Abstract
In Bacillus subtilis, carbon catabolite repression (CCR) of many genes is mediated at cis-acting carbon repression elements (cre) by the catabolite repressor protein CcpA. Mutations in transcription-repair coupling factor (mfd) partially relieve CCR at cre sites located downstream of transcriptional start sites by abolishing the Mfd-mediated displacement of RNA polymerase stalled at cre sites which act as transcriptional roadblocks. Although the acsA cre is centered 44.5 bp downstream of the acsA transcriptional start site, CCR of acsA expression is not affected by an mfd mutation. When the acsA cre is centered 161.5 bp downstream of the transcriptional start site for the unregulated tms promoter, CCR is partially relieved by the mfd mutation. Since CCR mediated at an acsA cre centered 44.5 bp downstream of the tms start site is not affected by the mfd mutation, the inability of Mfd to modulate CCR of acsA expression most likely results from the location of the acsA cre. Higher levels of CCR were found to occur at cre sites flanked by A+T-rich sequences than at cre sites bordered by G and C nucleotides. This suggests that nucleotides adjacent to the proposed 14-bp cre consensus sequence participate in the formation of the CcpA catabolite repression complex at cre sites. Examination of CCR of acsA expression revealed that this regulation required the Crh and seryl-phosphorylated form of the HPr proteins but not glucose kinase.
In Bacillus subtilis, carbon catabolite repression (CCR) of many genes is mediated at a cis-acting site called a carbon repression element (cre) (20). The cre sites for the acsA (15), xyl (22), gnt (11), and hut (31, 43) genes are downstream of the transcriptional start site, while the cre sites for the lev (26), bglPH (23), acu (15), amyE (42), and mmg (3) genes lie within or adjacent to the promoter region (Table 1). While the cre is generally considered to be a 14-bp sequence with dyad symmetry (20, 42), most cre sites are flanked by A+T-rich sequences (Table 1). CCR mediated at all known B. subtilis cre sites is relieved by inactivation of the ccpA gene, which encodes the CCR repressor protein (3, 7, 10, 15, 19, 23, 26, 43). CcpA is a member of the LacI/GalR family of regulatory proteins (19, 41). The cre sites have significant sequence similarity to the operators for other proteins belonging to the LacI/GalR family (20). Recently, CCR of gnt and xyl expression was reported to be partially relieved by inactivation of the ccpB gene, which encodes a CcpA homolog (5). Interestingly, CcpB-dependent regulation was observed only in cells grown in liquid cultures with “low aeration” or on solid medium.
TABLE 1.
Sequence alignment and positions of established carbon repression elementsa
| Gene or operonb | DNA Sequencec | Position (bp)d |
|---|---|---|
| lev | TAACAA TGAAAACGCTTAAC ACAACT | −45.5 |
| gnt (creup) | TAGAAA TGAAAGTGTTTGCA TAAAAG | −37.5 |
| bglPH | CAAAAA TGAAAGCGTTGACA TCTCAC | −36.5 |
| acu | CATTGT TGAAAACGCTTTAT AATTTG | −26.5 |
| amyE | TTTAAA TGTAAGCGTTAACA AAATTC | +4.5 |
| mmg | AGAAAT TGTAAGCGCTGTCT ATCTTC | +21.5 |
| acsA | TGAACT TGAAAGCGTTACCA GCAATA | +44.5 |
| xyl | CTATTT TGGAAGCGTAAACA AAGTGG | +140.5 |
| gnt (credown) | TCTGAT TGAAAGCGGTACCA TTTTAT | +147.5 |
| hut | CGCAAT TGAAACCGCTTCCA AAAAGA | +209.5 |
| Consensus | WWWW TGWAARCGYTWNCW WWWW |
References for each cre are given in the text.
creup and credown denote gnt cre upstream and downstream sites, respectively.
Symbols for ambiguous nucleotides in the consensus sequence are as follows: W represents A or T; R represents A or G; Y represents C or T; and N represents A, C, G, or T.
The location of each cre is given as a center position relative to the transcriptional start site.
Multiple factors, including the protein homologs HPr and Crh as well as glucose-6-phosphate (Glc-6-P), have been proposed to function as corepressors for CcpA binding to cre sites (11, 12, 14, 27). The HPr protein is a signal transduction component of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) (33). In gram-positive bacteria, HPr can be phosphorylated at two residues, His-15 by enzyme I of the PTS and Ser-46 by the ATP-dependent HprK kinase (13, 34, 35). It has been demonstrated in vitro that the seryl-phosphorylated form of HPr (HPr-ser-P) enhances binding of CcpA to the downstream cre sites in the B. subtilis gnt and the Bacillus megaterium xyl operons (11, 14, 27). The in vivo role of HPr in B. subtilis CCR has been studied in strains that contain a mutation in the HPr-encoding ptsH gene that replaces the Ser-46 codon with an alanine codon. This allele, ptsH1, produces a mutant form of HPr that cannot be phosphorylated by HprK (8, 13, 34, 35). The ptsH1 mutation relieves CCR of the bglPH (23), gnt (8, 27), iol (12), lev (26), and xyl (7) genes but does not affect glucose repression of the amyE gene (40). Crh (crh), an HPr homolog, has also been found to be involved in B. subtilis CCR (12). It was reported that complete relief of CCR for inositol dehydrogenase, levanase, and β-xylosidase expression required both ptsH1 and crh mutations (12). Since Crh is phosphorylated at a seryl residue by the HprK kinase, the phosphorylated form of Crh has also been proposed to function as a corepressor for the binding of CcpA (12, 13).
Glc-6-P enhances the in vitro binding of CcpA to multiple cre sites within the B. megaterium xyl and B. subtilis gnt operons (14, 27). The primary cre in the B. megaterium xyl operon is centered 130.5 bp downstream of the xyl transcriptional start site. In the presence of Glc-6-P, CcpA binds cooperatively to this downstream xyl cre and two auxiliary cre sites, one of which is located within the xyl promoter region (14). In contrast, HPr-ser-P enhances noncooperative binding to the primary xyl cre site and the downstream gnt cre site (14, 27). In vivo evidence that Glc-6-P is directly involved in mediating CCR has been suggested by the observation that a loss-of-function mutation in the B. megaterium gene encoding glucose kinase (glk) partially relieves CCR of xyl operon expression (38).
Mutations in mfd, which encodes transcription-repair coupling factor, partially relieve CCR mediated at the downstream cre sites in the hut and gnt operons but not at cre sites located in promoter regions of the bglPH, gnt, and amyE genes (44). Mfd promotes strand-specific DNA repair by displacing RNA polymerase stalled at a nucleotide lesion and recruiting the (A)BC exinuclease to the DNA damage site (36). These results suggest that the downstream cre sites in the hut and gnt operons act as transcriptional roadblocks for RNA polymerase and that Mfd enhances CCR mediated at these sites by displacing RNA polymerase stalled at CcpA-cre complexes in the hut and gnt operons.
Surprisingly, CCR of acsA expression, which requires a cre centered 44.5 bp downstream of the acsA transcriptional start site, is not altered in the mfd mutant (15). In this report, we demonstrate that the location of the acsA cre is most likely responsible for the inability of Mfd to modulate CCR mediated at this cre. Interestingly, nucleotides adjacent to the proposed 14-bp cre consensus sequence were found to contribute to the level of CCR mediated at cre sites.
MATERIALS AND METHODS
Bacterial strains.
Table 2 lists B. subtilis strains used in this study. All lacZ transcriptional fusions were transformed into strain 168 (trpC2) by using plasmid DNA, as previously described (43). The mfd22::Tn10 insertion was transferred by transformation with selection for transposon-encoded chloramphenicol resistance. Transformation with selection for spectinomycin (spc) resistance was used to transfer the glcK::spc, ccpB::spc, and crh::spc mutations. The ptsH1 mutation was transferred by transformation with selection for the genetically linked chloramphenicol resistance gene. Transformants containing the ptsH1 mutation were identified by lack of growth on mannitol minimal medium plates containing ammonium as the nitrogen source (8).
TABLE 2.
B. subtilis strains used in this study
| Strain | Genotypea | Reference, source, and/or derivationb |
|---|---|---|
| 168 | trpC2 | This laboratory |
| QB7097 | trpC2 crh::spc | I. Martin-Verstraete |
| GM1222 | trpC2 pheA1 Δ(bgaX) ptsH1(cat) ΔamyE::(gntRK′-lacZ) | J. Deutscher (8) |
| JZ6 | trpC2 glcK::spc | 168 × pGLK6 |
| JZ7 | trpC2 ccpB::spc | 168 × pCCP105 |
| SF13CDH | trpC2 ccpA::Tn10 | This laboratory |
| SF22CDH | trpC2 mfd22::Tn10 | 44 |
The lacZ α-complementation Escherichia coli strain TOP10 (Invitrogen Corp.) was used as the host for DNA cloning experiments with plasmid pMTL21P (4). E. coli MC1061 contains a deletion of the chromosomal lac genes and was used for the construction of lacZ fusions. A derivative of MC1061 containing the plasmid copy number mutation pcnB80 (25) was used as the host for plasmids containing the B. subtilis glcK gene.
Cell growth, media, and enzyme assays.
The methods used for bacterial cultivation have been previously described (1). Minimal liquid cultures were grown in the morpholinepropanesulfonic acid (MOPS) minimal medium of Neidhardt et al. (29). Glucose and glutamine were added to final concentrations of 0.5 and 0.2%, respectively, to MOPS minimal medium.
Extracts for enzyme assays were prepared as previously described (1). Cells grown in minimal medium were harvested during exponential growth (75 to 85 Klett units). β-Galactosidase was assayed as described previously (1). One unit of β-galactosidase activity produced 1 nmol of o-nitrophenol per min. β-Galactosidase activity was always corrected for endogenous β-galactosidase activity present in B. subtilis 168 cells containing the promoterless lacZ gene from pSFL6 or pSFL7 integrated at the amyE site.
Plasmids and lacZ fusions.
pSFL6 and pSFL7 are neomycin resistance lacZ transcriptional fusion vectors that integrate into the amyE locus and contain promoterless trpA-lacZ and spoVG-lacZ genes, respectively (44). The TMS922 lacZ fusion contains the tms promoter (28) cloned into pSFL7 (44). The HUT924 lacZ fusion is a derivative of pSFL7 that contains a DNA fragment with the hut cre located downstream of the tms promoter (44).
B. subtilis SF13CDH contains a Tn10 transposon insertion in the ccpA gene (9). Chromosomal DNA adjacent to the transposon insertion was cloned by plasmid rescue (39) and used for directed chromosomal walking to obtain DNA downstream of ccpA that contained the acsA gene (16). Plasmid pACS6 was constructed by inserting a SalI-EcoRV DNA fragment containing the acsA promoter and cre into pJDC9 (6). The ACS7 lacZ fusion contains the acsA DNA fragment from pACS6 inserted into pSFL6.
Inactivation of the chromosomal ccpB and glcK genes.
A 1,680-bp DNA fragment containing the ccpB gene was obtained by PCR amplification of B. subtilis chromosomal DNA with Pwo DNA polymerase (Boehringer Mannheim) by using primers CCPB1 (5′-GAAAAAAGGATATTCCGGCACAG) and CCPB2 (5′-TCATTCGCTCTAAATCATTGACCC). Plasmid pCCP104 contains a 1,537-bp PstI-NsiI DNA fragment from the PCR-generated DNA cloned into pMTL21P (4). The ccpB coding sequence contains an HpaI site that overlaps codons 78 and 79. pCCP105 contains a spectinomycin resistance gene cassette (18) inserted into this HpaI site of pCCP104. The chromosomal ccpB gene was disrupted by transforming B. subtilis cells with linearized pCCP105 DNA.
PCR amplification with primers GLKA1 (5′-TTCGCCTTCACACCAGGAGTC) and GLKA2 (5′-CCGTTCATTTTTCGTTGAGGG) was used to obtain a 2,247-bp DNA fragment containing the B. subtilis glcK coding region (37). A 2,039-bp BclI-HindIII DNA fragment from the PCR-generated DNA was cloned into pJDC9 to construct pGLK3. Plasmid pGLK6 contains a spectinomycin resistance gene cassette (18) inserted into an EheI site located within the glcK coding sequence of pGLK3. The chromosomal glcK gene was disrupted by transforming B. subtilis cells with linearized pGLK6 DNA.
Oligonucleotide mutagenesis.
Plasmid pTMS902 contains the tms promoter cloned into the polylinker region of pALTER-1 (Promega Corp.). pTMS949 and pTMS950 were constructed by oligonucleotide-directed mutagenesis of pTMS902 by a protocol provided by the supplier of pALTER-1 (Promega Corp.) to place the acsA cre sequence at +44.5 relative to the tms promoter transcriptional start site. The EcoRI-HindIII fragments from pTMS949 and pTMS950 were cloned into pSFL7 to construct the TMS951 and TMS952 lacZ fusions. pHUT904 was constructed by inserting a hut cre MunI-BsrBI DNA fragment downstream of the tms promoter in pTMS902. Mutations in the hut cre of pHUT904 were generated by oligonucleotide mutagenesis. EcoRI-HindIII DNA fragments containing the tms promoter and mutated hut cre were cloned into pSFL7 to construct the TMS940, TMS942, and TMS948 lacZ fusions.
RESULTS
CCR of acsA expression in an mfd mutant strain.
To determine if the Mfd protein is involved in mediating CCR at the acsA cre, expression of an acsA-lacZ fusion (ACS7) was examined in wild-type and mfd mutant strains. Although the acsA cre site is centered 44.5 bp downstream of the acsA transcriptional start site (15), the mfd mutation does not alter CCR of acsA expression (Table 3). This result was unexpected because several lines of evidence suggested that Mfd would be involved in CCR at a cre located in this position. First of all, Mfd can stimulate transcription-dependent repair at nucleotide lesions located 15 nucleotides downstream of a promoter start site (36). Secondly, when the E. coli lac repressor functions as a transcriptional roadblock in vivo, transcription elongation by RNA polymerase terminates 16 nucleotides upstream of the center of the lac operator (17). If this observation for the lac operator is applied to the acsA cre, then transcription elongation would be expected to terminate 28 nucleotides downstream of the acsA transcriptional start site (assuming that the acsA cre functions as a roadblock). Taken together, these observations suggest that the Mfd protein should be able to recognize and dissociate RNA polymerase from a transcriptional roadblock at the acsA cre.
TABLE 3.
β-Galactosidase expression from lacZ fusions in wild-type and mfd mutant strains
| lacZ fusiona | Relevant genotype | cre position (bp)b | β-Galactosidase sp act (U/mg of protein) in cells grown onc:
|
Glucose repression ratiod | |
|---|---|---|---|---|---|
| Glucose | Citrate | ||||
| ACS7 | Wild type | +44.5 | 3.6 | 62.1 | 17 |
| mfd | +44.5 | 4.6 | 88.1 | 19 | |
| TMS922 | Wild type | 30.5 | 30.4 | 1.0 | |
| mfd | 51.2 | 49.7 | 1.0 | ||
| HUT924 | Wild type | +161.5 | 2.5 | 34.8 | 14 |
| mfd | +161.5 | 13.3 | 68.5 | 5.1 | |
| TMS940 | Wild type | +161.5 | 1.1 | 25.0 | 23 |
| mfd | +161.5 | 5.1 | 45.0 | 8.8 | |
| TMS942 | Wild type | +161.5 | 8.5 | 32.1 | 3.8 |
| mfd | +161.5 | 25.0 | 45.0 | 1.8 | |
| TMS948 | Wild type | +161.5 | 12.9 | 33.7 | 2.6 |
| TMS951 | Wild type | +44.5 | 0.5 | 15.9 | 32 |
| mfd | +44.5 | 0.2 | 35.4 | 177 | |
| TMS952 | Wild type | +44.5 | 5.9 | 30.7 | 5.2 |
| mfd | +44.5 | 8.0 | 53.7 | 6.7 | |
All strains are derivatives of 168, with the indicated lacZ fusion integrated as a single copy at the amyE locus.
Position relative to the transcriptional start site (+1).
Data are averages of three or more determinations which did not vary by more than 20%. Cultures were grown in MOPS minimal medium containing glutamine as the nitrogen source and the indicated carbon source.
The glucose repression ratio was calculated by dividing the enzyme activity found in cultures grown with citrate by the enzyme activity found in glucose-grown cultures.
CCR at cre sites centered 161.5 bp downstream of the transcriptional start site for an unregulated promoter.
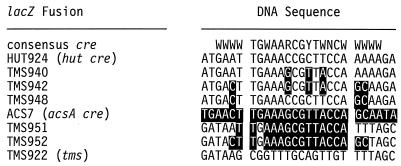
It is possible that the CcpA-acsA cre complex can function as a transcriptional roadblock but that some unique characteristics of the acsA cre, such as the proximity of the acsA cre to the acsA promoter, prevents Mfd from interacting with RNA polymerase. To test this hypothesis, the acsA cre was placed downstream of the tms promoter in a Ptms-lacZ transcriptional fusion and CCR of β-galactosidase expression was examined in wild-type and mfd mutant strains. Expression of the tms promoter is not regulated by CCR in wild-type or mfd mutant cells (TMS922 lacZ fusion [Table 3]). The HUT924 lacZ fusion (44) contains the hut cre site centered 161.5 bp downstream of the transcriptional start site for the tms promoter and is regulated by CCR (Table 3). Oligonucleotide-directed mutagenesis was used to convert the hut cre in HUT924 to an acsA cre by making three nucleotide changes within the 14-bp central hut cre. The resulting TMS940 lacZ fusion contains the central 14-bp sequence of the acsA cre (Fig. 1). β-Galactosidase expression from the TMS940 lacZ fusion was repressed 23-fold by glucose in the wild-type strain but only 8.8-fold in the mfd22 mutant (Table 3). These results indicate that Mfd can modulate CCR mediated at a 14-bp acsA cre centered 161.5 bp downstream of a transcriptional start site.
FIG. 1.
cre DNA sequences. The HUT924 sequence corresponds to the hut cre and flanking DNA present in the HUT924 lacZ fusion (Ptms-hut cre-lacZ). The ACS7 sequence corresponds to the acsA cre with its flanking DNA present in ACS7 (acsA-lacZ) and is shown in reverse text. HUT940, HUT942, and HUT948 were derived from HUT924; the nucleotides shown in reverse text denote the positions within the hut cre that were changed to match the sequence of the acsA cre (ACS7). The TMS922 sequence corresponds to DNA located 44.5 bp downstream of the tms promoter (30). TMS951 and TMS952 were derived from TMS922; the nucleotides shown in reverse text denote nucleotides downstream of the tms promoter that were changed to match the sequence of the acsA cre.
As noted previously, most cre sequences contain a 14-bp core sequence flanked by A or T nucleotides (Table 1). The acsA cre, which contains G or C nucleotides at three of the four positions directly adjacent to the 14-bp core cre sequence, is a notable exception to this generalization (Table 1). To examine the contribution of the flanking G and C nucleotides to the activity of the acsA cre (Table 1), six bases of the hut cre in the HUT924 lacZ fusion were altered so that they matched the acsA cre sequence. The resulting 18-bp acsA cre in the TMS942 lacZ fusion contains the 14-bp core acsA cre sequence and its flanking nucleotides (Fig. 1). Interestingly, β-galactosidase expression from the TMS942 lacZ fusion was repressed only 3.8-fold by glucose in the wild-type cells (Table 3). Expression of the TMS942 fusion was partially relieved by the mfd mutation (Table 3). Although the level of CCR observed with the TMS942 lacZ fusion is surprisingly low, Mfd modulates CCR observed with the TMS942 fusion.
The low level of CCR observed with the TMS942 lacZ fusion suggested that nucleotides flanking the 14-bp core cre sequence can affect the ability of the cre to mediate CCR. To test this hypothesis, three nucleotides flanking the 14-bp hut cre in the HUT924 lacZ fusion were altered so that they matched the flanking nucleotides present in the acsA cre. The resulting lacZ fusion, TMS948, contains the 14-bp hut cre flanked by the G and C nucleotides present in the acsA cre (Fig. 1). Expression of TMS948 was repressed only 2.6-fold by glucose in the wild-type strain (Table 3). Since the level of CCR observed with the TMS948 lacZ fusion (2.6-fold) is lower than that seen with the HUT924 fusion (14-fold), the activity of a cre appears to be dependent upon its flanking nucleotides.
CCR at cre sites centered 44.5 bp downstream of the transcriptional start site for an unregulated promoter.
To determine whether Mfd can modulate CCR mediated at an acsA cre centered 44.5 bp downstream of the transcriptional start site of a heterologous promoter, oligonucleotide-directed mutagenesis was used to place an acsA cre 44.5 bp downstream of the tms promoter. The TMS951 lacZ fusion contains the 14-bp core acsA cre sequence flanked by A+T-rich nucleotides (Fig. 1). The 14-bp core acsA cre sequence in the TMS952 lacZ fusion is flanked by the three G and C bases found adjacent to the acsA cre site (Fig. 1). In wild-type cells, lower levels of CCR were observed with the TMS952 lacZ fusion (5.2-fold) than with the TMS951 lacZ fusion (32-fold) (Table 3). CCR of neither the TMS951 nor the TMS952 lacZ fusion was relieved by the mfd mutation (Table 3).
Role of CcpB, glucose kinase, HPr, and Crh in acsA catabolite repression.
One possible explanation for why Mfd does not modulate acsA CCR is that a unique catabolite repressor complex binds to the acsA cre and functions as an Mfd-independent transcriptional roadblock. This hypothesis was tested by determining whether factors known to participate in CCR at other cre sites are involved in CCR of acsA expression.
The B. subtilis ccpB gene encodes a CcpA homolog which has been reported to mediate CCR of gnt and xyl expression in cells grown on solid medium and in liquid cultures grown with low aeration (5). Although CCR of acsA expression has been shown to be dependent upon CcpA in cells grown in nutrient sporulation medium (15), CcpB may participate in acsA CCR under our growth conditions. Similar levels of CCR for ACS7 were observed in liquid cultures of wild-type and ccpB mutant cells grown in minimal medium (Table 4). In addition, no difference in the color of colonies formed by wild-type and ccpB mutant cells containing the ACS7 fusion was observed on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) minimal plates containing glucose and glutamine as carbon and nitrogen sources (data not shown). Taken together, these results indicate that CcpB does not participate in CCR of acsA expression under these growth conditions.
TABLE 4.
β-Galactosidase expression from an acsA-lacZ fusion in wild-type and mutant strains
| Relevant genotypea | β-Galactosidase sp act (U/mg of protein) in cells grown onb:
|
Glucose repression ratioc | |
|---|---|---|---|
| Glucose | Citrate | ||
| Wild type | 3.6 | 57.8 | 16 |
| ptsH1 | 7.9 | 59.8 | 7.6 |
| crh | 4.0 | 58.5 | 15 |
| crh ptsH1 | 35.2 | 56.3 | 1.6 |
| glcK | 3.7 | 58.4 | 16 |
| ccpB | 4.2 | 55.3 | 13 |
All strains are derivatives of 168 containing the ACS7 lacZ fusion integrated as a single copy at the amyE locus.
Data are averages of three or more determinations which did not vary by more than 20%. Cultures were grown in MOPS minimal medium containing glutamine as the nitrogen source and the indicated carbon source.
The glucose repression ratio was calculated by dividing the enzyme activity found in cultures grown with citrate by the enzyme activity found in glucose-grown cultures.
Glc-6-P has been proposed to serve as a corepressor for the cooperative binding of CcpA to the B. megaterium xyl cre sites (14). There are two pathways for Glc-6-P synthesis in B. subtilis. Glc-6-P is the product of glucose transport by the PTS system (33). Alternatively, glucose can also be directly transported into the cell by the GlcP glucose permease (32) and phosphorylated by glucose kinase to produce Glc-6-P. Two observations indicate that GlcP plays a significant role in glucose transport in B. subtilis. First, the uptake rate of glucose is 30% lower in a glcP mutant than in wild-type cells (32). Second, CCR of gnt expression is partially relieved in B. subtilis glcP mutants (32). Since glcK encodes the only glucose kinase enzyme present in B. subtilis (37), Glc-6-P levels are likely to be reduced in glcK mutants because glucose transported by GlcP cannot be phosphorylated. Because CCR of xyl expression is partially relieved in a B. megaterium strain lacking glucose kinase (37), a B. subtilis glcK mutant strain was used to examine the possibility that Glc-6-P participates in CCR of acsA expression. No difference in the levels of β-galactosidase expression from the ACS7 fusion (acsA-lacZ) was seen in wild-type and glcK cells (Table 4).
The seryl-phosphorylated forms of HPr and Crh have been proposed to function as corepressors for the noncooperative binding of CcpA to cre sites (11, 12, 14). To determine whether HPr and Crh are required for acsA CCR, expression of the ACS7 fusion was examined in B. subtilis strains containing the ptsH1 and crh mutations. In cells grown in minimal medium, the ptsH1 mutation partially relieved CCR of acsA expression while the crh mutation did not affect acsA regulation (Table 4). In a ptsH1 crh double mutant, CCR of acsA expression was almost completely relieved (Table 4).
DISCUSSION
Mfd modulates CCR of gene expression when the acsA cre is centered 161.5 bp, but not 44.5 bp, downstream of the transcriptional start site. This suggests that the location of the cre relative to the transcriptional start site, rather than the promoter or cre sequence, determines whether Mfd participates in CCR mediated at a cre site. It is not clear why Mfd is unable to modulate CCR at a cre centered 44.5 bp downstream of the transcriptional start site. RNA polymerase stalled at a cre site located in this position may have a conformation that prevents Mfd-dependent displacement. Alternatively when the acsA cre is centered 44.5 bp downstream of the transcriptional start site, CcpA may inhibit the initiation of transcription by a cooperative interaction involving DNA looping between the downstream cre site and an unidentified upstream cre site in the promoter region. If the inability of Mfd to modulate CCR at cre sites centered 44.5 bp downstream of both the tms and acsA promoters results from the cooperative binding by CcpA, then these promoter regions must contain unrecognized CcpA binding sites that participate in this cooperative binding.
Both HPr and Crh participate in CCR of acsA expression. In cells grown in minimal medium, mutations in both the ptsH and crh genes are required to significantly relieve CCR of acsA expression. This agrees with previously published results showing that both Crh and HPr are required for wild-type levels of CCR of iol, lev, and β-xylosidase expression in cells grown in minimal medium (12). In vitro studies have shown that HPr-ser-P promotes noncooperative binding of CcpA to cre sites (14, 27), while Glc-6-P triggers cooperative binding of CcpA to the downstream xyl cre and an auxiliary cre site located within the xyl promoter region of the B. megaterium xyl operon (14). The observation that CCR of acsA expression is partially dependent upon HPr, but not glucose kinase, is consistent with the hypothesis that CcpA binds noncooperatively to the acsA cre and acts as a transcriptional roadblock.
The cre site is generally considered to be a 14-bp DNA sequence with dyad symmetry (20, 42). With the exception of the acsA cre site, all known B. subtilis cre sites are surrounded by A+T-rich sequences (Table 1). Mutational analysis of the nucleotides adjacent to the 14-bp cre site showed that the sequence context affects the level of CCR mediated at a cre site. When the hut and acsA cre sites are positioned downstream of the tms promoter, higher levels of CCR are seen at cre sites flanked by A+T-rich DNA regions than at sites containing the same 14-bp core sequence flanked by G and C nucleotides. This suggests that CcpA interacts with base pairs immediately adjacent to the 14-bp core cre sequence and argues that the cre consensus sequence should include these flanking sequences. Examination of the in vitro interactions between CcpA and the amyE cre revealed that CcpA contacts three phosphate groups at each end of the 14-bp core cre sequence (21). Our results suggest that optimal interactions between CcpA and these phosphate groups occur when the cre core sequence is flanked by A+T-rich sequences. Replacement of the A+T-rich sequences adjacent to the cre core sequence with G and C nucleotides could cause subtle DNA conformation changes that diminish these interactions and reduce the affinity of CcpA for the cre site. Similar observations have been made for the binding sites for other members of the LacI/GalR family of regulatory proteins. Analysis of the ideal lac operator revealed that nucleotides flanking the 14-bp operator can affect both the in vivo repression level and the in vitro binding affinity of lac repressor (24).
It is perplexing that the 18-bp acsA cre mediates higher levels of CCR when centered 44.5 bp downstream of the acsA start site in the ACS7 fusion (17-fold) than when this sequence is centered either 44.5 bp downstream of the tms start site in the TMS952 fusion (5.2-fold) or 161.5 bp downstream of the tms start site in TMS942 (3.8-fold). One explanation for this result is that the acsA promoter region contains sequence determinants which enhance CCR at the cre site and that these sequence determinants are not present in the tms promoter region. It is also possible that in addition to CcpA, other trans-acting factors may regulate expression of the acsA promoter in response to carbon availability. Indeed, we have observed that CodY contributes to the regulation of acsA expression in response to carbon availability (9).
ACKNOWLEDGMENTS
We are grateful to Isabelle Martin-Verstraete for strain QB7097 and to Joseph Deutscher for strain GM1222. We thank James Park for providing technical assistance.
This research was supported by Public Health Service research grant GM51127 from the National Institutes of Health.
REFERENCES
- 1.Atkinson M R, Wray L V, Jr, Fisher S H. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J Bacteriol. 1990;172:4758–4765. doi: 10.1128/jb.172.9.4758-4765.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biaudet V, Samson F, Anagnostopoulos C, Ehrlich S D, Bessières P. Computerized genetic map of Bacillus subtilis. Microbiology. 1996;142:2669–2729. doi: 10.1099/13500872-142-10-2669. [DOI] [PubMed] [Google Scholar]
- 3.Bryan E M, Beall B W, Moran C P., Jr A ςE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J Bacteriol. 1996;178:4778–4786. doi: 10.1128/jb.178.16.4778-4786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers S P, Prior S E, Barstow D A, Minton N P. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 5.Chauvaux S, Paulsen I T, Saier M H., Jr CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J Bacteriol. 1998;180:491–497. doi: 10.1128/jb.180.3.491-497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Morrison D A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong antiterminators. Gene. 1987;55:179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- 7.Dahl M K, Hillen W. Contributions of XylR, CcpA and HPr to catabolite repression of the xyl operon in Bacillus subtilis. FEMS Microbiol Lett. 1995;132:79–83. [Google Scholar]
- 8.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, S. H., J. M. Zalieckas, and L. V. Wray, Jr. 1997. Unpublished data.
- 10.Fujita Y, Miwa Y. Catabolite repression of the Bacillus subtilis gnt operon is mediated by the CcpA protein. J Bacteriol. 1994;176:511–513. doi: 10.1128/jb.176.2.511-513.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 12.Galinier A, Haiech J, Kilhoffer M-C, Jaquinod M, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gösseringer R, Deutscher J, Galinier A, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 15.Grundy F J, Turinsky A J, Henkin T M. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 17.Guérin M, Leng M, Rahmouni A R. High resolution mapping of E. coli transcription elongation complex in situ reveals protein interactions with the non-transcribed strand. EMBO J. 1996;15:5397–5407. [PMC free article] [PubMed] [Google Scholar]
- 18.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 19.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 20.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim J-H, Chambliss G H. Contacts between Bacillus subtilis catabolite regulatory protein CcpA and amyO target site. Nucleic Acids Res. 1997;25:3490–3496. doi: 10.1093/nar/25.17.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraus A, Hueck C J, Gärtner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüger S, Hecker M. Regulation of the putative bglPH operon for aryl-β-glucoside utilization in Bacillus subtilis. J Bacteriol. 1995;177:5590–5597. doi: 10.1128/jb.177.19.5590-5597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehming N, Sartorius J, Niemöller M, Genenger G, Wilcken-Bergemann B, Müller-Hill B. The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 1987;6:3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 26.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levenase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miwa Y, Nagura K, Eguchi S, Fukuda H, Deutscher J, Fujita Y. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 28.Moran C P, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;182:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 29.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsson D, Hove-Jensen B, Arnvig K. Primary structure of the tms and prs genes of Bacillus subtilis. Mol Gen Genet. 1989;218:565–571. doi: 10.1007/BF00332425. [DOI] [PubMed] [Google Scholar]
- 31.Oda M, Katagai T, Tomura D, Shoun H, Hoshino T, Furukawa K. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated from the site of transcription. Mol Microbiol. 1992;6:2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 32.Paulsen I T, Chauvaux S, Choi P, Saier M H., Jr Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: identification of a novel hexose:H+ symporter. J Bacteriol. 1998;180:498–504. doi: 10.1128/jb.180.3.498-504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 35.Reizer J, Sutrina S L, Wu L, Deutscher J, Reddy P, Saier M H., Jr Functional interactions between proteins of the phosphoenolpyruvate:sugar phosphotransferase systems of Bacillus subtilis and Escherichia coli. J Biol Chem. 1992;267:9158–9169. [PubMed] [Google Scholar]
- 36.Selby C P, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 37.Skarlatos P, Dahl M K. The glucose kinase of Bacillus subtilis. J Bacteriol. 1998;180:3222–3226. doi: 10.1128/jb.180.12.3222-3226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spath C, Kraus A, Hillen W. Contribution of glucose kinase to glucose repression of xylose utilization in Bacillus megaterium. J Bacteriol. 1997;179:7603–7605. doi: 10.1128/jb.179.23.7603-7605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinmetz M, Richter R. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J Bacteriol. 1994;176:1761–1763. doi: 10.1128/jb.176.6.1761-1763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voskuil M I, Chambliss G H. Significance of HPr in catabolite repression of α-amylase. J Bacteriol. 1996;178:7014–7015. doi: 10.1128/jb.178.23.7014-7015.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weickert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 42.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repressor operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wray L V, Jr, Pettengill F K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zalieckas J M, Wray L V, Jr, Fisher S H. Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Mol Microbiol. 1998;27:1031–1038. doi: 10.1046/j.1365-2958.1998.00751.x. [DOI] [PubMed] [Google Scholar]