Abstract
Most Listeria monocytogenes virulence genes are positively regulated by the PrfA protein, a transcription factor sharing sequence similarities with cyclic AMP (cAMP) receptor protein (CRP). Its coding gene, prfA, is regulated by PrfA itself via an autoregulatory loop mediated by the upstream PrfA-dependent plcA promoter. We have recently characterized prfA* mutants from L. monocytogenes which, as a result of a single amino acid substitution in PrfA, Gly145Ser, constitutively overexpress prfA and the genes of the PrfA virulence regulon. Here, we show that about 10 times more PrfA protein is produced in a prfA* strain than in the wild type. Thus, the phenotype of prfA* mutants is presumably due to the synthesis of a PrfA protein with higher promoter-activating activity (PrfA*), which keeps its intracellular levels constantly elevated by positive feedback. We investigated the interaction of PrfA and PrfA* (Gly145Ser) with target DNA. Gel retardation assays performed with a DNA fragment carrying the PrfA binding site of the plcA promoter demonstrated that the PrfA* mutant form is much more efficient than wild-type PrfA at forming specific DNA-protein complexes. In footprinting experiments, the two purified PrfA forms interacted with the same nucleotides at the target site, although the minimum amount required for protection was 6 to 7 times lower with PrfA*. These results show that the primary functional consequence of the Gly145Ser mutation is an increase in the affinity of PrfA for its target sequence. Interestingly, similar mutations at the equivalent position in CRP result in a transcriptionally active, CRP* mutant form which binds with high affinity to target DNA in the absence of the activating cofactor, cAMP. Our observations suggest that the structural similarities between PrfA and CRP are also functionally relevant and support a model in which the PrfA protein, like CRP, shifts from transcriptionally inactive to active conformations by interaction with a cofactor.
Virulence genes in the gram-positive, facultative intracellular pathogen Listeria monocytogenes are regulated by the pleiotropic transcriptional activator PrfA, encoded by the prfA gene (6, 8, 21, 25, 27). An ambient temperature of 37°C is necessary for the transcriptional activation of prfA and PrfA-dependent genes (24). This is, however, not sufficient for the full activation of the PrfA regulon. Wild-type strains express PrfA-regulated genes to a very low level in rich media (e.g., brain-heart infusion medium [BHI]) at 37°C (30), but strongly activate their transcription if cultured in BHI treated with activated charcoal (28–30) or if transferred from BHI to minimal essential medium (5). This requirement for a suitable combination of environmental signals of a physical and chemical nature may be a fail-safe mechanism used by L. monocytogenes to prevent the expression of virulence genes in situations in which they are not required, i.e., when the bacteria are outside an appropriate host niche. Recent observations have suggested that there is also a mechanism of negative regulation in L. monocytogenes which abolishes the expression of virulence genes in the presence of readily fermentable carbon sources, such as glucose or cellobiose (26, 28). The molecular basis and biological relevance of this repression mechanism are unknown.
The primary structure of PrfA has significant similarities to that of Escherichia coli cyclic AMP (cAMP) receptor protein (CRP) and other members of the CRP-FNR family of bacterial transcription factors (21, 23). PrfA has, for example, a helix-turn-helix (HTH) motif in the C-terminal region, at the same position as in CRP and related proteins. This HTH motif has been shown to interact specifically with target DNA sequences called “PrfA-boxes,” which are 14-bp-long palindromes centered at position −41 relative to the transcription start site in PrfA-dependent promoters (3, 9, 11, 33). Binding to these PrfA-boxes is affected by the number of nucleotide mismatches they carry, becoming weaker as the sequence diverges from the perfect palindrome (4, 12, 34). The symmetrical structure of PrfA-boxes suggests that like CRP, PrfA binds to target DNA as a dimer, and there is experimental evidence that PrfA forms a homodimer in solution (9).
Evidence that PrfA and CRP are functionally related has been provided by our recent characterization of prfA* mutants from L. monocytogenes (28, 29, 31). Mutatis mutandis, these prfA* strains are analogous to the crp* mutants of E. coli in that they constitutively overexpress prfA and PrfA-dependent genes under culture conditions in which the PrfA regulon is normally downregulated (e.g., at 37°C in BHI), to levels reached by wild-type strains only if cultured in charcoal-treated BHI (28–30). These prfA* mutants carry a Gly→Ser substitution in residue 145 of PrfA that seems to increase the transcriptional activity of the regulator, releasing it from a variety of repressor signals including low temperature and growth on glucose or cellobiose (28–30). This mutation is located in a PrfA region of 11 amino acids (residues 141 to 151) with a sequence very similar (70% similarity) to that of the D α-helix of CRP (29). Several crp* mutations in E. coli that allow CRP to function in the absence of cAMP, the cofactor required for its allosteric activation, also map in this region (13, 15a, 20). One such CRP* mutation, Ala144Thr, which presumably mimics the conformational change caused by the cofactor (19, 20), maps in the aligned proteins to the position equivalent to that of the Gly→Ser PrfA mutation (29). These observations led us to hypothesize that PrfA functions via a cofactor-mediated allosteric transition mechanism similar to that of CRP, and that the Gly145Ser mutation is a cofactor-independent PrfA* form that is “frozen” in an active conformation (29).
In this study, we investigated the interaction of wild-type PrfA and mutant PrfA* (Gly145Ser) with target DNA. As for CRP* altered forms (2, 32, 35), the Gly145Ser mutant protein bound with higher affinity to specific DNA than did the wild-type protein, further supporting the notion that PrfA is a structural and functional homolog of CRP.
MATERIALS AND METHODS
L. monocytogenes strains and culture conditions.
P14, an L. monocytogenes wild-type strain of serovar 4b, and its prfA* mutant, P14-A, have been described in detail elsewhere (28–31). L. monocytogenes EGD, a wild-type strain of serovar 1/2a, and its prfA deletion mutant, ΔprfA, have also been previously described (3, 5, 29). They were grown in BHI broth at 37°C with shaking.
General DNA techniques.
Restriction enzymes were purchased from Pharmacia and used as recommended by the manufacturer. The Expand high-fidelity PCR system (Boehringer Mannheim) was used to amplify specific DNA fragments. PCR products were purified from gels with the Qiaquick (Qiagen) gel extraction kit. Plasmid DNA was extracted from E. coli with a plasmid purification kit from Qiagen. DNA sequencing was performed with an Applied Biosystems 377 apparatus.
L. monocytogenes cell protein extracts, SDS-PAGE, and anti-PrfA immunoblotting.
Soluble protein extracts from L. monocytogenes were prepared and stored as described by Böckmann et al. (3). Total protein concentration was determined with the Bio-Rad Protein-Microassay. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 12% acrylamide slab gels as described by Laemmli (22). For immunoblotting, proteins were electrotransferred from the gels to nitrocellulose sheets (Schleicher & Schuell), and PrfA was detected by using a previously described anti-PrfA polyclonal hyperimmune serum (3), peroxidase-conjugated secondary antibodies, and 4-chloro-1-naphthol.
Expression in E. coli and purification of wild-type and Gly145Ser mutant PrfA proteins.
The prfA and prfA* alleles from P14 and P14-A, respectively, were amplified by PCR with the oligonucleotide pair N-PR1 (5′-ATGACTCGAGAACGCTCAAGCAGAAGAA-3′) and C-PR1 (5′-CTGTAGATCTTTTAATTTAATTTTCCCCA-3′), which contain XhoI (N-PR1) and BglII (C-PR1) restriction sites (underlined). The resulting prfA-containing DNA fragments were cloned in E. coli with the pMOSBlue T-vector kit (Amersham), and then transferred to pFLAG-MAC expression vector (Sigma) by using the XhoI and BglII sites, giving rise to the plasmids pF-PrfA and pF-PrfA*(G145S). A fusion was created in these plasmids, resulting in a sequence that encodes a recombinant PrfA protein with an N-terminal tag of 14 amino acids including an 8-mer peptide marker (FLAG epitope). The whole open reading frame was checked by sequencing both strands in each expression plasmid. Recombinant PrfAs were overproduced in E. coli DH5α. Host bacteria were grown at 37°C in 500 ml of Luria-Bertani medium containing ampicillin (50 μg/ml) until the optical density at 600 nm was 1.0, and expression was induced by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 3 h, the induced bacteria were pelleted and lysed by suspension in 20 ml of lysis buffer A (50 mM Tris-HCl [pH 8], 5 mM EDTA, 50 μg of sodium azide per ml, 0.25 mg of lysozyme per ml) and addition of 2 ml of lysis buffer B (1.5 M NaCl, 100 mM CaCl2, 100 mM MgCl2, 0.02 mg of DNase I per ml, 50 μg of phenylmethylsulfonyl fluoride). Recombinant PrfAs, which did not form inclusion bodies in E. coli, were purified from the bacterial soluble extract by column affinity chromatography with anti-FLAG M2 monoclonal antibody resin (Sigma), according to the manufacturer’s instructions. The eluted fractions were analyzed by SDS-PAGE with Coomassie blue staining. The fractions containing >98% pure PrfA (which migrated as a band of 28.5 kDa, 1.5 kDa larger than predicted from the prfA sequence due to the presence of the extra 14 N-terminal amino acids) were collected, concentrated by Centricon devices (Amicon), and preserved at −20°C with 20% glycerol.
DNA mobility shift and footprinting assays.
A 136-bp double-stranded PCR fragment containing the plcA-hly promoter region was used as target DNA. It was amplified from strain P14 with primers YV3 (5′-TCCTATCTAGAAGTTACTTTTATGTC-3′) and YV4 (5′-TATTGGATCCATTCGCTTCTAAAGATG-3′), which contain XbaI and BamHI restriction sites (underlined). Previously described protocols were used for electrophoretic mobility shift assays with L. monocytogenes protein extracts or purified PrfA proteins (3). DNase I footprinting experiments were performed as previously described (9).
RESULTS
Amounts of PrfA in the wild-type and the prfA* (Gly145Ser) mutant of L. monocytogenes.
The levels of expression of prfA are primarily controlled by the PrfA-dependent plcA promoter, from which a bicistronic transcript covering the plcA-prfA operon is generated. This plcA-prfA mRNA creates an autoregulatory loop that is essential for the normal function of the PrfA regulon, presumably because it ensures the synthesis of sufficient quantities of the PrfA protein (5, 7, 12, 24, 25, 29) (see Fig. 6). Even if the prfA gene remains intact, any interruption of this autoregulatory loop (e.g., by insertional mutagenesis in plcA or in the plcA-prfA intergenic region) leads to a PrfA− phenotype (7, 11, 25, 28, 29). trans-complementation experiments have suggested that the mutant form of PrfA synthesized from prfA* (Gly145Ser), PrfA*, is more effective than the wild-type protein at activating PrfA-dependent promoters (28, 29). This would result in PrfA* constantly switching on the autoregulatory loop such that more PrfA protein was produced in the mutant prfA* background than in the wild type. We tested this by analyzing cell extracts of the L. monocytogenes wild-type strain, P14, and its prfA* mutant, P14-A, grown in BHI at 37°C, by Western blotting with anti-PrfA antibodies.
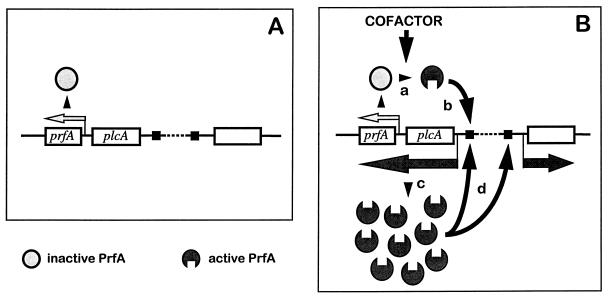
FIG. 6.
Model for PrfA-mediated gene regulation (29). Central to this model is the assumption that PrfA has two functional conformations, inactive and active, and shifts from one to the other on interaction with a hypothetical activating cofactor, the intracellular concentrations of which depend on the environmental conditions. A key element is also that prfA can be expressed in two different ways: (i) constitutively and at low levels, from monocistronic transcripts driven by promoters in the plcA-prfA intergenic region (represented by a small light-gray arrow above prfA on panels A and B); and (ii) dependent on PrfA, from bicistronic transcripts originating from the plcA promoter (dark gray arrow below plcA and prfA on panel B), thereby creating an autoregulatory circuit. The regulation mechanism would be as follows. Under normal conditions, there is no cofactor, and, thus, the PrfA protein is synthesized at low, basal levels from the monocistronic transcripts (A). However, if L. monocytogenes senses a suitable combination of activating environmental signals (a temperature of 37°C and a particular composition of the extracellular medium), the intracellular concentration of the hypothetical cofactor increases (B). This cofactor interacts with the inactive PrfA protein synthesized from monocistronic transcripts (a), causing a conformational change that results in a significant increase in the binding affinity of PrfA for its target DNA (b) (PrfA sites are indicated by black squares). The transcriptionally active PrfA causes the synthesis of more PrfA (in active conformation) by positive feedback (c), which boosts the transcription of all the PrfA-dependent genes (d) (dark gray arrows; the empty rectangle represents any PrfA-dependent gene). The PrfA regulon remains switched on as long as there are sufficiently high levels of the cofactor in the bacterial cytoplasm, but the system is rapidly switched off if the activating environmental signals cease and the concentration of the cofactor drops. A second level of regulation is provided by the differential response of the PrfA-dependent promoters according to the structure of the PrfA target site, which affects the binding affinity of PrfA. (See references 4, 7, 11, 12, and 34 for details about this cis-acting control mechanism.) Evidence for a negative autoregulation mechanism involving a putative PrfA-binding site in the plcA-prfA intergenic region has been also presented (11, 12), which would add complexity to the transcriptional control mediated by PrfA. The proposed regulatory model is highly versatile and makes possible an immediate, fine-tuned adaptive response to rapidly changing environmental conditions, such as those encountered by the soil bacterium L. monocytogenes during its transition from free to parasitic life and within the various compartments and tissues of the infected host.
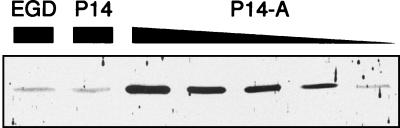
There was clearly more PrfA protein in P14-A than in P14 (Fig. 1), suggesting that PrfA* did indeed activate its own synthesis by a positive feedback mechanism. The constitutive overexpression of PrfA-dependent virulence genes in prfA* mutants is therefore presumably due to the sustained production of high levels of a transcriptionally active PrfA* form. Densitometric analysis of the blots showed that the intracellular levels of PrfA* were around 10 times higher in P14-A than in P14.
FIG. 1.
Determination of PrfA protein in L. monocytogenes P14 (wild type), P14-A (prfA* [Gly145Ser] mutant from P14), and EGD (control wild-type strain). Total cell extracts from these strains were subjected to SDS-PAGE in a 12% acrylamide gel (protein amounts loaded: P14 and EGD, 30 μg; P14-A [from left to right], 30, 15, 10, 5, and 2.5 μg) and analyzed by Western immunoblotting with an anti-PrfA hyperimmune serum. The PrfA protein is detected as a 27-kDa band. Note that equivalent amounts of PrfA protein are present in 30 μg of the P14 and EGD extracts and 2.5 μg of the P14-A extract.
DNA-protein complex formation by PrfA and PrfA* (Gly145Ser) in cell extracts of L. monocytogenes.
We investigated whether the differences in virulence gene transcription in the wild type and prfA* mutants (29, 30) were due to differences in the DNA-binding activity of the corresponding PrfA and PrfA* proteins. Electrophoretic mobility shift assays were carried out with cell extracts from P14 and P14-A and a 136-bp PCR fragment containing the plcA promoter and its PrfA-box, used as the target DNA. This PrfA-box is shared with the divergently transcribed hly gene and represents the “perfect” palindrome, to which PrfA presumably binds with maximal affinity (4, 34). Extracts from L. monocytogenes EGD, a wild-type strain in which the deduced amino acid sequence of PrfA is identical to that of P14 (29), and its prfA deletion mutant (EGD ΔprfA) were used as controls.
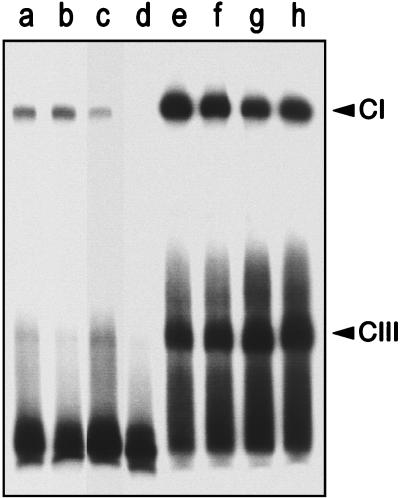
Specific protein-DNA complexes (CI) (3, 9) were formed with the cell extracts from all of the PrfA-proficient strains used (Fig. 2). However, there was a higher level of complex formation with the PrfA*-containing extract than with that containing wild-type PrfA, as determined from the intensities of the CI PrfA-dependent complexes formed (see lanes b and h, in which the amounts of PrfA protein are equivalent). The level of PrfA-dependent complex formation with extracts from EGD was identical to and as low as that with the extract from the P14 wild-type strain (lanes a and c). As expected, there was no binding activity observed with the EGD ΔprfA extract (lane d). Therefore, the higher transcriptional activity of the PrfA* (Gly145Ser) mutant form correlates with a higher affinity for target DNA.
FIG. 2.
Electrophoretic mobility shift assays with a 136-bp DNA fragment containing the PrfA-box of the plcA-hly promoter region and L. monocytogenes cell extracts. Lanes: a and b, P14 (30 and 60 μg, respectively); c, EGD (30 μg); d, ΔprfA mutant from EGD (30 μg); e to h, P14-A (30, 15, 10, and 5 μg, respectively). Lanes b and h contain equal amounts of PrfA protein (Fig. 1). CI and CIII, respectively, low- and high-mobility specific PrfA-DNA complexes. (See text and references 3 and 9 for details.)
Interaction of purified PrfA and PrfA* (Gly145Ser) with target DNA.
We characterized the differential interaction of PrfA and PrfA* (Gly145Ser) with the target DNA in more detail by gel retardation assays with the purified proteins. The PrfA proteins from strains P14 and P14-A were produced in E. coli with a FLAG epitope fused to the N terminus, which allowed them to be purified by affinity chromatography with an anti-FLAG-peptide monoclonal antibody (see Materials and Methods). The recombinant purified proteins were called F-PrfA and F-PrfA*, respectively. Addition of an N-terminal tag to PrfA has been shown to have no major effect on the DNA-binding function of the protein (3, 9). We used a recently described protocol with which direct, specific binding of purified PrfA can be observed in the absence of additional factors from the listerial cytoplasm (9). In this case, a high-mobility DNA-PrfA complex (CIII) is formed (9).
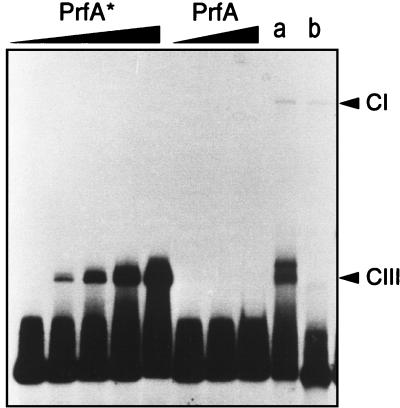
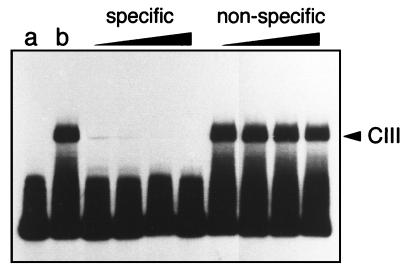
As little as 1.5 ng of F-PrfA* was sufficient to produce a visible CIII complex with the 136-bp DNA fragment containing the plcA-hly PrfA-box (Fig. 3). This interaction was specific, as shown by the ability of the unlabeled specific probe and the inability of nonspecific DNA to compete out CIII complex formation (Fig. 4). In contrast, no mobility shift was detectable with F-PrfA, even at high protein concentrations (Fig. 3). Addition of PrfA-free L. monocytogenes extract (from EGD ΔprfA) led to CI complex formation by F-PrfA* and, also, by F-PrfA (Fig. 3). Therefore, although it cannot directly interact with DNA to form a visible CIII complex, purified wild-type PrfA is able to bind to its target site in the presence of additional factors from the listerial extract. These results show that the PrfA* form is clearly more efficient than the wild-type protein at establishing direct interaction with the PrfA-specific target site.
FIG. 3.
Binding of the purified PrfA proteins to the plcA-hly promoter fragment. Various amounts of PrfA preparation were used (from left to right: PrfA* [Gly145Ser], 0.5, 1.5, 3, 6, and 18 ng; PrfA, 120, 240, and 480 ng). Lanes a and b: low-mobility CI-specific protein-DNA complex formation by purified PrfA* and PrfA proteins (50 ng each), respectively, in the presence of a PrfA-free L. monocytogenes cell extract (30 μg) from EGD ΔprfA. CIII, high-mobility specific PrfA-DNA complexes. (See text and references 3 and 9 for details.)
FIG. 4.
Specificity of the interaction of PrfA* (8 ng) with target DNA resulting in CIII complex formation. Competition assays with (from left to right) 50-, 100-, 200-, and 400-fold molar excess of specific (unlabeled 136-bp plcA-hly promoter fragment) and nonspecific DNA (from herring sperm). Lanes: a, control with the labeled 136-bp plcA-hly promoter fragment alone; b, control with the labeled probe plus 8 ng of purified PrfA*.
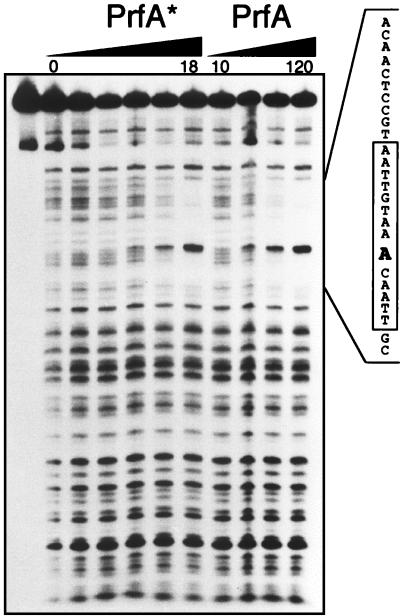
We investigated whether the higher DNA-binding activity of PrfA* was associated with a different pattern of interaction at the PrfA site by footprinting analysis of the DNA sequence protected by F-PrfA and F-PrfA* in the plcA-hly promoter region (Fig. 5). Various amounts of the purified PrfAs were used to further evaluate the differential binding affinity of the two proteins for the target site. In contrast to the results obtained with gel retardation assays, we did detect direct interaction of F-PrfA with target DNA. This probably results from the different experimental conditions for the two techniques, particularly the relatively high concentration of poly(dI-dC), used in the mobility shift experiments as nonspecific competitor to minimize nonspecific or low-affinity protein-DNA interactions (9). However, the amount of purified wild-type protein required for complete protection was significantly higher (6 to 7 times) than that for F-PrfA*. The DNA region protected from DNase I digestion was exactly the same for both purified proteins (positions −58 to −33 relative to the transcriptional start site of hly, including the PrfA-box palindrome and 10 bp upstream and 2 bp downstream from it) (Fig. 5) and was concordant with that previously reported for PrfA (9).
FIG. 5.
DNA footprinting experiments with various amounts of the purified PrfA proteins (PrfA*, 0, 0.5, 1.5, 3, 6, and 18 ng; PrfA, 10, 20, 40, and 120 ng) and the plcA-hly promoter fragment. The protected sequences, identical for the two PrfA proteins, are indicated on the right. The palindromic PrfA binding site is boxed, and numbers indicate the nucleotide position with respect to the transcription start site of the hly mRNA. The hypersensitive nucleotide (A) at position −40, close to the center of the palindrome, is in boldface. To the left is shown the uncleaved probe.
DISCUSSION
We have shown, by electrophoretic mobility shift and footprinting assays, that (i) wild-type PrfA interacts weakly with the specific target DNA, and (ii) a mutant PrfA form, PrfA* (Gly145Ser), has a much higher DNA-binding activity than the wild-type protein. There was no difference between the DNA sequences footprinted by the mutant and wild-type PrfA proteins, demonstrating that the primary functional consequence of the Gly145Ser substitution is a very significant increase in binding affinity for the target DNA. Since prfA is positively regulated by its own product, PrfA (24, 25, 28, 29) (Fig. 6), the expected outcome of the mutation in vivo is an increase in the levels of the PrfA protein. This has been also demonstrated herein. Our results are consistent with the physiological properties of wild-type and prfA* (Gly145Ser) backgrounds of L. monocytogenes, which in normal culture media express low and constitutively high levels of PrfA-dependent genes, respectively (28–30).
PrfA and CRP exhibit significant similarities at the level of primary structure (21, 23, 33). The observations here reported with PrfA are also very similar to those for the structure-function relationships of CRP, supporting the notion that PrfA acts via a regulatory mechanism similar to that of the E. coli transcription factor. The inactive form of CRP binds to specific DNA with very low affinity, so the interaction is not normally detectable in gel retardation assays. However, if complexed with the activating cofactor, cAMP, CRP undergoes a conformational change that is associated with a dramatic increase in affinity for the target DNA (2, 16, 17, 35). Binding of the cofactor is thought to alter intersubunit alignment and interdomain orientation in CRP, ultimately resulting in the protrusion of the F α-helix which is part of the HTH DNA-binding motif, thereby facilitating productive specific protein-DNA interaction (1, 14, 16, 19, 20). CRP* mutations in the D α-helix, which spans residues 139 to 150, close to the hinge region connecting the N-terminal cAMP-binding domain and the C-terminal DNA-binding domain of the CRP subunit (13, 15a), are thought to evoke the conformational change caused by cAMP, resulting in transcriptional activation in the absence of significant amounts of the cofactor (14, 19, 20, 32). In this D α-helix, any amino acid substitution introducing a larger side chain at position 144, which aligns with PrfA residue 145 (the site of our PrfA* mutation) (29), results in a cAMP-independent CRP* phenotype (19). According to the determined crystal structure of CRP, amino acid 144 faces residue 190 in the F α-helix (20, 37). Therefore, the cAMP-independent phenotype is presumably due to the larger side chain pushing the DNA-binding sequence outward (19, 20). The PrfA* mutation studied, mapping to a region that is remarkably homologous to the D α-helix of CRP (29), is similar to the CRP* mutation previously characterized at position 144, Ala to Thr (14, 15a, 20), and involves the replacement of a small amino acid (Gly) with a larger one (Ser). A second CRP* mutation described at position 141 in the D α-helix is also a Gly→Ser substitution, and we recently characterized another PrfA* mutation that maps nearby within the same D α-helix-homologous region, which also involves a replacement by a bulkier amino acid (36). Except for the fact that PrfA has an extra 25 amino acids at the extreme C terminus, the C-terminal domain of CRP is very similar (45% identity, 60% similarity from amino acid 128 to amino acid 201) to the corresponding region in PrfA (21, 23, 36). These observations suggest that the mutations resulting in the PrfA* phenotype are associated with conformational changes in the DNA-binding domain similar to those that are thought to occur in CRP* mutant proteins.
Our findings provide support for our model of PrfA-dependent regulation (29). In this model, similar to that proposed for CRP, PrfA undergoes an allosteric transition from an inactive to an active conformation upon interaction with an environmentally regulated low-molecular-weight cofactor. A key element of this model is the positive autoregulatory circuit of prfA, an aspect in which the listerial regulatory gene also resembles the transcriptional control mediated by crp in E. coli (15). (See Fig. 6 for a detailed description of the model.)
Two typical features of CRP are particularly well conserved in PrfA. One is the HTH motif in the C-terminal region, for which the functional similarity between the two proteins has been already documented (33). The other is a series of short antiparallel β-strands delimited by glycine residues, which may form a β-roll structure involving most of the N-terminal half of the protein (21, 23). The prediction of such a structure in PrfA is quite intriguing, because in CRP it forms the pocket in which the activating cofactor, cAMP, is buried in the N-terminal domain of the protein (20). However, cAMP is undetectable, and it is not known to function as an effector molecule in gram-positive bacteria (18). In fact, most of the residues in CRP that are important for cAMP binding are not conserved in PrfA (21, 36), and addition of exogenous cAMP does not result in PrfA activation (36). It is, however, unknown whether cAMP is taken up by Listeria. Preliminary studies with the extrinsic fluorescence probe 8-anilino-1-naphthalenesulfonic acid (ANS) have shown that as for CRP (16), the addition of cAMP to purified PrfA results in a significant fluorescence quenching (36). This indicates that cAMP induces a conformational change, but not necessarily that it allosterically activates PrfA. The cyclic nucleotide cGMP, for example, does not functionally activate CRP, but it does bind to it with an affinity similar to that of cAMP, and there are cAMP analogs that bind to CRP and cause a conformational change similar to that elicited by cAMP but do not activate transcriptional function (10). It is therefore possible that the putative cofactor for PrfA is a cyclic nucleotide similar to cAMP. We are currently working toward the identification of this putative PrfA cofactor and the genetic characterization of the signal transduction machinery that connects the PrfA system with the extracellular environment.
ACKNOWLEDGMENTS
We thank V. de Lorenzo and G. Bertoni for expert advice on the production of recombinant proteins in E. coli, J. Kreft for helpful discussions, F. Gómez-Gallego and J. M. Bautista for their help in PrfA conformational studies, and S. Rodríguez-Malvar for technical assistance.
This investigation was supported by grants from the European Commission (BMH4-CT96-0659), the Dirección General de Investigación Cientifica y Técnica (DGICYT PB94-0330-C01), the Deutsche Forschungsgemeinschaft (SFB 165-B4), and the Fonds der Chemischen Industrie. Y.V. received a Ph.D. studentship from the Universidad Complutense de Madrid.
REFERENCES
- 1.Baichoo N, Heyduk T. Mapping conformational changes in a protein: application of a protein footprinting technique to cAMP-induced conformational changes in cAMP receptor protein. Biochemistry. 1997;36:10830–10836. doi: 10.1021/bi970714v. [DOI] [PubMed] [Google Scholar]
- 2.Blazy B, Ullmann A. Properties of cyclic AMP-independent catabolite gene activator proteins of Escherichia coli. J Biol Chem. 1986;261:11645–11649. [PubMed] [Google Scholar]
- 3.Böckmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol. 1996;22:643–653. doi: 10.1046/j.1365-2958.1996.d01-1722.x. [DOI] [PubMed] [Google Scholar]
- 4.Bohne J, Kestler H, Uebele C, Sokolovic Z, Goebel W. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol Microbiol. 1996;20:1189–1198. doi: 10.1111/j.1365-2958.1996.tb02639.x. [DOI] [PubMed] [Google Scholar]
- 5.Bohne J, Sokolovic Z, Goebel W. Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol Microbiol. 1994;11:1141–1150. doi: 10.1111/j.1365-2958.1994.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 6.Brehm K, Kreft J, Ripio M T, Vázquez-Boland J A. Regulation of virulence gene expression in pathogenic Listeria. Microbiología SEM. 1996;12:219–236. [PubMed] [Google Scholar]
- 7.Camilli A, Tilney L, Portnoy D. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty T, Leimeister-Wächter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickneite C, Böckmann R, Spory A, Goebel W, Sokolovic Z. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol Microbiol. 1998;27:915–928. doi: 10.1046/j.1365-2958.1998.00736.x. [DOI] [PubMed] [Google Scholar]
- 10.Ebright R H, Le Grice S F J, Miller J P, Krakow J S. Analogs of cyclic AMP that elicit the biochemically defined conformational change in catabolite gene activator protein (CAP) but do not stimulate binding to DNA. J Mol Biol. 1985;182:91–107. doi: 10.1016/0022-2836(85)90030-0. [DOI] [PubMed] [Google Scholar]
- 11.Freitag N E, Rong L, Portnoy D A. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell-spread. Infect Immun. 1993;61:2537–2544. doi: 10.1128/iai.61.6.2537-2544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitag N E, Portnoy D A. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol Microbiol. 1994;12:845–853. doi: 10.1111/j.1365-2958.1994.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 13.Garges S, Adhya S. Sites of allosteric shift in the structure of cyclic AMP receptor protein. Cell. 1985;41:745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- 14.Garges S, Adhya S. Cyclic AMP-induced conformational change of cyclic AMP receptor protein (CRP): intragenic suppressors of cyclic AMP-independent CRP mutations. J Bacteriol. 1988;170:1417–1422. doi: 10.1128/jb.170.4.1417-1422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanamura A, Aiba H. A new aspect of transcriptional control of the Escherichia coli crp gene: positive autoregulation. Mol Microbiol. 1992;6:2489–2497. doi: 10.1111/j.1365-2958.1992.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 15a.Harman J G, McKenney K, Peterkofsky A. Structure-function analysis of three cAMP-independent forms of the cAMP receptor protein. J Biol Chem. 1986;261:16332–16339. [PubMed] [Google Scholar]
- 16.Heyduk T, Lee J C. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry. 1989;28:6914–6924. doi: 10.1021/bi00443a021. [DOI] [PubMed] [Google Scholar]
- 17.Heyduk T, Lee J C. Application of fluorescence energy transfer and polarization to monitor Escherichia coli cAMP receptor protein and lac promoter interaction. Proc Natl Acad Sci USA. 1990;87:1744–1748. doi: 10.1073/pnas.87.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Adhya S, Garges S. Allosteric changes in the cAMP receptor protein of Escherichia coli: hinge reorientation. Proc Natl Acad Sci USA. 1992;89:9700–9704. doi: 10.1073/pnas.89.20.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 21.Kreft J, Bohne J, Gross R, Kestler H, Sokolovic Z, Goebel W. Control of Listeria monocytogenes virulence genes by the transcriptional regulator PrfA. In: Rappuoli R, Scarlato V, Arico B, editors. Signal transduction and bacterial virulence. R. G. Austin, Tex: Landes Company; 1995. pp. 129–142. [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lampidis R, Gross R, Sokolovic Z, Goebel W, Kreft J. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcriptional regulators. Mol Microbiol. 1994;13:141–151. doi: 10.1111/j.1365-2958.1994.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 24.Leimeister-Wächter M, Domann E, Chakraborty T. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J Bacteriol. 1992;174:947–952. doi: 10.1128/jb.174.3.947-952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mengaud J, Dramsi S, Gouin E, Vázquez-Boland J A, Milon G, Cossart P. Pleiotropic control of Listeria monocytogenes virulence factors by a gene which is autoregulated. Mol Microbiol. 1991;5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 26.Millenbachs A A, Brown D P, Moors M, Youngman P. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol Microbiol. 1997;23:1075–1085. doi: 10.1046/j.1365-2958.1997.2711634.x. [DOI] [PubMed] [Google Scholar]
- 27.Portnoy D A, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripio M-T, Brehm K, Lara M, Suárez M, Vázquez-Boland J-A. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J Bacteriol. 1997;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ripio M-T, Domínguez-Bernal G, Lara M, Suárez M, Vázquez-Boland J-A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol. 1997;179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripio M T, Domínguez-Bernal G, Suárez M, Brehm K, Berche P, Vázquez-Boland J A. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res Microbiol. 1996;147:371–384. doi: 10.1016/0923-2508(96)84712-7. [DOI] [PubMed] [Google Scholar]
- 31.Ripio M-T, Vázquez-Boland J A, Vega Y, Nair S, Berche P. Evidence for expressional crosstalk between the central virulence regulator PrfA and the stress response mediator ClpC in Listeria monocytogenes. FEMS Microbiol Lett. 1998;158:45–50. doi: 10.1111/j.1574-6968.1998.tb12798.x. [DOI] [PubMed] [Google Scholar]
- 32.Ryu S, Kim J, Adhya S, Garges S. Pivotal role of amino acid at position 138 in the allosteric hinge reorientation of cAMP receptor protein. Proc Natl Acad Sci USA. 1993;90:75–79. doi: 10.1073/pnas.90.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheehan B, Klarsfeld A, Ebright R, Cossart P. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol Microbiol. 1996;20:785–797. doi: 10.1111/j.1365-2958.1996.tb02517.x. [DOI] [PubMed] [Google Scholar]
- 34.Sheehan B, Klarsfeld A, Msadek T, Cossart P. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J Bacteriol. 1995;177:6469–6476. doi: 10.1128/jb.177.22.6469-6476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan G-S, Kelly P, Kim J, Wartell R M. Comparison of cAMP receptor protein (CRP) and a cAMP-independent form of CRP by Raman spectroscopy and DNA binding. Biochemistry. 1991;30:5076–5080. doi: 10.1021/bi00234a034. [DOI] [PubMed] [Google Scholar]
- 36.Vega, Y., M. T. Ripio, and J. A. Vázquez-Boland. Unpublished data.
- 37.Weber I T, Steitz T A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]