Summary
We introduce an all-optical technique that enables volumetric imaging of brain-wide calcium activity and targeted optogenetic stimulation of specific brain regions in unrestrained larval zebrafish. The system consists of three main components: a 3D tracking module, a dual-color fluorescence imaging module, and a real-time activity manipulation module. Our approach uses a sensitive genetically encoded calcium indicator in combination with a long Stokes shift red fluorescence protein as a reference channel, allowing the extraction of Ca2+ activity from signals contaminated by motion artifacts. The method also incorporates rapid 3D image reconstruction and registration, facilitating real-time selective optogenetic stimulation of different regions of the brain. By demonstrating that selective light activation of the midbrain regions in larval zebrafish could reliably trigger biased turning behavior and changes of brain-wide neural activity, we present a valuable tool for investigating the causal relationship between distributed neural circuit dynamics and naturalistic behavior.
Subject areas: Neuroscience, Behavioral neuroscience, Biocomputational method
Graphical abstract

Highlights
-
•
All-optical method enables whole-brain imaging and optogenetics in free-moving zebrafish
-
•
Dual-color imaging and adaptive filtering correct neural activity
-
•
Real-time algorithm enables targeted optogenetics in unrestrained zebrafish
Neuroscience; Behavioral neuroscience; Biocomputational method
Introduction
One of the central questions in systems neuroscience is understanding how distributed neural activity over space and time gives rise to animal behaviors.1,2 This relationship is confounded by recent recordings in several model organisms, which reveal that brain-wide activity is pervaded by behavior-related signals.3,4,5,6,7 All-optical interrogation, which enables simultaneous optical readout and manipulation of activity in brain circuits, opens a new avenue to investigate neural dynamics that are causally related to behaviors and neural representation of behaviors that are involved in different cognitive processes.8,9,10,11
All-optical neurophysiology has been successfully applied to probe the functional connectivity of the neural circuit in vivo and the impact of a genetically or functionally defined group of neurons on the behaviors of head-fixed animals.12,13,14 Here, we extend this technique to freely swimming larval zebrafish, which allows simultaneous targeted stimulation of the brain region of interest and readout of whole-brain Ca2+ activity during naturalistic behavior, such that all sensorimotor loops remain intact and active. Our approach leverages recent advancements in volumetric imaging and machine learning: (1) with the advent of light-field microscope (LFM), brain-wide neural activity can be captured rapidly and simultaneously; (2) deep neural network-based image detection and registration algorithms enable robust real-time tracking and brain region selection for activity manipulation.
There have been several reports on whole brain imaging of freely swimming zebrafish.15,16,17,18,19,20 However, a serious problem can hinder the wide use of this technique: swimming itself causes substantial fluctuations in the brightness of the neural activity indicator. These fluctuations can interfere with the accurate interpretation of true neural activity in zebrafish.19 In an effort to overcome this issue, we have integrated an imaging channel designed for simultaneous pan-neuronal imaging of a long Stokes shift and activity-independent red fluorescence protein (Figure S1). This protein shares the same excitation laser as the Ca2+ indicator. The incorporation of a reference channel, alongside the implementation of an adaptive filter algorithm, enables us to correct activity signals tainted by motion artifacts resulting from the zebrafish’s swift movements.
Figure 1 shows a schematic of our system that integrates tracking, dual-color volumetric fluorescence imaging, and optogenetic manipulation. We performed simultaneous brain-wide Ca2+ signal and reference signal recording using the fast eXtended LFM (XLFM).16 An optogenetic module was incorporated into the imaging system to enable real-time activity manipulation in defined brain regions in unrestrained larval zebrafish.
Figure 1.
Schematics of the dual-color whole-brain imaging and optogenetic system
The system integrated tracking, dual-color light-field imaging, and optogenetic stimulation. A convolutional neural network (CNN) was used to detect the positions of fish head from dark-field images captured by the near-infrared (NIR) tracking camera. A tracking model converted the real-time positional information into analog signals to drive the high-speed motorized stage and compensate for fish movement. The neural activity-dependent green fluorescence signal and the activity-independent red fluorescence signal were split into two beams by a dichroic mirror before entering the two sCMOS cameras separately. The dichroic mirror was placed just before the micro-lens array. Both the red and green fluorophores can be excited by a blue laser (488 nm). An xy galvo system deflected a yellow laser (588 nm) to a user-defined ROI in the fish brain for real-time optogenetic manipulation with the aid of a fast whole-brain image reconstruction and registration algorithm. See also Figure S1.
Results
Tracking system
To reliably maintain the head of a swimming fish within the field of view (FoV) of the microscope, we redesigned our high-speed tracking system16 with two major changes. First, to correctly identify the position of the fish head and yolk from a complex background, we replaced the conventional computer vision algorithms for object detection (that is, background modeling and adaptive threshold) with a U-Net21 image processing module (STAR Methods). This approach greatly improves the accuracy and robustness of the tracking in a complex environment while ensuring a high image detection speed (<3 ms). Second, we combined the current position of the fish and its historical motion trajectory17 to predict fish’s motion (Figure 2A). This allows the system to preemptively adjust the stage position to keep the fish in view, even when it is swimming quickly.
Figure 2.
Tracking system
(A) Flowchart of the tracking system.
(B) A near-infrared (NIR) image captured by the tracking camera, with the tracking error highlighted. The cyan circle indicates the field of view (FoV) of the XLFM. The tracking error is the distance between the center of the fish head and the center of the FoV. The scale bar is 2.5 (left) and 1 mm (right).
(C) The cumulative distribution of the tracking error, based on all time points or when the fish was in motion. “In motion” is defined as a tracking error that lasts for at least 30 ms and exceeds 50 μm in maximum magnitude. The data come from 34 fish, each of which was tracked for more than 10 min. The red dashed line is the maximum tolerable error, the distance beyond which the fish brain is not completely visible in the FoV. (D–G) Tracking example of a freely swimming larval zebrafish stimulated by water flow.
(D) Swimming distance during example trajectory.
(E) The example trajectory. The yellow line indicates the example trajectory of the fish, and the gray lines indicate all other movements in the microfluidic chip.
(F) Top, NIR tracking video images. Scale bar, 2.5 mm. Bottom, reconstructed whole-brain fluorescence images obtained during this trajectory. Scale bar, 300 μm.
(G) Tracking error during this example trajectory. Note that the applied water flow (arrow in E) forced the fish to move backward (Video S1), an unexpected movement pattern for the motion prediction model. As a result, the system shows a larger tracking error in the blue-shaded period. See also Figures S2 and S3.
Example NIR tracking video. A zebrafish was swimming in the presence of water flow. Our tracking algorithm was able to accurately identify the fish head and complete the tracking despite interference from the distracting background.
To quantify the tracking performance, we define the tracking error as the distance between the center of the fish head and the center of the microscope FoV (Figure 2B right). The FoV of the XLFM is 800 μm in diameter, and the size of the brain along the rostrocaudal axis is about 600 μm. Therefore, a tracking error less than 100 μm is sufficient to capture the image of the entire brain. We tested our tracking system in several experimental paradigms, including spontaneous behavior, swimming in the presence of water flow, and during optogenetic stimulation. We found that about 92.6% of the frames fell within 100 μm tracking error at all times (a total of 13,801,838 frames, Figure 2C). During locomotion, 58.69% of the frames were within this range of tracking error (n = 26,930 bouts, 2,293,258 frames). As a comparison, the old tracking system based on threshold segmentation and PID control was able to keep 25.4% of the frames within 100 μm tracking error (Figure S2A) in the presence of large background noise. On the z axis, the depth of field (DOF) of our XLFM is 400 μm.16 The brain of larval zebrafish is about 300 μm thick. When the centers of the fish brain are less than 50 μm away from the focal plane, it is within the range of the DOF of the XLFM. Our current autofocus module operates at a speed of 100 fps and was able to bring 93.29% images within the error tolerance (Figure S2B, n = 15, total 2,113,097 frames).
Figures 2D–2G show the performance of the tracking system when a water flow stimulus was applied to the fish. On this occasion, a large bubble appeared in the microfluidic chamber (Figure 2F, top). Despite a distracting background, the image detection module was still able to accurately identify the position of the fish and keep the head of the fish within the microscope FoV (Figure 2F, bottom). Taken together, these results demonstrate that our tracking system is highly reliable and can be used in a variety of behavior experiments.
Dual-color volumetric image alignment
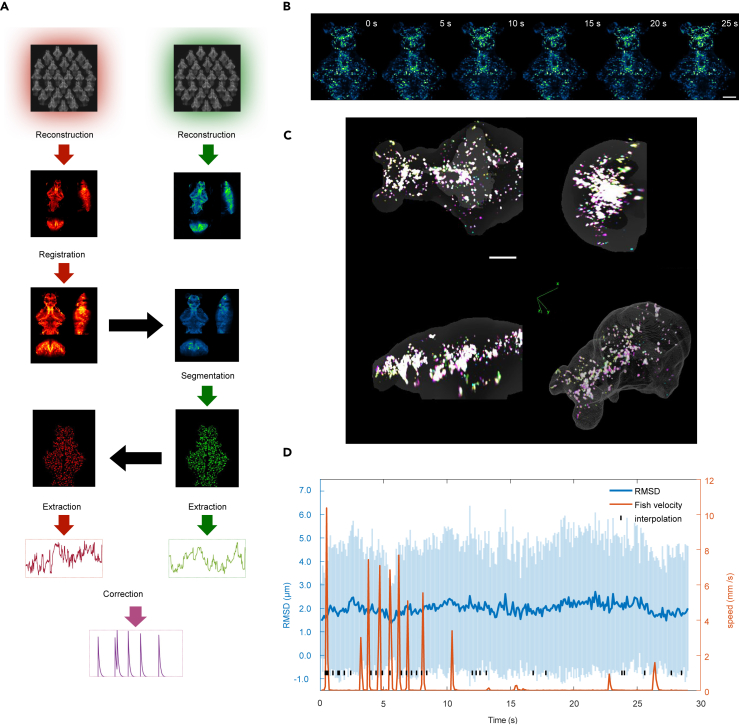
Two-color fluorescence imaging (Figure 1) enables us to use a reference signal (Figure S1) to correct for Ca2+ signal artifacts caused by zebrafish movements. Figure 3A shows each step in the image processing pipeline. Briefly, the reconstructed 3D volumetric image frames of the activity-independent red channel were registered and aligned with a template. The pairwise transformation matrix was then applied to the 3D images of the green channel. The aligned green channel images were segmented into region of interest (ROI) based on the spatiotemporal correlation of the intensity of the voxel (STAR Methods), and the Ca2+ signal in each ROI was extracted and corrected.
Figure 3.
Dual-color image registration
(A) Dual-color image processing pipeline (STAR Methods). The flow chart highlights the following steps: 3D light-field reconstruction; multi-scale image alignment; region of interest (ROI) segmentation; and Ca2+ signal extraction and correction. Black arrows indicate that identical operations can be applied directly to a different channel.
(B) Aligned images of sparsely labeled EGFP zebrafish, guided by the reference channel (Video S2). Scale bar, 100 μm.
(C) 3D visualization of the alignment of (B). Scale bar, 100 μm.
(D) The root-mean-square displacement (RMSD) of the positions of the neuronal center of mass over time. This displacement was measured in relation to their coordinates within a specified reference frame and averaged across all neurons that could be matched within a frame; the shaded region indicates SD. The red curve shows the instantaneous swimming speed of larval zebrafish. See also Figure S4.
Aligning whole-brain image frames is one of the critical steps in extracting the Ca2+ signal accurately. However, the brain regions could deform significantly and the fluorescence intensity of most ROIs could change significantly in swimming zebrafish. These factors make whole-brain image registration a challenging task.
Here, we used CMTK toolkit22 and imregdemons function in MATLAB23 to complete the rigid and nonrigid registration of a 3D image, respectively (STAR Methods). To test our alignment algorithm, we injected the Tol2-elavl3:h2b-EGFP plasmid into fertilized eggs of elavl3:h2b-LSSmCrimson zebrafish. Due to the uneven distribution of the plasmid in the eggs, EGFP did not achieve whole-brain neuronal expression in this generation of zebrafish, but rather showed expression patterns with different degrees of sparseness. We selected zebrafish with moderately sparse expression of EGFP at 6 days post-fertilization (dpf) and recorded their dual-channel images during fish movement. Due to the sparse expression of EGFP, individual neurons randomly distributed in different brain regions can be seen in the 3D reconstructed images. After registering red channel frames with pan-neuronal expression of LSSmCrimson (Figure 3A), we applied the same transformation to the sparse EGFP images (Figure 3B). This allowed us to view the alignment results for each neuron in the green channel (STAR Methods) (Video S2), and to test and optimize our registration algorithm.
Example alignment video. Sparsely EGFP labeled zebrafish were used to test the effectiveness of our alignment method. Left, the fish brain MIP after 3D reconstruction; right, the post-registered fish brain MIP guided by the LSSmCrimson channel.
Figure 3C represents the aligned EGFP multi-frame images (Figure 3B) in different colors, overlaid to visualize the alignment effect. Whiter colors indicate better alignment (Figure 3C). The sparsity of EGFP expression allows us to track about 160 neurons over time according to the spatiotemporal continuity of an object (STAR Methods and Figure S4). We quantified the root-mean-square displacement (RMSD), namely , of the center of mass of a neuron relative to its coordinates in the reference frame (Figure 3D), where the blue line indicates an average over all the neurons whose correspondents could be identified. The RMSD (Figure 3D) is much smaller than the ROI size (8.8–18.3 μm, 25%–75% quantile) resulting from our segmentation algorithm (STAR Methods). Together, these results suggest that our data processing pipeline is effective in aligning whole-brain images of larval zebrafish and thus can extract Ca2+ signals from most ROIs accurately.
Motion artifact correction
Rapid movements of larval zebrafish (translation and tilt) within FoV can cause significant changes in fluorescence intensity in both green and red channels, even when neural activity does not change.19 Here, we introduce an adaptive filter (AF) algorithm (Figure 4A) to correct for motion artifacts (STAR Methods). We use the AF algorithm to dynamically predict the green signal from the signal in the red channel so that the difference between the predicted and the actual green signal in the current time frame , , is as small as possible. In the absence of neural activity, is expected to fluctuate around 0. When there is neural activity, the resulting Ca2+ signal would rise rapidly and would have a large positive value. We identified the predicted as the time-dependent baseline of the signal in the green channel due to movements of zebrafish. The Ca2+ activity was inferred from the normalized signal difference .
Figure 4.
AF algorithm for signal correction
(A) Schematics of the AF algorithm. The AF algorithm works by first estimating the motion artifacts in the green channel using a weighted sum of the red channel signal r(n) over a recent history. The weights w are dynamically updated so that the residual error, , is minimized.
(B) Top, representative raw fluorescence signals from an ROI in freely swimming zebrafish with panneuronal expression of EGFP and LSSmCrimson. Bottom, inferred activity traces. The AF method is more accurate than the conventional ratiometric method (Ratio) in removing motion artifacts.
(C) The histogram shows the distribution of the inferred ROI activity level, defined as the standard deviation of activity over time. Related to (B).
(D) Top, a raw EGFP fluorescence signal with randomly added synthetic neural activity (purple). Bottom, inferred activity trace (yellow). See also Figures S5–S7.
The AF algorithm uses the history-dependent correlation between the green fluorescence signals (jGCaMP8s24) and the red fluorescence signals (LSSmCrimson) to correct for changes in motion-induced signals. These changes are caused by two major factors.
Inhomogeneous light field
When a larval zebrafish moves, the intensity of the excitation light varies across the FoV. In this case, the change of fluorescence intensity in the green channel and that in the red channel differ by a proportionality constant, namely . A simple ratiometric division25,26 between the green and red channels can largely correct for this effect.
Scattering and attenuation
When the larval zebrafish tilts its body, the fluorescence emitted from the same neuron is scattered and obscured by the brain tissues and body pigments. This complicated time-varying process is likely to have a chromatic difference, leading to disproportionate changes in the intensity between the green and red signals. The latter effect cannot be easily corrected for by a direct division. The history dependence of the signal comes from the observation that motion-induced fluorescence changes can persist over multiple frames.
To verify the effectiveness of the motion correction algorithm, we constructed a transgenic zebrafish line with pan-neuronal nucleus expression of EGFP and LSSmCrimson. We then simultaneously recorded the green and red fluorescence signals in a freely swimming larval zebrafish. LSSmCrimson was used to remove the fluctuation of the EGFP signal caused by animal movements. The ideal corrected EGFP signal, which does not change due to neural activity, would be close to 0. Figure 4B shows representative fluorescence signals from an ROI before and after correction. The AF algorithm significantly reduced the motion-induced green fluorescence change compared to the ratiometric method. The improvement over the conventional method was quantified by plotting the distribution of the corrected signal fluctuation (i.e., the standard deviation) across all ROIs (Figure 4C).
After demonstrating that our dual-channel motion correction algorithm can largely eliminate signal changes due to animal movements, we next show that the same algorithm can extract true Ca2+ signal due to neural activity. First, we overlay computer-generated randomly timed neural activity on EGFP signals (Figure 4D, top), namely , where is synthetic neural activity. We then asked whether the AF algorithm could correctly identify these synthetic signals (Figure 4D, bottom) by examining the correlation coefficient r between and the inferred signal. We identified three factors that contribute to the precision of the AF algorithm (Figure S7): a higher amplitude of Ca2+ activity, a higher correlation between the green and red channel signals, and a lower coefficient of variation (CV) of the red channel signal. We established a criterion (see STAR Methods) for screening ROIs using the two-channel signal correlation and the CV of the LSSmCrimson signal, such that the inferred signal and the synthetic signal would exhibit a high correlation. Inferred results from brain regions that met this criterion were considered reliable and used for further analysis.
Second, we wanted to investigate whether we could identify brain regions with similar stimulus-triggered Ca2+ activity patterns in freely swimming larval zebrafish and in the same animal that was immobilized in agarose. We decided to use a spatially invariant external stimulus: the blue excitation laser (488 nm, Figure 1). In other words, we suddenly turned on the blue excitation light when the zebrafish was in the dark, thus inducing brain activity. The main advantage of this approach is that the blue excitation light bathes on the zebrafish head in a cylindrical shape, so the blue laser remains a relatively invariant stimulus for the animal even if its body orientation changes. Furthermore, the sudden onset of blue light in the dark is a powerful stimulus that can easily trigger brain activity.
Figure 5A shows the procedure of our blue light stimulation experiment. First, we presented a freely swimming larval zebrafish with a 20-s blue-light stimulation (1 ms pulsed illumination at 25 Hz) followed by a 30-s dark period. The same animal was then immobilized with agarose and the same visual stimuli were applied. Brain-wide Ca2+ activity was recorded in both trials. We identified ROIs in immobilized zebrafish that showed prominent activity in response to the onset of repeated blue light stimulation (Figure 5B, right). We then examined neural responses in the same ROIs when the animal was swimming freely. The similarity of the Ca2+ dynamics between different experimental conditions (Figure 5A) was quantified by correlation analysis (Figure 5D, left), where each data point represents a single trial from a single ROI. Many trials exhibited high correlation and similarity improved after we applied the AF algorithm to remove motion artifacts. However, we observed a clear difference in the activities of several brain regions between the immobilized state and the freely behaving state. In the freely moving state, significant jGCaMP8 activity did not manifest immediately after blue light stimulation, but emerged later when the zebrafish began to move (e.g., the third trial of the red trace in Figure S8). The late appearance of activity in the green channel and a concurrent drop of the red reference signal led to several large negative correlations in the AF-inferred result (Figure 5D, right).
Figure 5.
Blue light stimulation
(A) Experimental paradigm of the blue light stimulation. A larval zebrafish was freely swimming under our tracking microscope when the blue excitation light (1 ms pulsed stimulation, 2.5% duty cycle) was suddenly turned on. The light was turned on and off at 20/30 s intervals. The same animal was then immobilized in low melting point agarose and an identical pattern of light stimulation was applied. Brain-wide Ca2+ activity was recorded in both conditions.
(B) Top: jGCaMP8s and LSSmCrimson raw fluorescence signals in 5 representative ROIs in which neurons showed prominent Ca2+ activity after light stimulus onset. Shaded regions indicate the dark period. Bottom: inferred calcium activity using the AF algorithm. Left panels are recordings from the free-swimming condition, while the right panels are from the immobilized condition.
(C) The spatial location of the brain regions in (B). Scale bar, 100 μm.
(D) Violin charts of trial-to-trial pairwise correlation between Ca2+ activity in freely swimming and immobile conditions. Left, correlations of raw jGCaMP8s signals; right, correlations of AF inferred signals. Each violin chart represents the distribution of r for each of the 4 trials (see B) in 18 selected ROIs (a total of 72 data points, see Figures S8–S10). See also Figures S8–S10.
Last but not least, we wanted to determine the direction of zebrafish’s turn—left or right—from its brain-wide neural activity recorded during spontaneous movements (see STAR Methods). Using AF-inferred activity data from 50 widely distributed anatomical brain regions, we could discriminate the direction of the zebrafish’s turn with a high degree of precision: 93.6% of classifications were correct on the test set (Figure 6). In particular, while population activity can be used to decode a turning direction on a per-trial basis, individual ROI activities exhibited remarkably high variability from one trial to the next (Figure 6C), making it hard to determine the turning behavior of larval zebrafish. We will consider the implications of this observation in discussion.
Figure 6.
Population activity across multiple brain regions can be used to decode turning direction of zebrafish
(A) Confusion matrix of linear discriminant analysis (LDA).
(B) Spatial distribution of the 50 brain regions on zebrafish brain browser (ZBB). Scale bar, 100 μm.
(C) Activities in 8 brain regions labeled in (B). The thick line indicates the average activity over multiple trials while the thin line shows the AF-inferred activity in a single trial. Green: left turn; Red: right turn. The vertical dashed line indicates the onset of the turn. See also Figure S12.
Real-time optogenetic manipulation of unrestrained larval zebrafish
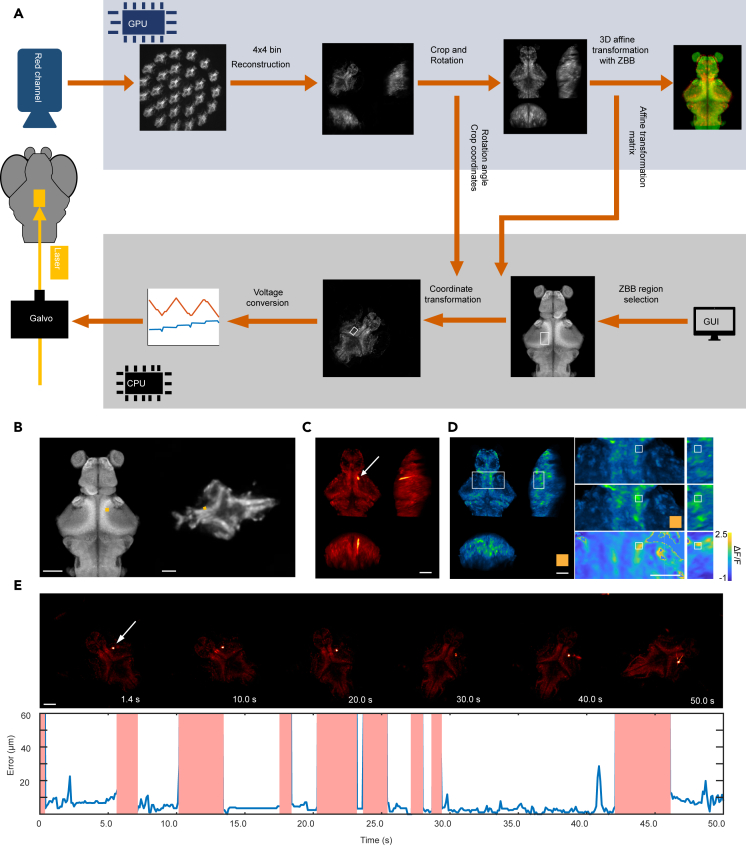
After introducing the dual-color AF method to extract brain-wide Ca2+ activity, we now describe the optogenetic system that enables real-time manipulation of user-defined brain regions in unrestrained larval zebrafish (Figure 1). A user first selects the region to be stimulated on the zebrafish brain browser (ZBB) atlas (Figure 7B, left).27 The system then translates the region into actual locations on the fish brain and delivers photostimulation through real-time image processing and coordinate transformation.
Figure 7.
Real-time optogenetic system
(A) Schematics of the optogenetic stimulation workflow.
(B) Transformation of a user-defined brain region on a ZBB atlas (left) to a real-time position in unrestrained zebrafish (right). The yellow rectangle shows the stimulation pattern. Scale bar, 100 μm.
(C) Pseudo-color fish brain pixel averaged across 200 registered red channel fluorescence images. The white arrow indicates the location of the red fluorescence excited by the laser beam (STAR Methods). Scale bar, 100 μm.
(D) Left, green channel image. Scale bar, 100 μm. Right, a zoomed-in image around the stimulated region (white rectangle) before (top) and during (middle) yellow light stimulation. The color map (bottom) indicates the change in fluorescence intensity (STAR Methods). Scale bar, 50 μm.
(E) Top, images of the fish brain during optogenetic stimulation. Scale bar: 100 μm. Bottom, the displacement between the actual position of the light beam and the targeted position. We deflected the laser out of the FoV during periods of rapid fish movements, indicated by the shaded red regions. See also Figure S11.
Figure 7A shows the workflow of the optogenetic module. We used the red channel fluorescence image for brain region selection. To achieve online image processing, we speed up the reconstruction and registration algorithm by resizing the images, reducing the number of iterations in the reconstruction algorithm,16 and using a deep neural network model to compute the affine transformation matrix.28 These optimizations reduce the image processing time to 80 ms, faster than the acquisition speed of fluorescence imaging (10 Hz). After the coordinates of the user-selected region on the ZBB atlas are translated into the coordinates on the real-time image, the position is converted into a two-dimensional analog voltage signal. This signal is used to control the rapid deflection of the galvo mirror in the X and Y directions, which completes the optical stimulation of a specified brain region. Note that the movement of larval zebrafish is characterized by bout and bout intervals: a pattern of intermittent rapid motion (300 ms in duration) followed by pauses. Our current optogenetic system is designed to target specific regions of the brain during these intervals (STAR Methods and Discussion). Once desired brain activity is triggered, zebrafish can exhibit natural and unrestricted behavioral responses. Importantly, while the zebrafish remains stationary during bout intervals, the orientation of its head varies between bouts. Our algorithm aims to consistently and accurately target the designated brain regions in different orientations.
Here, we used transgenic zebrafish with pan-neuronal expression of jGCaMP8s,24 LSSmCrimson, and the light-sensitive protein ChrimsonR29 (elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson × elavl3:ChrimsonR-tdTomato, 7 dpf) to test the capability of our system. A minimal area was selected in the ZBB atlas (Figure 7B, left) and after coordinate transformation, a yellow laser beam was applied to the corresponding area on the fish head (Figure 7B, right). The full x width at half-maximum (FWHM) of the photostimulation intensity profile is 7.8 μm and the y FWHM is 6.7 μm (Figure S11D). We could identify the activation of the corresponding region in the green channel before and after the onset of optogenetic stimulation (Figures 7D, S11A, and S11B). During stimulation, our aim was to consistently target the same area (Figure S11C) and Figure 7E shows the displacement (or error) between the actual position of the light spot and the target position during a 50-s experiment. The shaded red regions indicate periods of swift fish motion during which the yellow laser was deflected out of the FoV to avoid targeting the wrong area.
Optogenetic manipulation of nMLF
Finally, we demonstrate how our integrated system, which combines 3D tracking, brain-wide Ca2+ imaging, and optogenetic stimulation of a defined brain region, can be used to probe the relationship between neural activity and behavior. We performed a transient (1.5 s) unilateral optogenetic stimulation (Figure 8A) of the tegmentum region including nMLF,30,31 which quickly induced an ipsilateral turn within 2 s in the vast majority of cases (Video S3). Figure 8B shows example bouts from 1 fish and Figure 8C plots the turning angle distribution from 6 fish. Spontaneous turns in the absence of light stimulation did not show directional bias, and the magnitude of a turn was smaller (Figures 8B and 8C). As a control, when the same light stimulation was applied to zebrafish that did not express the light-sensitive protein ChrimsonR, the animals’ movements did not show directional preference: they exhibited more forward runs and when they turned, the turning amplitude was much smaller (Figures 8D and 8E).
Figure 8.
Optogenetic activation of the unilateral nMLF region induced ipsilateral turning behavior and activity changes
(A) Stimulation regions and paradigm. The left and right midbrain regions including nMLF were alternatively stimulated (Video S3). Scale bar, 200 μm.
(B) Example bout trajectories from a elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson × elavl3:ChrimsonR-tdTomato fish.
(C) Histogram of bout angle from all elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson × elavl3:ChrimsonR-tdTomato fish (n = 6).
(D) Example bout trajectory from a elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson fish without the expression of opsin (ChrimsonR) in neurons.
(E) Histogram of bout angle from all elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson fish (n = 5, STAR Methods).
(F) Activity appeared after optogenetic stimulation of the unilateral nMLF region. Active regions were color coded according to their response onset time when the corrected signal intensity reached 20% of their maximum after optogenetic manipulation. Scale bar, 100 μm.
(G) Pairwise Pearson’s r of brain-wide activity between time frames, defined as . Time frames include 500-ms periods (5 frames in each trial and 20 trials) after activating the unilateral nMLF region, as well as randomly selected 500-ms epochs when no optogenetic manipulation was performed. See also Figure S13.
Example optogenetics video. On the left, after unilateral optogenetic activation of the tegmentum region containing nMLF, the zebrafish turned ipsilaterally. On the right, changes in whole-brain activity were recorded simultaneously.
All-optical interrogation enabled us to investigate how local optogenetic manipulation impacts brain-wide activity in unrestrained zebrafish. Figure 8F reveals the appearance of neural activity in brain circuits after optogenetic activation of nMLF. The trial-averaged active regions were colored according to their response onset time, when the activity amplitude reached 20% of its maximum after optogenetic manipulation. We found that evoked neural activity appeared in different regions of the brain, including the tegmentum, the optic tectum, the torus semicircularis, and the cerebellum (Figure S13). In particular, a total of n = 1788 ROIs exhibited prominent Ca2+ activity during optogenetic stimulation of the left or right nMLF region. The population neural activity pattern, which can be viewed as an n-dimensional vector , exhibited higher Pearson’s correlation coefficient in trials when unilateral stimulation was applied to the same side of the brain than when stimulation was applied to the opposite side (Figure 8G and legends).
Discussion
Freely swimming zebrafish exhibit different internal states and behavioral responses to sensory inputs than head-fixed zebrafish.17,31 By combining robust tracking in different conditions, Ca2+ signal correction based on dual-color fluorescence imaging, and optogenetic manipulation of unrestrained zebrafish, our method enables more accurate readouts of brain activity associated with different behaviors, and all-optical interrogation of the brain-wide circuit in a freely swimming larval zebrafish (Figure 9).
Figure 9.
Summary of the all-optical interrogation method
Robust tracking, accurate extraction of Ca2+ activity, as well as manipulation of neural activity in specific brain regions enable us to investigate the neural mechanisms underlying different behaviors of freely swimming zebrafish.
Our new tracking system keeps 92.6% frames within the tolerance range of tracking errors at all times, and 58.69% frames within this range during locomotion (Figure 2C). We found that missing a small number of frames has a minimal effect on the extraction of Ca2+ activity. This is because the movements of zebrafish are punctuated by intervals between bouts, and the actual movement only accounts for a small fraction of the total recording time (16.6%, n = 34 fish). Furthermore, Ca2+ fluorescent activity is typically slow, allowing effective interpolation of lost frames to recapture real neural activity. We verified this conclusion by deleting the same fraction of frames during head-fixed and tail-swing recordings (STAR Methods). The interpolation error was less than 0.1 in almost all brain regions of head-fixed fish (98.6%) and freely swimming fish (95.1%). Looking ahead, we will relax the tracking error tolerance by enlarging the FoV of the fluorescence imaging module. Moreover, we will enhance the tracking speed by improving the algorithms for predicting zebrafish movements and a more responsive moving stage.
Extracting neural activity from Ca2+ signals in the presence of strong noise is a challenging task. Here, we tackle this problem using the AF method. Adaptive filter algorithms in the biomedical field primarily target noise removal in various physiological signals. Examples include the extraction of fetal ECG signals from overlaid maternal ECG signals and the separation of electroencephalogram signals from electrooculogram signals.32,33,34,35 These applications use reference signals to estimate noise signals by dynamically adjusting the parameters for optimal noise removal.
Ca2+ imaging in freely moving small animals such as C. elegans, Drosophila, and zebrafish (where head mounting equipment is impractical) has been approached differently. Some studies omitted signal correction entirely and others used the direct ratiometric method. Our innovation lies in the integration of the AF algorithm into the calcium signal correction procedure. Compared to other motion correction methods,36 the AF algorithm is fast and does not require prior knowledge. To enhance the performance of the algorithm, we constructed a zebrafish line that achieves brain-wide expression of the long Stokes shift fluorescent protein. This ensures that the red emission reference channel and jGCaMP8s, essential for calcium imaging, use an identical excitation light source. This shared excitation maximizes the correlation of the change in fluorescence intensity due to the movement of zebrafish, allowing for a more accurate estimate of the motion artifact in the signal of jGCaMP8s. In our method, motion-induced interference signals contain a factor that is multiplied with the targeted signals (Equation 3), a distinction from earlier techniques where interference was only additive.
Improving the correlation and signal-to-noise ratio (SNR) of the green and red signals allows our AF algorithm to obtain more accurate signal correction results (Figure S7). Using less pigmented and more transparent zebrafish, such as the casper line37 for brain imaging, helps to reduce the chromatic aberration caused by pigmentation and tissue scattering, and could thus improve the correlation between red and green signals. Using brighter red fluorescent proteins and Ca2+ indicators with a larger dynamic range38 would further improve SNR and thus make the extraction of the activity signal more accurate. Another possibility is to use SomaGCaMP39 instead of nuclear-localized GCaMP, which would increase the amount of fluorophore expression. The AF algorithm is not always accurate for all brain regions, especially when the correlation and SNR of dual channel signals are low. To address this issue, we may need to develop refined models that use the statistics of dual-channel signals. One possible improvement is to perform an in-depth statistical analysis of the data collected from the freely swimming EGFP × LSSmCrimson zebrafish. We could also extract features of neural activity-induced signal changes in jGCaMP8s × LSSmCrimson zebrafish paralyzed by bungarotoxin. This prior information, when combined with the location of each brain region, could be used to develop a more accurate model for dual-channel signals.
Previous optogenetic experiments in zebrafish can be divided into two categories: (1) optogenetic manipulation of specific brain regions with spatially patterned illumination in head-fixed zebrafish12,14 and (2) manipulation of freely swimming zebrafish with spatially localized photosensitive proteins using full-field light.40,41 The first method can provide spatially accurate stimulation, but behavior responses are restricted and not natural. The second approach allows optogenetic manipulation of naturalistic behavior, but generating fish lines expressing photosensitive proteins in desired brain regions is challenging and only one spatial pattern of stimulation can be applied to the fish. Our system aims to overcome the limitations of both approaches: it allows selective light stimulation of specific brain regions and rapid switching of stimuli between multiple regions in unrestrained zebrafish.
The current optogenetic module uses a laser without beam expansion, and manipulation of a selected brain region is achieved by high-speed 2D galvo mirror scanning, a design that greatly reduces the loss of laser energy and allows for ultra-high intensity of light projection. An alternative method is to generate patterned illumination using the digital micromirror device,13,42 which could be less energy efficient. However, our design results in optogenetic manipulation without Z-resolution (Figure 7C). A set of lenses could be added before the galvo mirror for beam expansion. This would allow the beam to converge only near the focal plane, and the rapid decrease in light intensity away from the focal plane would enable neurons to be activated only near the focal plane. The position of the focal plane could be adjusted by moving the lens with a piezo.
Our real-time optogenetic system is tailored to target specific brain regions during zebrafish bout intervals. Although we have the potential to enhance our closed-loop speed 10-folds (from 10 to 100 Hz), the zebrafish’s swift movements—with an instantaneous velocity of up to 100 mm/s and accelerations between 1 and 2 g—make it virtually impractical to precisely target a single brain region within one bout without compromising the spatial resolution and accuracy. However, for many of our research interests, this limitation is not detrimental. Unlike C. elegans or Drosophila larva, where constant movement is characteristic, larval zebrafish exhibit more intermittent bouts of activity. We are confident that our system offers a versatile tool for optogenetic manipulation in zebrafish with dense and spatially distributed light-sensitive channels, ensuring intervention in a more natural state of movement.
We use visible light for single-photon optogenetic manipulation, an approach that allows us to manipulate a wide range of brain regions nearly simultaneously and has minimal thermal effects compared to infrared light. However, visible light is easily scattered by brain tissue, making manipulation less spatially accurate, especially for deep brain regions. Recently developed two-photon optogenetics and the holographic technique have enabled the manipulation of multiple neurons at different locations in a fixed 3D volume.9,12 Two-photon microscopy has also demonstrated its ability to track and stimulate a single neuron in a freely moving Drosophila larva.43 The inherent nature of two-photon excitation can significantly reduce tissue scattering and improve manipulation accuracy.44 Combining two-photon optogenetics with one-photon volumetric imaging in unrestrained zebrafish is a promising future direction if the spatial accuracy of optogenetic manipulation at the single-neuron level is critical; if speed and cost are critical, then single-photon optogenetics would be the better choice.
Using linear discriminant analysis, we identified 50 brain regions whose population activity could accurately decode the turning direction of zebrafish (Figure 6). This result does not necessarily imply that the activity of the individual region is a good indicator of turning direction (Figure 6C); nor does it suggest a causal relationship between its activity and a specific turning behavior. Indeed, by selectively activating each of the 50 brain regions, we found that most stimulations did not trigger behavior responses, except for brain regions 1 and 6 (Figures 6B and S12A) that are located near the left and right torus semicircularis correspondingly. Optogenetic stimulation of either region induced more vigorous swimming behavior (Figure S12B) with disproportionately large turning angles (Figure S12C). In particular, stimulating either region could trigger a left or right turn (Figure S12C). On the contrary, we did not observe such behavior changes in control animals that did not express light-sensitive channels ChrimsonR in neurons (Figures S12B and S12C). Together, our results highlight the potential of our integrated system to probe the functional relationship between specific brain regions and behaviors.
Molecular biology approaches can be used to further improve the accuracy and adaptability of optical readouts and the manipulation of defined neural populations. For example, the integration of nuclear localization sequences optimizes the confinement of calcium indicators and RFP within the cell nucleus.45 This optimization could intensify the sparsity of fluorescence expression, thereby elevating the resolution of the reconstructed images. With the aid of suitable promoters and the GAL4/UAS system, opsins can be expressed in defined brain regions or cell types.46,47,48 This capability would enable us to explore the influence of anatomically and/or genetically defined cell assemblies on brain-wide activity and animal behavior.
In conclusion, we anticipate that our all-optical technique, when combined with recent development in volumetric imaging methods,17,19,49,50,51,52,53,54,55 would significantly advance the investigation of neural mechanisms underlying various naturalistic behaviors in zebrafish and other model organisms.25,56,57,58,59
Limitations of the study
Our current optogenetic module does not possess a Z-resolution.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| MAXIscript T7 kit | Thermo Fisher Scientific | Cat #: AM1334 |
| Deposited data | ||
| Datasets and analysis files | This manuscript | https://doi.org/10.6084/m9.Figureshare.24032118.v3 |
| Experimental models: Organisms/strains | ||
| Zebrafish: Tg(elavl3:H2B- EGFP) | This manuscript | N/A |
| Zebrafish: Tg(elavl3:H2B- LSSmCrimson) | This manuscript | N/A |
| Zebrafish: Tg(elavl3: ChrimsonR-tdTomato) | Jiulin Du | N/A |
| Zebrafish: Tg(elavl3: H2B- jGCaMP8s) | This manuscript | N/A |
| Recombinant DNA | ||
| Plasmid: elavl3:H2B- EGFP | This manuscript | N/A |
| Plasmid: elavl3:H2B- LSSmCrimson | This manuscript | N/A |
| Plasmid: elavl3: H2B- jGCaMP8s | This manuscript | N/A |
| Software and algorithms | ||
| MATLAB | Mathworks | www.mathworks.com |
| OptoSwim | This manuscript | github.com/Wenlab/OptoSwim |
| CMTK | NITRC | www.nitrc.org/projects/cmtk |
| Python | Python Software Foundation | www.python.org/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Quan Wen (qwen@ustc.edu.cn).
Materials availability
All unique transgenic lines and plasmids generated in this study are available from the lead contact without restriction.
Data and code availability
-
•
Calcium imaging and behavior data have been deposited at Figureshare and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
All original code has been deposited at GitHub and Figureshare and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
All larval zebrafish were raised in 0.5 × E2 Embryo Media at 28.5°C and a 14/10 hr light/dark cycle. The 0.5 × E2 Embryo Media consisting of 7.5 mM NaCl, 0.25 mM KCl, 0.5 mM MgSO4, 75 μM KH2PO4, 25 μM Na2HPO4, 0.5 mM CaCl2, 0.35 mM NaHCO3. Larval zebrafish aged 6–11 days post-fertilization (dpf) were used for all experiments. Sex discrimination was not included since the sex of zebrafish is not specified at this stage.
All the transgenic fish lines used in this article were generated as follows. The corresponding genes were amplified and cloned into elavl3 promoter and H2B sequence-containing vector. The corresponding plasmid (dosage of 20 ng/μl) was co-injected with Tol2 transposase mRNA (dosage of 50 ng/μl, transcribed using MAXIscript T7 toolkit according to standard procedures) into one-cell stage nacre embryos to make the corresponding transgenic line.
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Science and Technology of China (USTC).
Method details
Hardware
The new system was an update of XLFM.16 The upgraded system consists of three main components: a 3D tracking module, a dual-color fluorescence imaging module, and an optogenetic manipulation module.
The 3D tracking module used a high-speed camera (0.8 ms exposure time, 340 fps, Basler aca2000-340kmNIR, Germany) to capture the lateral motion of the fish. We developed a U-Net21 based system that could rapidly identify the position of the head and the yolk. The error signal between the actual head position and the set point was then fed into the tracking model to generate output signals and control the movement of a high-speed custom stage. The autofocus camera (100 fps, Basler aca2000-340kmNIR) behind a 5-microlens array captured 5 images of the fish from different perspectives. The z position of the fish can be estimated by calculating the inter-fish distance based on the principle of LFM. The error signal between the actual axial position of the fish head and the set point was then fed into the PID to generate an output signal to drive a piezo (PI P725KHDS, 400 μm travel distance) coupled to the fish container.
In the dual-color fluorescence imaging module, a blue excitation laser (Coherent, Sapphire 488 nm, 400 mW) was expanded and collimated into a beam with a diameter of ∼25 mm. It was then focused by an achromatic lens (focal length: 125 mm) and reflected by a dichroic mirror (Semrock, Di02-R488-25X36, US) in the back pupil of the imaging objective (Nikon N25X-APO-MP, 25X, NA 1.1, WD 2 mm, Japan), resulting in an illumination area of ∼1.44 mm in diameter near the objective focal plane. This 488 nm laser was used to simultaneously excite jGCaMP8s and LSSmCrimson. Along the fluorescence imaging light path, the fluorescence collected by the objective was split into two beams by a dichroic mirror (Semrock, FF556-SDi01-25X36) before the microlens arrays and entered the two sCMOS cameras (Andor Zyla 4.2, UK) separately. Each lenslet array consisted of two groups of microlenses with different focal lengths (26 mm or 24.6 mm) to extend the axial field of view while maintaining the same magnification for each subimage. Both lenslet arrays were conjugated to the objective back pupil by a pair of achromatic lenses (focal lengths: F1 = 180 mm and F2 = 160 mm). Two bandpass filters (Semrock FF01-525/45 and Semrock FF02-650/100) were placed before 2 cameras, respectively, to block light from other wavelengths.
A 588 nm laser (CNI MGL-III-588, China) reflected by a 2D galvo mirror system (Thorlabs GVS002, USA) was used for optogenetic manipulation. The midpoint of two galvo mirrors was conjugated onto the back pupil of the imaging objective by a pair of achromatic lenses (focal lengths: F1 = 180 mm and F2 = 180 mm). A dichroic mirror (Semrock, Di01-R405/488/594-25X36) reflected the 588 nm laser and transmitted the green and red fluorescence.
A U-Net neural network to track fish position and orientation
We used a simplified U-Net21 model to detect the head and yolk of the fish. The model contains only two downsampling layers and two upsampling layers, which improves the detection speed. The heat map is used as the output of the model, showing the location of the target point and the confidence level. To create a training dataset, we used custom MATLAB tools (Natick, MA). First, we used k-means to extract the key frames in the video. The key frames covered the variety of fish movements and the complexity of the background. Second, we read the first three keyframes of each video and manually marked the position of the fish’s head and yolk. We rotated the three keyframes until the fish’s head was facing the same direction, calculated the average image, and rotated the average images at 5-degree intervals to create 72 templates. Then, the positions of the head and yolk of the fish in all keyframes were determined using template matching. We manually checked the position marked on each keyframe and corrected for the wrong label. Finally, we created a training dataset by combining all labeled frames and dividing them into subsets of test and training frames.
We used pytorch to build, train and test the model. For training, we used the Adam optimizer and MSE loss function, with a batch size of 32, an initial learning rate of 0.001 and a gradual decay. We saved the model with the lowest loss in the test set and stopped training when the loss was no longer decreasing. We implemented the models in our tracking system using TensorRT. We reduced the model precision to Float16 to improve inference speed without sacrificing inference accuracy.
Model predictive control (MPC)
We adopted the MPC method in17 to control the X-Y motorized stage. We modeled the motion of the stage and the fish, and then selected the optimal stage input by minimizing future tracking error. The stage was modeled as a linear time-invariant system, whose velocity was predicted by convolving the input with the impulse response function of the system. The motion of the fish during a bout was modeled as a uniform linear motion. Instead of directly predicting the trajectory of the fish brain, as in,17 we first predicted the trajectory of the fish yolk, which is much straighter, especially at the beginning of a bout. We then predicted the fish brain position by shifting along the current heading vector. The loss function to be minimized is the sum of squares of tracking error over six times steps into the future, plus an L2 penalty for stage input. We replaced the L2 penalty on the future planned acceleration vector17 with the L2 penalty on stage inputs, which empirically reduced stage vibration.
Assessment of interpolation
Given that a single bout typically concludes within 300 ms and our volumetric imaging rate was 10 Hz, the fish head could be partially or completely out of the FoV in 1 or 2 frames during a rapid movement. Therefore, we performed two types of procedures to assess the impact of missing frames during fish movement: In the context of head-immobilized zebrafish with tail-free movements, we deleted 1 or 2 frames each time the tail swings. From the calcium imaging data of freely swimming zebrafish, we arbitrarily selected a single frame or two successive frames, deleted them, and subsequently replaced them with interpolated data (7.9% of the total frames were interpolated).
To assess the interpolation error, we calculated the mean relative difference between the interpolated signal and the actual jGCaMP8s signal (Figure S3):
| (Equation 1) |
where is the total number of interpolated frames, is the index of interpolated frames, and are the interpolated and actual fluorescence intensity of an ROI, respectively.
Volumetric image registration
Accurate alignment of volumetric images of freely swimming zebrafish larvae is essential for subsequent signal correction and neural activity analysis. Because the green and the red fluorescence signals are derived from splitting a single beam of light, the raw reconstructed volumetric images between the two channels differ only by a simple affine transformation. The red channel is activity-independent and the intensity difference between frames is relatively smaller than that in the green channel. Therefore, we first performed multistep alignment on the red channel and then applied identical operations on the green channel (Figure 3A). Direct alignment of raw images is time-consuming and data-intensive. To address these challenges, we designed and implemented the following four-step registration pipeline.
Crop
We first rotated the reconstructed original 3D images (600 × 600 × 250 voxels) based on the orientation of the fish head recorded by the behavior camera. This rotation aligned the rostrocaudal axis of all frames in the same direction. We then cropped and removed the black space surrounding the fish. Finally, the images were resized to 308 × 380 × 210 voxels, which are the dimensions of the ZBB atlas template.27
Approximate registration
We selected one cropped image and aligned it with the ZBB atlas to generate a unified template for the whole sequence. Next, we use the CMTK toolbox to register each frame with the template using the affine transformation.22,60 Using the ’correlation ratio, CR’ as the registration metric, we achieved the best result.61 With "OpenMP" multithread optimization, we were able to finish one frame of approximate registration in 25 seconds.62
Remove fish eyes
Because the rotation of fish eyes could seriously affect the next step of non-rigid registration, we used a U-net neural network to automatically remove fish eyes.
Diffeomorphic registration
To handle the minute changes in an image caused by breathing, heartbeat, and body twisting, we found it necessary to refine our approximate registration. We used the optical flow algorithm Maxwell’s demons to implement non-rigid registration in the final phase.23 To increase the precision of the optical flow method with our data, we generated a fresh alignment template. This was achieved by taking an average from every tenth frame across every hundred frames within the image sequence. Subsequently, we lined up each segment of the sequence with the newly created alignment templates. This algorithm was developed using MATLAB and can be speeded up via a GPU. In practice, the execution time for each frame was averaged at 72 seconds on a single RTX 3090 GPU. Efficiency can be further optimized by deploying multiple GPUs.
To evaluate alignment accuracy, we first implemented our multistep registration pipeline in the red and green channels of a swimming larval zebrafish. The green channel in this case contained the EGFP signals that were sparsely expressed in the neuronal nuclei. We used the MATLAB toolkit "CellSegm"63 to perform cell segmentation and obtain the centroid coordinates of neurons expressed by EGFP in every frame. We then matched each neuron’s positions in a post-registered time frame to its correspondent in a fixed template frame using the Hungarian algorithm. We identified 164 ± 15 (mean ± SD) neurons in all time frames and 195 neurons in the template. Due to uneven distribution of excitation light in the FoV, scattering, and attenuation of fluorescence by brain tissue and body pigments, not all neurons’ cell bodies were visible in every frame. We introduced the searching radius, a hyperparameter in the Hungarian algorithm that controls how far the algorithm will look for matches. The root mean square displacement (RMSD) between the positions of pairs of matched neurons and the matching ratio both increased with the searching radius and plateaued at a large radius (Figure S4C). We selected the result with a 5-voxel search radius as our best estimate of alignment accuracy (Figure 3D; Figure S4C), because it was large enough to find most of the true matches.
Segmentation
After applying multistep registration to the activity-dependent green channel, we performed cell segmentation based on temporal correlation in the green channel, following the approach introduced in.64 We calculated the average correlation between each voxel and its 14 neighbors to obtain a "correlation map". We then implemented the watershed algorithm on this correlation map to obtain a preliminary segmentation result. For each voxel, we further analyzed its correlation with the average activity of that specific segmented region and obtained the ’coherence map’. Finally, we used a threshold filter on the coherence map to obtain the final segmentation results, which was also applied to the red channel.
Correlation-based segmentation was not suitable for the elavl3:H2B – EGFP × elavl3:H2B - LSSmCrimson fish data because the green channel lacks the neural activity signals. Instead, we divided the entire image into 8 × 8 × 6 voxel regions. The grid size was close to the mean segmented ROI size of our Ca2+ imaging data.
Normalized least-mean-square (NLMS) adaptive filter
We start by presenting a phenomenological model of the fluctuation of fluorescence signals in the activity-dependent green channel and the activity-independent red channel. Noise in the red channel has two major contributions: motion-induced fluctuation of fish and independent noise introduced along the optical pathway and by the sCMOS camera. Here we model the red LSSmCrimson signal from a given ROI at time frame as
| (Equation 2) |
where is independent noise, is the local excitation light intensity and is the baseline fluorescence, which in theory only depends on the number of fluorophores expressed in the neuron. However, fish movements in 3 dimensions make both and time dependent.
We model the green jGCaMP8s signal from the same ROI in a similar way by incorporating the Ca2+ activity .
| (Equation 3) |
where is independent noise and is the baseline. If the emission light fields of the green and red fluorophores captured by the imaging objective and the camera are identical, then we can identify , where is a proportionality constant. Therefore, a simple division between the green and red signals is sufficient to extract Ca2+ activity, provided that the independent noise is small. In practice, due to various scattering of brain tissue and obstruction of body pigments, we are agnostic about the relationship between and ; but it is reasonable to assume that they are strongly correlated in a complicated and time-dependent way, which is consistent with our observation and analysis of the raw EGFP and LSSmCrimson signals.
Under the assumption that motion-induced fluctuations are strongly correlated between the green and the red channels, and such fluctuation is history dependent, we decided to use an adaptive filter (AF) to extract Ca2+ activity corrupted by motion artifacts. We aim to predict the green channel signal from the red channel using the normalized least mean squares (NLMS) AF algorithm.65 The NLMS algorithm continuously subtracts the predicted signal from so that the residue is minimized. The predicted is a weighted sum of the history-dependent red signal:
| (Equation 4) |
where is the filter length and here we set . We iteratively update the weight (i.e., filter) vector using gradient descent:
| (Equation 5) |
We follow66 to determine the iteration step size . Once was computed, we made the following assumption that:
| (Equation 6) |
As a result, the Ca2+ activity is given by
| (Equation 7) |
provided that , namely the independent noise is much smaller than motion-induced fluctuation.
Performance of AF algorithm
To identify the factors that affect the inference ability of the AF algorithm, we added randomly timed synthetic signals with fixed amplitude to the EGFP signals of freely swimming zebrafish. We then evaluated the AF inference performance by calculating the cross-correlation (Pearson’s r) between the AF inferred signal and the synthetic signal for each ROI. We visually represented each ROI’s correlation as colored scattered points in two dimensions with the correlation between dual-channel signals on the y-axis and the coefficient of variation (CV) of the red channel on the x-axis for different synthetic signal amplitudes (Figure S7, left column). ROIs with a higher correlation between dual-channel signals and smaller CV typically exhibit better AF inference performance. As the amplitude of the synthetic signal increases (defined as the CV of an ROI’s EGFP signal multiplied by a constant), the overall inference performance also increases (Figure S7, right column).
To establish a criterion for the screening of ROIs with a potential high AF inference performance, we binarized the r between AF inferred signal and synthetic signal with a threshold value of 0.5. We considered ROIs with a r greater than 0.5 to have a good inference performance (Figure S6). We then used polynomial logistic regression to perform a binary classification task, which can be modeled as:
where is a cubic polynomial function with fitted coefficients, and , are CV of the red channel and Pearson’s r of dual-channel signals respectively. For visualization purposes, the pink regime in Figure S7 corresponds to while the blue one corresponds to ; the dashed line dividing the two regimes can be viewed as a decision boundary. The area under the ROC curve (AUC) was used to quantify the classification performance.
We found that the measured medium amplitude of jGCaMP8s in head-fixed larval zebrafish was about 4 times the CV of EGFP signals in freely swimming zebrafish (Figure S5). We therefore selected the fitted model when the amplitude of synthetic neural activity is four times (4 ×) the CV of the EGFP signals (Figure S7C). Note that when the same model (Figure S7C) was applied to synthetic signals whose amplitudes were sampled from a defined statistical distribution instead of a fixed amplitude, we found a similar classification performance.
Decoding turning direction through linear discriminant analysis
We simultaneously recorded spontaneous locomotion and whole brain Ca2+ activity of elavl3:H2B-jGCaMP8s zebrafish. Taking the head orientation before the bout as 0°, a turn angle >20° after the bout was defined as a right turn bout, and a turn angle <20° after the bout was defined as a left turn bout. We attempted to perform a linear discriminant analysis of these two turn directions using brain activity. First, we used a one-way ANOVA to identify 566 brain regions with significant differences in activity during the two types of bouts. We then used the activity of these 566 brain regions to perform LDA analysis (using the MATLAB function fitcdiscr). We divided the brain activity data during the left-turn bout and the right-turn bout into five equal parts in chronological order, respectively, and then took one part of each to form the test set and the remaining data to form the training set. This yielded 25 unique combinations of training and test sets. The average accuracy performed on the test set was used as the evaluation criterion for the LDA model. The accuracy of LDA using the 566 brain regions was 87.9%. We then selected the 50 brain regions with the highest absolute values of the coefficients in the model and redid the linear discriminant analysis with an accuracy of 93.6%. The results of the 25 tests were pooled to obtain the confusion matrix chart in Figure 6A.
Galvo voltage matrix
We placed a fluorescence plate under the microscope objective and adjusted the stage height and laser intensity to maximize the brightness and minimize the size of the fluorescence spot excited by the optogenetic laser. The galvo input is a voltage pair (GalvoX, GalvoY). We varied the galvo voltage input with an interval of 0.1 V and a range of −1.5 V to +1.5 V in both the X and Y directions. We recorded 10 frames per voltage pair. The raw recorded images were resized to 512 × 512 pixels and reconstructed. We took the coordinates of the brightest point on the image of each voltage pair. If there was no bright spot in the FoV, the voltage pair was deleted. As a result, the correspondence between some of the galvo input voltage pairs and image coordinates was known. Assuming a linear transformation relationship between the voltage pairs and the coordinates, we found the affine transformation matrix using the known points. Then, we calculated the galvo voltage pair corresponding to each point in the image and stored it as the GalvoX and GalvoY voltage matrices.
Online optogenetics pipeline
A custom C++ program was used to implement the optogenetic system. After the images were captured by the red fluorescence camera, the image reconstruction and alignment process were implemented in the GPU, while the coordinate transformation and the control of the galvo were implemented in the CPU.
The image processing algorithm was run on an RTX 3080 Ti GPU using CUDA 11. We resized an image from 2048 × 2048 pixels to 512 × 512 pixels using the AVIR image resizing algorithm designed by Aleksey Vaneev (https://github.com/avaneev/avir). Due to the reduced image size and memory consumption, we could use the PSF of the whole volume to do the deconvolution with a total of 10 iterations. The size of the reconstructed 3D image is 200 × 200 × 50 voxels. It took about 75 ms to reconstruct one frame.
We used TCP to communicate between the tracking system and the optogenetic system. We rotated the fish head orientation of the 3D image to match that of the ZBB atlas using the fish heading angle provided by the tracking system. We then found the maximum connected region by threshold segmentation and removed redundant pixels outside the region. The size of the image after cropping was 95 × 76 × 50 pixels, which is the same as the ZBB atlas. Finally, we aligned the 3D image with the standard brain by affine transformation using a transformer neural network model. Rotation, cropping, and affine alignment took about 10 ms.
The coordinate transformation first calculated the inverse of the affine matrix and the rotation matrix. The user-provided coordinates of the region on the ZBB atlas were then multiplied by the transformation matrix. Finally, the transformed coordinates were shifted by the upper left corner coordinates of the cropped image. This converted the coordinates of the specified region selected in the ZBB atlas to the coordinates of the actual fish brain.
The voltage pairs to be applied to Galvo were read from the GalvoX and GalvoY voltage matrices. The voltage signals were then delivered to the 2D galvo system (Thorlabs GVS002, USA) using an I/O Device (National Instruments PCIe-6321, USA). The galvo system converted the voltage signals into angular displacements of two mirrors, allowing rapid scanning of a specified area.
Zebrafish movements consist of bout and bout intervals, namely rapid swimming periods separated by pause periods. To avoid targeting the wrong brain region, we decided to deliver light stimulation only during the bout interval. We maintained a queue of length 50 that stored the fish heading angle from the tracking system. The average of the heading angle of the fish in the queue was calculated. If the difference between the received fish heading angle and the average in the queue was greater than 5 degrees, the fish was considered to have entered a bout during the delay, and the laser beam was deflected out of the field of view.
Spatial accuracy of optogenetic stimulation
We selected an ROI of 1 × 1 pixel to test spatial precision during continuous optogenetic stimulation in unrestrained zebrafish. A notch filter (Thorlabs NF594-23) was used to exclude the scattering light from the 588 nm laser in Figures 7C and 7D. This filter was removed in order to accurately observe the position of the laser in Figure 7E. It is important to point out that our current optogenetic module does not possess a Z-resolution (see Discussion for potential improvement).
We computed pairwise voxel intensity difference (ΔF/F) between the average green channel image before stimulation and that during stimulation. Voxels with a value less than 20 were considered noisy background and set to 0. We show a single X-Y and Y-Z plane in Figure 7D.
Bout detection and behavior analysis
We use the displacement of the tracking stage δs(t) between consecutive frames to detect the movements of the zebrafish, and create a binary motion sequence by thresholding δs(t) at 20 μm. To identify the bouts, we merged adjacent binary segments to obtain a complete bout sequence. To characterize bouts, we calculated the angle between the heading direction at the beginning of a bout and the heading direction of each frame during a bout (Figures 8B and 8D). The bout angle (Figures 8C and 8E) was defined as the angle between the heading direction at the beginning of a bout and at the end of the bout. Positive values represent right turns.
Brain activity analysis during optogenetic stimulation
Whole brain neural activity were obtained from inferred signals using the AF algorithm, and unreliable regions of the brain were excluded from further analysis.
To characterize brain-wide activity evoked by optogenetic stimulation, we first identified brain regions with significantly elevated neural activity immediately after optogenetic activation of the ipsilateral nMLF. We compared Ca2+ activity in these regions with activity at times without optogenetic manipulation and used a rank sum test with multiple comparison correction to identify significant differences. We then calculated the mean activity trace for each ROI by averaging over multiple trials. Finally, we calculated the time (latency) it took for each ROI to reach 20% of its maximum activity. The latency for each ROI is colored in Figure 8F.
To characterize brain-wide activity and its differences during optogenetic-induced turns, we used a rank sum test with multiple comparison correction to compare Ca2+ activity from 3 frames before to 3 frames after the start of a bout and Ca2+ activity at other moments. We identified 1788 ROIs that exhibited significantly elevated calcium activity during turns.
Next, we selected 5 frames of data after the onset of unilateral nMLF photostimulation: 10 trials during left stimulation and 10 trials during right stimulation, for a total of 100 frames. We also selected 10 time segments, each of which contains 5 frames (50 frames in total), randomly selected from the remaining frames without optogenetic manipulation. We calculated the Pearson’s r between every pair of the population activity vectors. The results were represented by the similarity matrix (Figure 8G) using the frame index.
Optogenetic activation of the brain regions decoding turning direction
To test the relationship between the 50 brain regions identified by LDA and the behavior of zebrafish, we sequentially activated each brain region optogenetically, with a maximum 2.5-second illumination period per trial, and each brain region was tested at least five times. The presence or absence of zebrafish movement during light exposure as well as latency were used as screening criteria for further experiments. Regions 1 and 6 were selected for experiments in 7 elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson × elavl3:ChrimsonR-tdTomato fish. As a control group, the same brain regions were illuminated on 10 elavl3:H2B-jGCaMP8s - elavl3:H2B-LSSmCrimson fish. Finally, the frequency of the bout, that is, the total number of bouts during stimulation divided by the total activation time, the fraction of turning bouts (with a steering angle |θ| > 20°), and the steering angle of each bout were calculated under different conditions. A fish in the control group did not move during each optogenetic activation trial and was excluded from the analysis.
Quantification and statistical analysis
Mann Whitney U test with Bonferroni-Holm adjusted p value was used and data are expressed as the mean ± SEM in Figures S12B and S12C right. A two-sample Kolmogorov-Smirnov test was used in Figure S12C left. Data are expressed as the root mean square displacement (RMSD) ± standard deviation (SD) in Figures 3D and S4B. Data are expressed as the mean ± SD in Figure S13.
Acknowledgments
Q.W. was supported by the National Natural Science Foundation of China (NSFC) under grant NSFC-32071008 and STI2030-Major Projects 2022ZD0211900. We thank Dr. Kai Wang, Lin Cong, Zhenkun Zhang, and Zeguan Wang for their assistance with the imaging hardware design. The authors thank Dr. Jiulin Du for kindly sharing the elavl3: ChrimsonR-tdTomato fish line. The authors thank Dr. Jie He and Xiaoying Qiu for their assistance in generating and rearing the transgenic fish lines.
Author contributions
Q.W. and Y.C. conceived the study. Y.C. built the hardware of tracking, imaging, and optogenetic system. K.Q., D.L., and G.T. developed the tracking software. C.S., Y.C., and Q.W. modified the adaptive filtering algorithm for correction of calcium signals. K.Q. and C.S. developed the real-time optogenetic software. Y.W. constructed the zebrafish strains. Y.C. and K.Q. performed all imaging and optogenetic experiments. C.S., D.L., and G.T. built the calcium imaging data preprocessing pipeline. Y.C. and K.Q. completed the statistics and analysis of the results of the experiment. Y.G. and J.C. developed the LSSmCrimson protein. Y.M. directed the optogenetic experiments. Q.W., Y.C., K.Q., and C.S. wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108385.
Contributor Information
Yuming Chai, Email: ymchai@ustc.edu.cn.
Chen Shen, Email: cshen@ustc.edu.cn.
Quan Wen, Email: qwen@ustc.edu.cn.
Supplemental information
References
- 1.Urai A.E., Doiron B., Leifer A.M., Churchland A.K. Large-scale neural recordings call for new insights to link brain and behavior. Nat. Neurosci. 2022;25:11–19. doi: 10.1038/s41593-021-00980-9. [DOI] [PubMed] [Google Scholar]
- 2.Vyas S., Golub M.D., Sussillo D., Shenoy K.V. Computation Through Neural Population Dynamics. Annu. Rev. Neurosci. 2020;43:249–275. doi: 10.1146/annurev-neuro-092619-094115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker P.R.L., Brown M.A., Smear M.C., Niell C.M. Movement-Related Signals in Sensory Areas: Roles in Natural Behavior. Trends Neurosci. 2020;43:581–595. doi: 10.1016/j.tins.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan H.S., Zimmer M. Brain-wide representations of ongoing behavior: a universal principle? Curr. Opin. Neurobiol. 2020;64:60–69. doi: 10.1016/j.conb.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Stringer C., Pachitariu M., Steinmetz N., Reddy C.B., Carandini M., Harris K.D. Spontaneous behaviors drive multidimensional, brainwide activity. Science. 2019;364:255. doi: 10.1126/science.aav7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musall S., Kaufman M.T., Juavinett A.L., Gluf S., Churchland A.K. Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci. 2019;22:1677–1686. doi: 10.1038/s41593-019-0502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinmetz N.A., Zatka-Haas P., Carandini M., Harris K.D. Distributed coding of choice, action and engagement across the mouse brain. Nature. 2019;576:266–273. doi: 10.1038/s41586-019-1787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emiliani V., Cohen A.E., Deisseroth K., Häusser M. All-Optical Interrogation of Neural Circuits. J. Neurosci. 2015;35:13917–13926. doi: 10.1523/JNEUROSCI.2916-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Russell L.E., Packer A.M., Gauld O.M., Häusser M. Closed-loop all-optical interrogation of neural circuits in vivo. Nat. Methods. 2018;15:1037–1040. doi: 10.1038/s41592-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grosenick L., Marshel J.H., Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan L.Z., Kim D.K., Jennings J.H., Tian H., Wang P.Y., Ramakrishnan C., Randles S., Sun Y., Thadhani E., Kim Y.S., et al. All-optical physiology resolves a synaptic basis for behavioral timescale plasticity. Cell. 2023;186:543–559.e19. doi: 10.1016/j.cell.2022.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dal Maschio M., Donovan J.C., Helmbrecht T.O., Baier H. Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging. Neuron. 2017;94:774–789.e5. doi: 10.1016/j.neuron.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 13.Jiao Z.F., Shang C.F., Wang Y.F., Yang Z., Yang C., Li F.N., Xie J.Z., Pan J.W., Fu L., Du J.L. All-optical imaging and manipulation of whole-brain neuronal activities in behaving larval zebrafish. Biomed. Opt Express. 2018;9:6154–6169. doi: 10.1364/BOE.9.006154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vladimirov N., Wang C., Höckendorf B., Pujala A., Tanimoto M., Mu Y., Yang C.T., Wittenbach J.D., Freeman J., Preibisch S., et al. Brain-wide circuit interrogation at the cellular level guided by online analysis of neuronal function. Nat. Methods. 2018;15:1117–1125. doi: 10.1038/s41592-018-0221-x. [DOI] [PubMed] [Google Scholar]
- 15.Muto A., Ohkura M., Abe G., Nakai J., Kawakami K. Real-time visualization of neuronal activity during perception. Curr. Biol. 2013;23:307–311. doi: 10.1016/j.cub.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 16.Cong L., Wang Z., Chai Y., Hang W., Shang C., Yang W., Bai L., Du J., Wang K., Wen Q. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio) Elife. 2017;6:e28158. doi: 10.7554/eLife.28158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D.H., Kim J., Marques J.C., Grama A., Hildebrand D.G.C., Gu W., Li J.M., Robson D.N. Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat. Methods. 2017;14:1107–1114. doi: 10.1038/nmeth.4429. [DOI] [PubMed] [Google Scholar]
- 18.Symvoulidis P., Lauri A., Stefanoiu A., Cappetta M., Schneider S., Jia H., Stelzl A., Koch M., Perez C.C., Myklatun A., et al. NeuBtracker-imaging neurobehavioral dynamics in freely behaving fish. Nat. Methods. 2017;14:1079–1082. doi: 10.1038/nmeth.4459. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z., Bai L., Cong L., Yu P., Zhang T., Shi W., Li F., Du J., Wang K. Imaging volumetric dynamics at high speed in mouse and zebrafish brain with confocal light field microscopy. Nat. Biotechnol. 2021;39:74–83. doi: 10.1038/s41587-020-0628-7. [DOI] [PubMed] [Google Scholar]
- 20.Marques J.C., Li M., Schaak D., Robson D.N., Li J.M. Internal state dynamics shape brainwide activity and foraging behaviour. Nature. 2020;577:239–243. doi: 10.1038/s41586-019-1858-z. [DOI] [PubMed] [Google Scholar]
- 21.Ronneberger O., Fischer P., Brox T. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation; pp. 234–241. [Google Scholar]
- 22.Rohlfing T.J.N.P., SRI International. Menlo Park C.A. Vol. 3. 2011. (User Guide to the Computational Morphometry Toolkit). [Google Scholar]
- 23.Vercauteren T., Pennec X., Perchant A., Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009;45:S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Rózsa M., Liang Y., Bushey D., Wei Z., Zheng J., Reep D., Broussard G.J., Tsang A., Tsegaye G., et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature. 2023;615:884–891. doi: 10.1038/s41586-023-05828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grover D., Katsuki T., Li J., Dawkins T.J., Greenspan R.J. Imaging brain activity during complex social behaviors in Drosophila with Flyception2. Nat. Commun. 2020;11:623. doi: 10.1038/s41467-020-14487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen J.P., Shipley F.B., Linder A.N., Plummer G.S., Liu M., Setru S.U., Shaevitz J.W., Leifer A.M. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2016;113:E1074–E1081. doi: 10.1073/pnas.1507110112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquart G.D., Tabor K.M., Horstick E.J., Brown M., Geoca A.K., Polys N.F., Nogare D.D., Burgess H.A. High-precision registration between zebrafish brain atlases using symmetric diffeomorphic normalization. GigaScience. 2017;6:1–15. doi: 10.1093/gigascience/gix056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wodzinski M., Müller H. DeepHistReg: Unsupervised Deep Learning Registration Framework for Differently Stained Histology Samples. Comput. Methods Progr. Biomed. 2021;198:105799. doi: 10.1016/j.cmpb.2020.105799. [DOI] [PubMed] [Google Scholar]
- 29.Klapoetke N.C., Murata Y., Kim S.S., Pulver S.R., Birdsey-Benson A., Cho Y.K., Morimoto T.K., Chuong A.S., Carpenter E.J., Tian Z., et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiele T.R., Donovan J.C., Baier H. Descending control of swim posture by a midbrain nucleus in zebrafish. Neuron. 2014;83:679–691. doi: 10.1016/j.neuron.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Severi K.E., Portugues R., Marques J.C., O’Malley D.M., Orger M.B., Engert F. Neural control and modulation of swimming speed in the larval zebrafish. Neuron. 2014;83:692–707. doi: 10.1016/j.neuron.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poungponsri S., Yu X.-H. An adaptive filtering approach for electrocardiogram (ECG) signal noise reduction using neural networks. Neurocomputing. 2013;117:206–213. [Google Scholar]
- 33.Correa A.G., Laciar E., Patiño H.D., Valentinuzzi M.E. Artifact removal from EEG signals using adaptive filters in cascade. J. Phys, Conf. Ser. 2007;90 [Google Scholar]
- 34.Yang B., Duan K., Zhang T. Removal of EOG artifacts from EEG using a cascade of sparse autoencoder and recursive least squares adaptive filter. Neurocomputing. 2016;214:1053–1060. [Google Scholar]
- 35.Graupe D., Zhong Y., Graupe M.H.J.I. Vol. 7. 2005. pp. 166–168. (Extraction of Fetal ECG from Maternal ECG Early in Pregnancy). [Google Scholar]
- 36.Creamer M.S., Chen K.S., Leifer A.M., Pillow J.W. Correcting motion induced fluorescence artifacts in two-channel neural imaging. PLoS Comput. Biol. 2022;18:e1010421. doi: 10.1371/journal.pcbi.1010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., Zon L.I. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Shang Z., Chen J.H., Gu W., Yao L., Yang X., Sun X., Wang L., Wang T., Liu S., et al. Engineering of NEMO as calcium indicators with large dynamics and high sensitivity. Nat. Methods. 2023;20:918–924. doi: 10.1038/s41592-023-01852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shemesh O.A., Linghu C., Piatkevich K.D., Goodwin D., Celiker O.T., Gritton H.J., Romano M.F., Gao R., Yu C.C.J., Tseng H.A., et al. Precision Calcium Imaging of Dense Neural Populations via a Cell-Body-Targeted Calcium Indicator. Neuron. 2020;107:470–486.e11. doi: 10.1016/j.neuron.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oikonomou G., Altermatt M., Zhang R.-w., Coughlin G.M., Montz C., Gradinaru V., Prober D.A. The serotonergic raphe promote sleep in zebrafish and mice. Neuron. 2019;103:686–701.e8. doi: 10.1016/j.neuron.2019.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antinucci P., Folgueira M., Bianco I.H. Pretectal neurons control hunting behaviour. Elife. 2019;8:e48114. doi: 10.7554/eLife.48114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu P., Fajardo O., Shum J., Zhang Schärer Y.P., Friedrich R.W. High-resolution optical control of spatiotemporal neuronal activity patterns in zebrafish using a digital micromirror device. Nat. Protoc. 2012;7:1410–1425. doi: 10.1038/nprot.2012.072. [DOI] [PubMed] [Google Scholar]
- 43.Karagyozov D., Mihovilovic Skanata M., Lesar A., Gershow M. Recording Neural Activity in Unrestrained Animals with Three-Dimensional Tracking Two-Photon Microscopy. Cell Rep. 2018;25:1371–1383.e10. doi: 10.1016/j.celrep.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adesnik H., Abdeladim L. Probing neural codes with two-photon holographic optogenetics. Nat. Neurosci. 2021;24:1356–1366. doi: 10.1038/s41593-021-00902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyssenko N.N., Hanna-Rose W., Schlegel R.A. Cognate putative nuclear localization signal effects strong nuclear localization of a GFP reporter and facilitates gene expression studies in Caenorhabditis elegans. Biotechniques. 2007;43 doi: 10.2144/000112615. 596, 598, 560, 598, 560. [DOI] [PubMed] [Google Scholar]
- 46.Scott E.K., Mason L., Arrenberg A.B., Ziv L., Gosse N.J., Xiao T., Chi N.C., Asakawa K., Kawakami K., Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 47.Antinucci P., Dumitrescu A., Deleuze C., Morley H.J., Leung K., Hagley T., Kubo F., Baier H., Bianco I.H., Wyart C. A calibrated optogenetic toolbox of stable zebrafish opsin lines. Elife. 2020;9:e54937. doi: 10.7554/eLife.54937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Förster D., Dal Maschio M., Laurell E., Baier H. An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nat. Commun. 2017;8:116. doi: 10.1038/s41467-017-00160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J., Lu Z., Jiang D., Guo Y., Qiao H., Zhang Y., Zhu T., Cai Y., Zhang X., Zhanghao K., et al. Iterative tomography with digital adaptive optics permits hour-long intravital observation of 3D subcellular dynamics at millisecond scale. Cell. 2021;184:3318–3332.e17. doi: 10.1016/j.cell.2021.04.029. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Lu Z., Wu J., Lin X., Jiang D., Cai Y., Xie J., Wang Y., Zhu T., Ji X., Dai Q. Computational optical sectioning with an incoherent multiscale scattering model for light-field microscopy. Nat. Commun. 2021;12:6391. doi: 10.1038/s41467-021-26730-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voleti V., Patel K.B., Li W., Perez Campos C., Bharadwaj S., Yu H., Ford C., Casper M.J., Yan R.W., Liang W., et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods. 2019;16:1054–1062. doi: 10.1038/s41592-019-0579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Z., Zhou Y., Liu B., He J., Zhao J., Cai Y., Fan J., Li X., Wang Z., Lu Z., et al. Two-photon synthetic aperture microscopy for minimally invasive fast 3D imaging of native subcellular behaviors in deep tissue. Cell. 2023;186:2475–2491.e22. doi: 10.1016/j.cell.2023.04.016. [DOI] [PubMed] [Google Scholar]
- 53.Lu R., Liang Y., Meng G., Zhou P., Svoboda K., Paninski L., Ji N. Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nat. Methods. 2020;17:291–294. doi: 10.1038/s41592-020-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demas J., Manley J., Tejera F., Barber K., Kim H., Traub F.M., Chen B., Vaziri A. High-speed, cortex-wide volumetric recording of neuroactivity at cellular resolution using light beads microscopy. Nat. Methods. 2021;18:1103–1111. doi: 10.1038/s41592-021-01239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nöbauer T., Zhang Y., Kim H., Vaziri A. Mesoscale volumetric light-field (MesoLF) imaging of neuroactivity across cortical areas at 18 Hz. Nat. Methods. 2023;20:600–609. doi: 10.1038/s41592-023-01789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson R.E., Linderman S., Panier T., Wee C.L., Song E., Herrera K.J., Miller A., Engert F. Probabilistic Models of Larval Zebrafish Behavior Reveal Structure on Many Scales. Curr. Biol. 2020;30:70–82.e4. doi: 10.1016/j.cub.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marques J.C., Lackner S., Felix R., Orger M.B. Structure of the Zebrafish Locomotor Repertoire Revealed with Unsupervised Behavioral Clustering. Curr. Biol. 2018;28:181–195.e185. doi: 10.1016/j.cub.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 58.Orger M.B., de Polavieja G.G. Zebrafish Behavior: Opportunities and Challenges. Annu. Rev. Neurosci. 2017;40:125–147. doi: 10.1146/annurev-neuro-071714-033857. [DOI] [PubMed] [Google Scholar]
- 59.Grover D., Katsuki T., Greenspan R.J. Flyception: imaging brain activity in freely walking fruit flies. Nat. Methods. 2016;13:569–572. doi: 10.1038/nmeth.3866. [DOI] [PubMed] [Google Scholar]
- 60.Studholme C., Hill D.L., Hawkes D.J. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med. Phys. 1997;24:25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- 61.Roche A., Malandain G., Pennec X., Ayache N. Springer; 1998. The Correlation Ratio as a New Similarity Measure for Multimodal Image Registration; pp. 1115–1124. [Google Scholar]
- 62.Chandra R. 2001. Parallel Programming in OpenMP (Morgan Kaufmann) [Google Scholar]
- 63.Hodneland E., Kögel T., Frei D.M., Gerdes H.-H., Lundervold A. CellSegm-a MATLAB toolbox for high-throughput 3D cell segmentation. Source Code Biol Med. 2013;8:1–24. doi: 10.1186/1751-0473-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith S.L., Häusser M. Parallel processing of visual space by neighboring neurons in mouse visual cortex. Nat. Neurosci. 2010;13:1144–1149. doi: 10.1038/nn.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haykin S.S. Pearson Education India; 2002. Adaptive Filter Theory. [Google Scholar]
- 66.Mader A., Puder H., Schmidt G.U. Step-size control for acoustic echo cancellation filters–an overview. Signal Process. 2000;80:1697–1719. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example NIR tracking video. A zebrafish was swimming in the presence of water flow. Our tracking algorithm was able to accurately identify the fish head and complete the tracking despite interference from the distracting background.
Example alignment video. Sparsely EGFP labeled zebrafish were used to test the effectiveness of our alignment method. Left, the fish brain MIP after 3D reconstruction; right, the post-registered fish brain MIP guided by the LSSmCrimson channel.
Example optogenetics video. On the left, after unilateral optogenetic activation of the tegmentum region containing nMLF, the zebrafish turned ipsilaterally. On the right, changes in whole-brain activity were recorded simultaneously.
Data Availability Statement
-
•
Calcium imaging and behavior data have been deposited at Figureshare and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
All original code has been deposited at GitHub and Figureshare and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.