than TE which indeed corresponds with the pathological findings. BCCs have variable histological subtypes (superficial, nodular, pigmented, infundibulocystic, micronodular, infiltrating and sclerosing, etc.), while TEs present with a relatively simple morphological pattern which consists of nests and strands of basaloid cells in a racemiform configuration intermingling with or without horn cysts). 5 The image complexity may associate with relatively poor differentiation because of rapid disorganized cells growth and formation of irregular organizational patterns, such as infiltrating BCC. 23 The indicative feature of image complexity is T‐T_Nsubgraph which can also be regarded as one of key diagnosis points of malignant tumor. Apart from texture, morphological, and topological features, we investigated the TME signature difference between BCC and TE and found more stromal nuclei were found within tumor regions than BCC. Correspondingly, TE pathologically has a delicate stroma with abundant spindled cells in comparison to BCC with a myxoid stroma. 5 The most indicative features which represent a stroma enriched spindle cells are S‐T_HarmonicCentrality and S‐S_Degrees.
Or when we confirm the diagnosis of BCC, the risk grading classifier can stratify BCC into HR and LR subtype with AUC 0.920 ± 0.0579 in internal dataset, and 0.839 ± 0.0176, 0.825 ± 0.0153 in two external cohorts. The performance of the risk stratification classifier varied when applied to external test datasets, showing instability of this classifier. The reason for this phenomena might be associated with high histologic or nuclei variations of HR‐BCCs and laboratory differences in terms of tissue preparation and staining processes. 24
Likewise, in our study, we also characterized the nuclear and TME signatures of HR‐BCC and LR‐BCC, and identified the indicator associated with risk grading of cancer which may be applied for prognosis prediction in the future. Discriminative nuclear texture features demonstrated nucleus staining in HR‐BCC may be deeper than that of LR‐BCC, since the nuclear staining may be correlated with the differentiation degree of the tumor cells, and the deeper the nucleus staining represents the relatively poor differentiation the tumor cells are. 25 We also found the nucleus in LR‐BCC had a regular shape in comparison to HR‐BCC which may also associate with differentiation ability of tumor. The more differentiated the tumor is, the more regular the nucleus shape is or the less the nucleus pleomorphism is. The indicator of a regular nuclear shape is Solidity and this feature may be a one of hallmarks of low grade cancer. 25 With regard to topological features, tumor nucleus in LR‐BCC was prone to cluster and well‐circumscribed, while the tumor nucleus in HR‐BCC tended to disperse which is consistent with the pathologic manifestation. 26 The indexes to evaluate the degree of tumor dispersion or aggregation are T‐T_minEdgeLength, T‐T_Coreness, T‐T_Degrees and T‐T_Betweenness which also can be evaluation indexes of tumor risk grading. In addition to nuclear texture, morphological and topological features, the TME feature difference between HR‐BCC and LR‐BCC were identified. One TME feature, S‐T_Degrees, indicated the tumor cells in LR‐BCC are more tightly connected to the fibroblasts. This TME feature suggested the more abundant stromal nuclei within the tumor regions suggests a lower tumor grade and supported the notion that cancer‐associated fibroblasts have tumor‐suppressive functions. 27
There are still several limitations to this deep learning pipeline before it can be safely applied in daily clinical routines. The applicability of the deep learning system remains theoretical. More basaloid cell tumors need be inclusion in the dataset to develop an AI‐based clinical application with wide indications. More slides of HR‐BCC and LR‐BCC should be trained to increase the stability of risk grading classifier.
In summary, the deep learning system could perform several routine pathologist tasks such as BCC differentiation and stratification. Most importantly, we have identified differences of nuclear and TME features within basaloid cell tumors and these discriminative features may be applied for routine pathologic diagnosis workflow in the future. Furthermore, this method could be easily extended to perform other essential tasks such as prognosis prediction and identification the TME signatures of BCC prone to recurrence.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICAL APPROVAL
Reviewed and approved by the institutional review board at Hospital for Skin Diseases and Institute of Dermatology, Chinese Academy of Medical Sciences & Peking Union Medical College, Nanjing.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
The authors have nothing to report. This work was supported by CAMS Innovation Fund for Medical Sciences (2021‐I2M‐C&T‐B‐087) and National Natural Science Foundation of China (Nos. 62171230, 92159301, 91959207, 62301265).
Lan X, Guo G, Wang X, et al. Differentiation and risk stratification of basal cell carcinoma with deep learning on histopathologic images and measuring nuclei and tumor microenvironment features. Skin Res Technol. 2024;30:e13571. 10.1111/srt.13571
Xuemei Lan and Guanchen Guo are considered as joint first authors.
Yiqun Jiang, Jun XU and Xiangxue Wang are considered as joint corresponding authors.
Contributor Information
Xiangxue Wang, Email: xwang@nuist.edu.cn.
Jun Xu, Email: jxu@nuist.edu.cn.
Yiqun Jiang, Email: yiqunjiang2017@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Heath MS, Bar A. Basal cell carcinoma. Dermatol Clin. 2023;41(1):13‐21. [DOI] [PubMed] [Google Scholar]
- 2. Tanese K. Diagnosis and management of basal cell carcinoma. Curr Treat Options Oncol. 2019;20(2):13. [DOI] [PubMed] [Google Scholar]
- 3. Stanoszek LM, Wang GY, Harms PW. Histologic mimics of basal cell carcinoma. Arch Pathol Lab Med. 2017;141(11):1490‐1502. [DOI] [PubMed] [Google Scholar]
- 4. Dika E, Scarfì F, Ferracin M, et al. Basal cell carcinoma: a comprehensive review. Int J Mol Sci. 2020;21(15):5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carrasquillo OY, Cruzval‐O'Reilly E, Sánchez JE, Valentín‐Nogueras SM. Differentiation of basal cell carcinoma and trichoepithelioma: an immunohistochemical study. Am J Dermatopathol. 2021;43(3):191‐197. [DOI] [PubMed] [Google Scholar]
- 6. Chiang E, Stafford H, Buell J, et al. Review of the tumor microenvironment in basal and squamous cell carcinoma. Cancers (Basel). 2023;15(9):2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herrera‐Espiñeira C, Marcos‐Muñoz C, López‐Cuervo JE. Diagnosis of breast cancer by measuring nuclear disorder using planar graphs. Anal Quant Cytol Histol. 1997;19(6):519‐523. [PubMed] [Google Scholar]
- 8. Kararizou EG, Davaki PT, Davou RG, Vassilopoulos D. Morphometric assessments for the prognostic evaluation of human gliomas. Acta Cytol. 2011;55(2):203‐204. [DOI] [PubMed] [Google Scholar]
- 9. Bilal M, Raza SEA, Azam A, et al. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit Health. 2021;3(12):e763‐e772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas SM, Lefevre JG, Baxter G, Hamilton NA. Interpretable deep learning systems for multi‐class segmentation and classification of non‐melanoma skin cancer. Med Image Anal. 2021;68:101915. [DOI] [PubMed] [Google Scholar]
- 11. Kimeswenger S, Tschandl P, Noack P, et al. Artificial neural networks and pathologists recognize basal cell carcinomas based on different histological patterns. Mod Pathol. 2021;34(5):895‐903. [DOI] [PubMed] [Google Scholar]
- 12. Luo Y, Zhang J, Yang Y, et al. Deep learning‐based fully automated differential diagnosis of eyelid basal cell and sebaceous carcinoma using whole slide images. Quant Imaging Med Surg. 2022;12(8):4166‐4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang YQ, Xiong JH, Li HY, et al. Recognizing basal cell carcinoma on smartphone‐captured digital histopathology images with a deep neural network. Br J Dermatol. 2020;182(3):754‐762. [DOI] [PubMed] [Google Scholar]
- 14. Fernández‐Figueras MT, Malvehi J, Tschandl P, et al. Position paper on a simplified histopathological classification of basal cell carcinoma: results of the European Consensus Project. J Eur Acad Dermatol Venereol. 2022;36(3):351‐359. [DOI] [PubMed] [Google Scholar]
- 15. Krakowski AC, Hafeez F, Westheim A, Pan EY, Wilson M. Advanced basal cell carcinoma: what dermatologists need to know about diagnosis. J Am Acad Dermatol. 2022;86(6s):S1‐S13. [DOI] [PubMed] [Google Scholar]
- 16. Bungardean RM, Serbanescu MS, Streba CT, Crisan M. Deep learning with transfer learning in pathology. Case study: classification of basal cell carcinoma. Rom J Morphol Embryol. 2021;62(4):1017‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Brien B, Zhao K, Gibson TA, et al. Artificial intelligence for basal cell carcinoma: diagnosis and distinction from histological mimics. Pathology. 2023;55(3):342‐349. [DOI] [PubMed] [Google Scholar]
- 18. Beiraghi Toosi A, Mohamadian Roshan N, Ghoncheh M. Evaluation of subclinical extension of basal cell carcinoma. World J Plast Surg. 2017;6(3):298‐304. [PMC free article] [PubMed] [Google Scholar]
- 19. Sharma AN, Shwe S, Mesinkovska NA. Current state of machine learning for non‐melanoma skin cancer. Arch Dermatol Res. 2022;314(4):325‐327. [DOI] [PubMed] [Google Scholar]
- 20. Lee G, Veltri RW, Zhu G, Ali S, Epstein JI, Madabhushi A. Nuclear shape and architecture in benign fields predict biochemical recurrence in prostate cancer patients following radical prostatectomy: preliminary findings. Eur Urol Focus. 2017;3(4‐5):457‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romo‐Bucheli D, Janowczyk A, Gilmore H, Romero E, Madabhushi A. A deep learning based strategy for identifying and associating mitotic activity with gene expression derived risk categories in estrogen receptor positive breast cancers. Cytometry A. 2017;91(6):566‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mercan C, Balkenhol M, Salgado R, et al. Deep learning for fully‐automated nuclear pleomorphism scoring in breast cancer. NPJ Breast Cancer. 2022;8(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peris K, Fargnoli MC, Garbe C, et al. Diagnosis and treatment of basal cell carcinoma: European consensus‐based interdisciplinary guidelines. Eur J Cancer. 2019;118:10‐34. [DOI] [PubMed] [Google Scholar]
- 24. Veta M, van Diest PJ, Kornegoor R, Huisman A, Viergever MA, Pluim JP. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS One. 2013;8(7):e70221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peregrina‐Barreto H, Ramirez‐Guatemala VY, Lopez‐Armas GC, Cruz‐Ramos JA. Characterization of nuclear pleomorphism and tubules in histopathological images of breast cancer. Sensors (Basel). 2022;22(15):5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niculet E, Craescu M, Rebegea L, et al. Basal cell carcinoma: comprehensive clinical and histopathological aspects, novel imaging tools and therapeutic approaches (Review). Exp Ther Med. 2022;23(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park D, Sahai E, Rullan A. SnapShot: Cancer‐associated fibroblasts. Cell. 2020;181(2):486‐486.e1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


