Abstract
In lung and prostate adenocarcinomas, neuroendocrine (NE) transformation to an aggressive derivative resembling small cell lung cancer (SCLC) is associated with poor prognosis. We previously described dependency of SCLC on the nuclear transporter exportin 1. Here, we explored the role of exportin 1 in NE transformation. We observed up-regulated exportin 1 in lung and prostate pretransformation adenocarcinomas. Exportin 1 was up-regulated after genetic inactivation of TP53 and RB1 in lung and prostate adenocarcinoma cell lines, accompanied by increased sensitivity to the exportin 1 inhibitor selinexor in vitro. Exportin 1 inhibition prevented NE transformation in different TP53/RB1-inactivated prostate adenocarcinoma xenograft models that acquire NE features upon treatment with the aromatase inhibitor enzalutamide and extended response to the EGFR inhibitor osimertinib in a lung cancer transformation patient-derived xenograft (PDX) model exhibiting combined adenocarcinoma/SCLC histology. Ectopic SOX2 expression restored the enzalutamide-promoted NE phenotype on adenocarcinoma-to-NE transformation xenograft models despite selinexor treatment. Selinexor sensitized NE-transformed lung and prostate small cell carcinoma PDXs to standard cytotoxics. Together, these data nominate exportin 1 inhibition as a potential therapeutic target to constrain lineage plasticity and prevent or treat NE transformation in lung and prostate adenocarcinoma.
Graphical Abstract

INTRODUCTION
Histologic transformation to neuroendocrine (NE) derivatives resembling small cell lung cancer (SCLC) occurs in up to 14% of metastatic epidermal growth factor receptor (EGFR)–mutant lung adenocarcinomas (LUADs) treated with tyrosine kinase inhibitors (1) and in more than 20% of metastatic androgen receptor (AR)–dependent prostate adenocarcinomas (PRADs) treated with enzalutamide (2). Similar NE transformation has been described in LUADs with alternative genomic contexts and may represent a general mechanism of acquired resistance to other targeted therapies (3–5). NE-transformed small cell carcinomas are typically rapidly progressive and treatment refractory, leading to prognoses similar to or even worse than de novo SCLC (2, 6, 7).
NE transformation is a manifestation of lineage plasticity (3, 4, 8), has been associated with epigenetic reprogramming (4, 9), and may be facilitated or selected for by exposure to potent targeted therapies (4, 8, 10). Little is known about the molecular mechanisms driving transformation, although concurrent mutational inactivation of the tumor protein P53 (TP53) and retinoblastoma transcriptional corepressor 1 (RB1) tumor suppressor genes is implicated as a prerequisite for NE transformation (6, 11). Even if patients with adenocarcinoma at high risk of NE transformation could be identified, to date, no therapies are available to effectively constrain plasticity and prevent NE transformation. The identification of pharmacologically targetable effectors of transformation is an unmet clinical need.
A CRISPR dependency screen and subsequent validation studies identified exportin 1, encoded by the exportin 1 (XPO1) gene, as a potential therapeutic target in SCLC (12). Exportin 1 is a nuclear exporter facilitating transport of proteins and mRNAs from the nucleus to the cytosol (13). Known substrates for exportin 1 include factors implicated in cancer growth and survival, which prompted development of potent targeted inhibitors, including selinexor, which demonstrates activity against several hematologic malignancies and has been approved by the U.S. Food and Drug Administration for clinical use in patients with refractory multiple myeloma (14). SCLC shows higher exportin 1 expression than any other tumor type (12), and its inhibition enhances sensitivity to chemotherapeutic agents in de novo (nontransformed) SCLC (12). Given these data suggesting a selective dependency of SCLC on exportin 1, we hypothesized that targeting exportin 1 might represent a viable strategy for constraining NE transformation.
Here, we provide evidence for the role of exportin 1 in NE transformation across both lung and prostate cancers. We demonstrate in both tumor types that exportin 1 expression is elevated at early stages of NE transformation, that it is induced by TP53 and RB1 inactivation, and that its inhibition by selinexor suppresses NE transformation in vivo, extending response to targeted therapies. Mechanistic studies demonstrate that exportin 1 inhibition prevents targeted therapy–induced up-regulation of the SRY-box transcription factor 2 (SOX2), a transcription factor promoting stemness and implicated in NE transformation (8). Exogenous SOX2 overexpression rescues NE transformation under selinexor exposure. Together, these data define a potential therapeutic target to inhibit NE relapse with a clinically available agent, which will facilitate repurposing of selinexor for treatment of these patients.
RESULTS
Exportin 1 is up-regulated during NE transformation
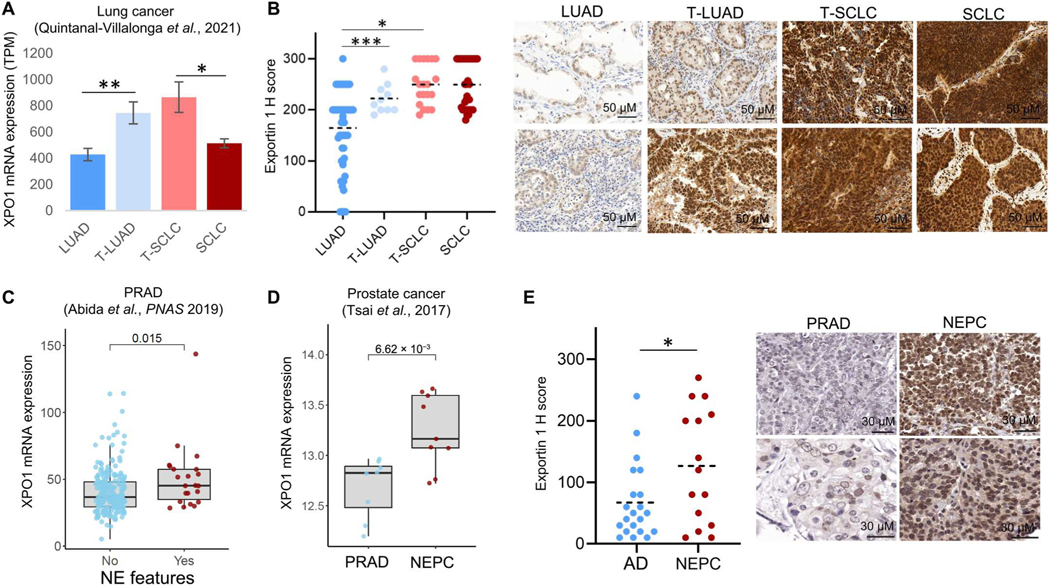
To evaluate the potential role of XPO1 in NE transformation, we first assessed expression across a previously published transcriptomic dataset on NE-transforming clinical specimens (4) including LUADs that never transformed (categorized as “LUAD”); LUADs obtained from pretransformation or microdissected combined histology cases, thought to be derived from lineage plasticity (categorized as “T-LUAD”); SCLCs from posttransformation or microdissected combined histology cases (“T-SCLC”); and de novo SCLCs (“SCLC”). XPO1 mRNA expression was elevated in T-LUAD relative to never-transforming LUAD and in T-SCLC relative to de novo SCLC (Fig. 1A) (4). Determination of exportin 1 protein abundance by immunohistochemistry (IHC) in an independent cohort of lung cancer clinical specimens and patient-derived xenografts (PDXs) confirmed increased exportin 1 protein expression in T-LUADs versus control never-transforming LUADs and further up-regulation in T-SCLC, similar to the high amounts in de novo SCLC (Fig. 1B). Although XPO1 mRNA expression was higher in T-SCLC as compared with de novo SCLC (Fig. 1A), protein abundance of exportin 1 was comparable in these cohorts (Fig. 1B). Leveraging previously published, publicly available prostate carcinoma clinical specimen datasets (15, 16), we observed that the presence of NE features was associated with increased XPO1 in PRAD (Fig. 1C) and that NE prostate carcinoma (NEPC) shows increased XPO1 expression relative to PRAD (Fig. 1D). In line with these results, assessment of exportin 1 protein abundance in an independent cohort of prostate cancers also revealed higher expression in NEPC than PRAD (Fig. 1E). The high expression of XPO1/exportin 1 at stages temporally proximal to NE transformation is consistent with a hypothesized role in promoting histologic transformation.
Fig. 1. Exportin 1 is up-regulated during NE transformation in lung and prostate tumors.
Exportin 1 mRNA expression (4) (A) and protein (B) abundance in lung cancer clinical specimens, categorized as control never-transformed adenocarcinomas (LUAD, RNA n = 11, protein n = 46), transforming adenocarcinomas (T-LUAD, RNA n = 11, protein n = 10), small cell carcinomas (T-SCLC, RNA n = 11, protein n = 20), and control de novo small cell carcinomas (SCLC, RNA n = 16, protein n = 32). For (B), H-score medians and SDs (right) and representative immunohistochemistry (IHC) images (left) are shown. (C) Exportin 1 mRNA expression in prostate adenocarcinoma (PRAD) tumors with (n = 22) or without (n = 210) NE features [data from Abida et al. (16)]. (D) Exportin 1 mRNA expression in PRADs (n = 8) and small cell NE prostate cancer (NEPC) (n = 9) [data from Tzelepi et al. (15)]. (E) Exportin 1 protein expression in PRAD (n = 21) and NEPC (n = 15) clinical specimens, as assessed by IHC. H-score medians and SD (right) and representative images (left) are shown. *P < 0.05, **P < 0.01, and ***P < 0.001. TPM, transcripts per million.
The combination of exportin 1 inhibition and chemotherapy is effective against NE-transformed carcinomas
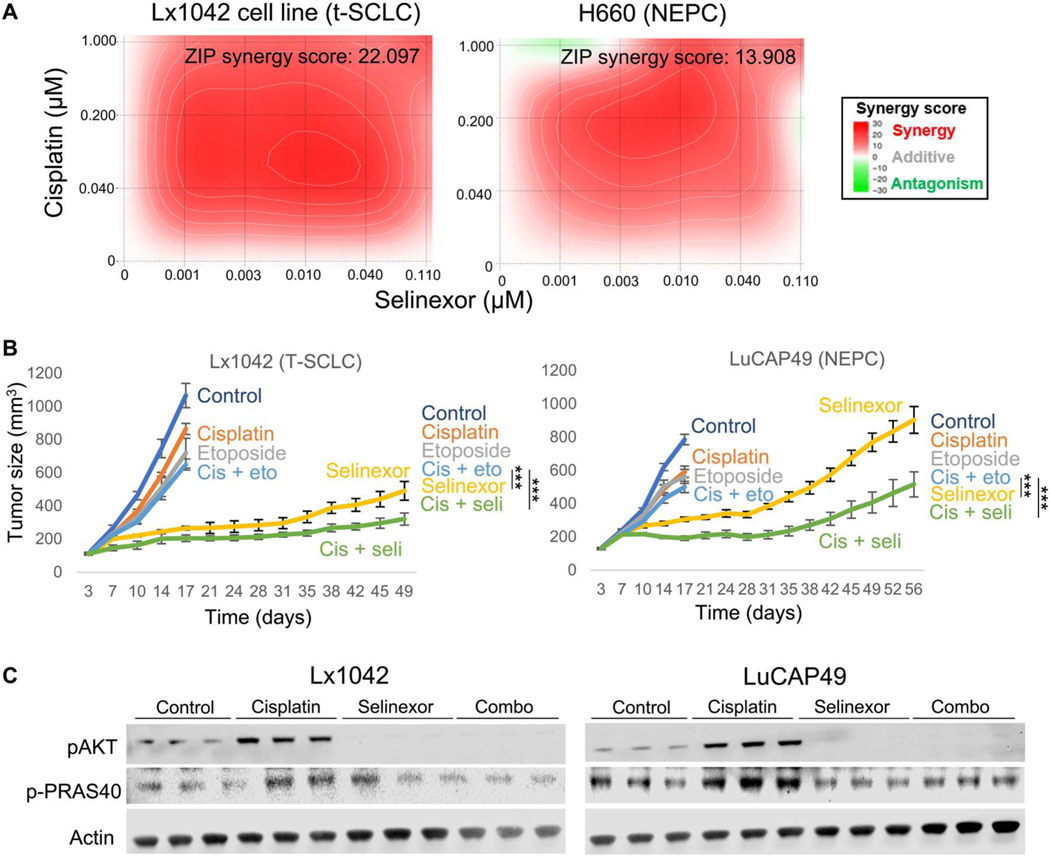
We have previously shown that inhibition of exportin 1 sensitizes PDXs derived from de novo SCLCs to chemotherapeutic agents used in the first- and second-line treatment of these tumors and that these effects are mediated by suppression of AKT/mammalian target of rapamycin (mTOR) signaling, a pathway induced after chemotherapy exposure (12). Recent data highlight some molecular and treatment response differences between de novo and T-SCLCs (4, 17) and suggest that T-SCLCs retain some molecular features of their previous LUAD state (4, 11). Thus, we aimed to confirm the capacity of selinexor to sensitize T-SCLCs and NEPCs to cisplatin, a key component of first-line treatment of de novo SCLC, T-SCLC, and NEPC. In vitro synergy assays in cell lines derived from a T-SCLC PDX (Lx1042) and an NEPC cell line (H660) demonstrated synergistic growth inhibition between selinexor and cisplatin (Fig. 2A). In vivo treatment of PDXs derived from a T-SCLC (Lx1042) and an NEPC [LuCAP49 (18)] engrafted subcutaneously in immunosuppressed nonobese diabetic (NOD).Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice confirmed that the combination of selinexor and cisplatin outperformed the combination of cisplatin and etoposide (Fig. 2B), with treatment/control (T/C) relative volume values of 14 and 19% (cisplatin + selinexor versus control) and of 27 and 32% (cisplatin + selinexor versus cisplatin + etoposide) for Lx1042 and LuCAP49, respectively. Similar to our previous observations of de novo SCLCs (12), these NE-transformed models showed increased activation of the AKT pathway after cisplatin treatment, which was suppressed by selinexor (Fig. 2C). These results extend the potential use of selinexor in combination with chemotherapy from de novo SCLC to T-SCLC and NEPC.
Fig. 2. Exportin 1 inhibition sensitizes NE-transformed lung and prostate cancers to chemotherapy.
(A) In vitro synergy assays in Lx1042 (T-SCLC, left) and H660 (NEPC, right) cell lines of the combination of selinexor and cisplatin with average synergy score displayed, as assessed by zero interaction potency (ZIP) and calculated using SynergyFinder. A representative plot is shown. (B) In vivo treatments of Lx1042 (T-SCLC) and LuCAP49 (NEPC) PDXs to compare the efficacy of the combination of cisplatin and selinexor versus that of cisplatin and etoposide. Four to eight female 6-week-old NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice (PDXs) were subcutaneously engrafted per treatment arm and were grown until tumors reached 100 to 150 mm3. At that point, mice were randomized into groups and treated with either vehicle (n = 8), cisplatin (2 mg/kg, i.p., once per week, n = 4 for Lx1042 and n = 5 for LuCAP49), etoposide (3 mg/kg, i.p., QDx3, n = 4 for Lx1042 and n = 5 for LuCAP49), selinexor (10 mg/kg, p.o., QDx3, n = 4 for Lx1042 and n = 5 for LuCAP49), or the combinations of cisplatin + etoposide (n = 8) or cisplatin + selinexor (n = 8). Mice weights and tumor volumes were measured twice a week, and mice were euthanized when tumors reached a humane end point (volume, 1000 mm3). P values were calculated using the Student’s t test (unpaired, heterogeneous variances, and two-tailed). (C) Representative Western blot images showing activation of the AKT/mTOR pathway in tumors derived from (B). ***P < 0.001.
TP53 and RB1 inactivation induce XPO1 expression and sensitivity to exportin 1 inhibition
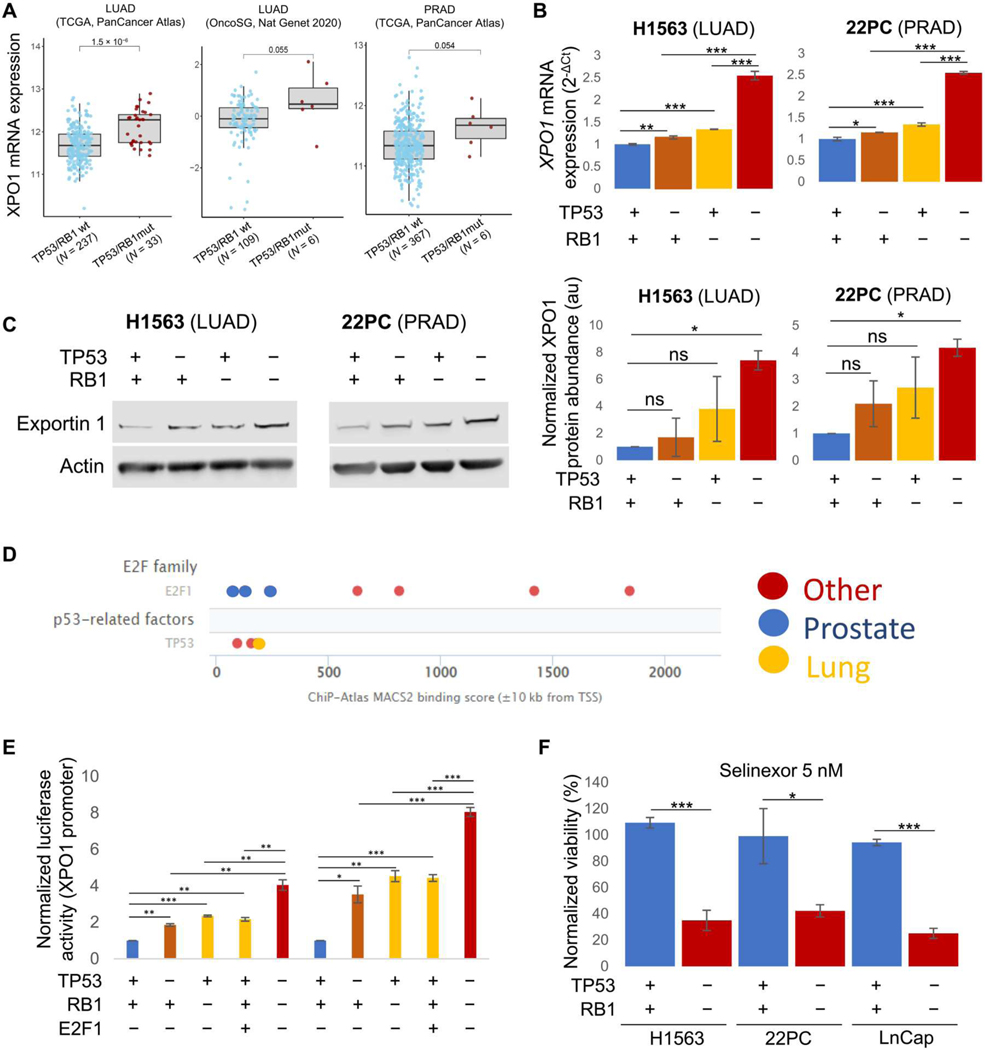
Our data suggest that exportin 1 up-regulation occurs early in the NE transformation process, with T-LUADs showing increased exportin 1 mRNA and protein expression relative to control never-transforming LUADs (Fig. 1, A and B). One of the hallmarks of NE transformation is functional inactivation of TP53 and RB1, through either genomic alterations or protein down-regulation (4, 6, 11). Inactivation of these two tumor suppressors typically occurs early in the transformation process and might serve as a licensing condition, necessary but not sufficient for histologic transformation (6, 11). Thus, we wondered whether loss of TP53/RB1 function might induce exportin 1 expression. Consistent with this hypothesis, we observed increased expression of XPO1 mRNA in LUADs and PRADs across multiple cohorts with concurrent TP53 and RB1 mutations relative to their double wild-type counterparts (Fig. 3A). Assessment of the potential contribution of loss of each factor individually was challenging because of the low number of samples showing RB1 genomic alterations without co-occurring TP53 mutations (fig. S1A), but the available data demonstrated an association of TP53 mutations with increased XPO1 expression relative to TP53/RB1–wild-type samples, and double TP53/RB1-mutated samples showed the highest XPO1 expression (fig. S1A). To test the contributions of each gene more directly, we generated TP53- and/or RB1-genetically inactivated LUAD (H1563, overexpression of dominant negative TP53 and short hairpin RNA against RB1) and PRAD (22PC, CRISPR-Cas9 inactivation) isogenic cell lines and assessed exportin 1 mRNA expression and protein abundance by quantitative polymerase chain reaction (qPCR) and Western blot, respectively (Fig. 3, B and C, and fig. S1B). Loss of function of either gene induced exportin 1 expression in both cell lines compared with the control condition, and further up-regulation was observed in the doubly inactivated lines (Fig. 3, B and C), consistent with the clinical specimen correlations (Fig. 3A). Assay for transposase-accessible chromatin sequencing (ATAC-seq) analysis of these TP53/RB1-inactivated cell lines did not reveal increased XPO1 gene accessibility after TP53/RB1 inactivation (fig. S1C). However, leveraging publicly available chromatin immunoprecipitation sequencing (ChIP-seq) datasets, we observed that both TP53 and E2F transcription factor 1 (E2F1), the latter being a primary transcription factor activated upon RB1 inactivation, appeared to bind proximal to the transcriptional start site of the XPO1 gene in datasets including prostate and lung cancers (Fig. 3D). This observation led us to the hypothesis that TP53 and E2F1 might be directly regulating XPO1 transcription by binding to the XPO1 gene promoter. To test this, we performed promoter reporter assays for the XPO1 gene promoter, in isogenic cell lines derived from H1563 and 22PC (Fig. 3E). In both cell lines, inactivation of TP53 led to increased XPO1 promoter activity, suggesting that TP53 binding could repress XPO1 gene expression. Similarly, inactivation of RB1 led to increased XPO1 promoter activity, at comparable amounts to E2F1 overexpression (Fig. 3D and fig. S1D), suggesting that E2F1 might be able to induce XPO1 gene expression after RB1 inactivation. Concurrent inactivation of TP53 and RB1 further increased XPO1 promoter activity (Fig. 3, A and B). Together, these data suggest that TP53 might directly repress XPO1 gene expression by directly binding the XPO1 promoter, whereas E2F1 might be able to directly bind the XPO1 gene promoter to induce XPO1 transcription after RB1 inactivation. In addition, treatment of matched isogenic cell lines, including an additional PRAD line (LnCap/AR), confirmed increased selinexor sensitivity in those cell lines with TP53/RB1 inactivation relative to either wild-type controls (Fig. 3F and fig. S1E) or single gene loss-of-function counterparts (fig. S1F). To study whether these effects may just be derived from TP53 and RB1 inactivation causing a more highly proliferative phenotype in these models, we performed proliferation assays in TP53/RB1-inactivated cell lines and studied the cell cycle profile in those being treated with selinexor (fig. S1, G and H). We observed a slight increase in proliferation after inactivation of TP53 and RB1, with no statistically significant shifts in cell cycle profile even at substantially cytotoxic concentrations of selinexor. Thus, the induced selinexor sensitivity by TP53 and RB1 inactivation did not appear to be a consequence of substantial cell cycle disruption as might occur with nonspecific cytotoxics. These results do suggest that loss of TP53 and RB1 activity in LUADs and PRADs induces expression of and greater dependency on exportin 1, associated with an increase in the therapeutic window for selinexor in this setting.
Fig. 3. Loss of TP53/RB1 function induces exportin 1 expression and sensitivity to selinexor.
(A) XPO1 mRNA expression in LUAD clinical specimens categorized by TP53/RB1 status. Data obtained from LUAD TCGA [PanCancer, n = 237 wild type (wt), 33 mutated], LUAD OncoSG (OncoSG, Nat Genetics 2020, n = 109 wt, 6 mutated), and PRAD TCGA (PanCancer, n = 367 wt, 6 mutated) (15, 16). (B) XPO1 mRNA expression in isogenic H1563 (LUAD) and 22PC (PRAD) cell lines with or without induced loss of function of TP53 and/or RB1 by short hairpin RNA against RB1 and dominant negative TP53 gene overexpression (H1563) or CRISPR-Cas9 knock out (22PC). (C) Western blot showing exportin 1 protein abundance in isogenic H1563 (LUAD) and 22PC (PRAD) cell lines with or without induced loss of function of TP53 and/or RB1 (left; see Materials and Methods) and Western blot quantification (n = 2, right). A representative Western blot image is shown. (D) Binding score for TP53 and E2F1 in the transcription start site (TSS) of the XPO1 gene in different experimental settings including specimens from lung, prostate, and other sites. Data obtained from the Signaling Pathways Project (ChIP-seq Atlas). (E) Barplot exhibiting data from XPO1 gene promoter reporter assays in isogenic H1563 (LUAD) and 22PC (PRAD) cell lines with or without induced loss of function of TP53 and/or RB, or with E2F1 overexpression. Normalized luciferase activity of a representative biological replicate is shown. (F) Barplot showing a representative biological replicate of an experiment assessing viability of control and TP53/RB1-inactivated H1563 (LUAD), 22PC, and LnCap/AR (PRAD) cells treated with 5 nM selinexor. Each of the conditions shown was normalized to their respective untreated condition and represented as a normalized viability percentage. For (B) and (C), P values were calculated using the Student’s t test (unpaired, heterogeneous variances, and two-tailed). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Exportin 1 inhibition interferes with NE relapse on targeted therapy
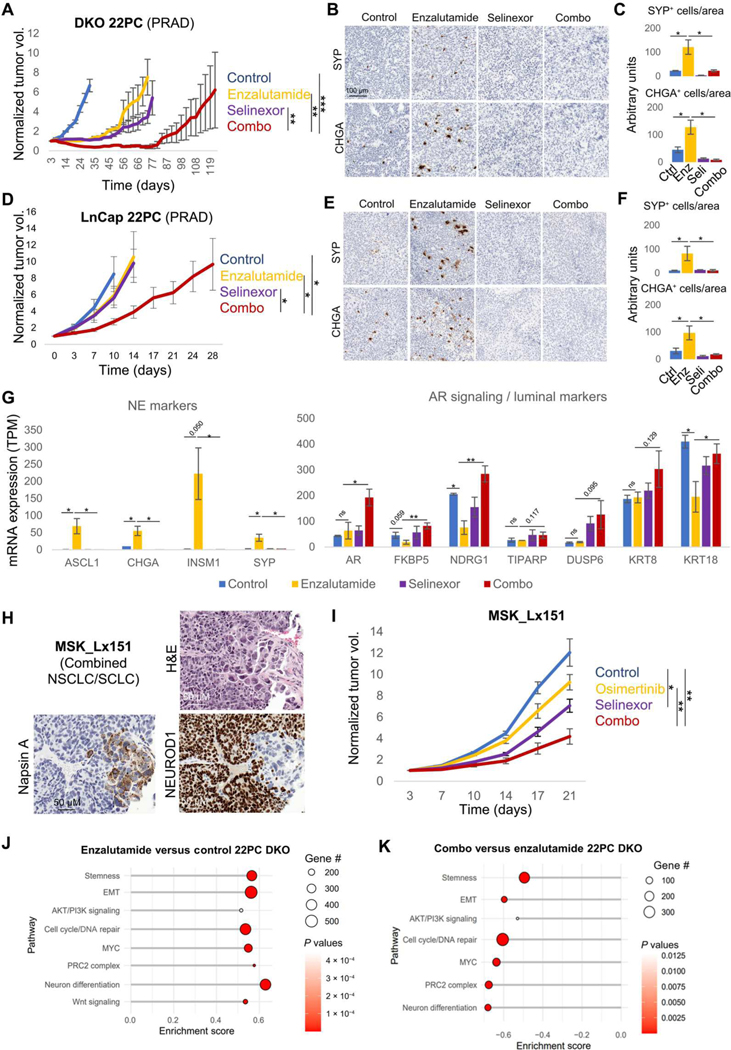
The observed induction of exportin 1 and selinexor sensitivity upon concurrent TP53/RB1 inactivation in both lung and prostate cancers (Fig. 3, A to F) provided a rationale to nominate exportin 1 as a potential therapeutic target to prevent NE transformation. To test this hypothesis, we leveraged previously described prostate NE transformation models (8), the PRAD cell lines 22PC and LnCap/AR, in which double knockout (DKO) of TP53 and RB1 induces AR-targeted therapy resistance in vivo, together with a loss of epithelial features and increased NE marker expression (8). We generated xenografts of using both cell lines in immunocompromised NSG mice, which were then treated with enzalutamide, selinexor, or their combination by oral gavage after tumors reached around 100 mm3 (Fig. 4, A and B). Although initially responsive to enzalutamide (T/C values of 18% at control arm end point, defining end point for a given treatment group as the time when group average size would reach 1000 mm3 of volume), the DKO 22PC tumors consistently demonstrated acquired resistance within the first 2 months of treatment (Fig. 4A). These tumors also showed initial sensitivity to selinexor monotherapy (T/C values of 17% at control arm end point), leading to resistance within a similar time frame to that observed for enzalutamide monotherapy (Fig. 4A). Combination treatment showed greater durability of response, approximately doubling time to tumor relapse relative to either drug alone (Fig. 4A). In the DKO LnCap/AR model, xenografts exhibited immediate resistance to either enzalutamide or selinexor monotherapies (Fig. 4B), with T/C values of 74 and 63%, respectively, at control arm end point. In this model, again, the combination of enzalutamide and selinexor exhibited the greatest effectivity at suppressing tumor growth (T/C value of 31% at control arm end point), doubling the time until relapse as compared with enzalutamide monotherapy arm (Fig. 4B). The combination treatment did not show increased toxicity relative to enzalutamide monotherapy as determined by mouse body weight over time (fig. S2A). Determination of protein expression of NE markers synaptophysin and chromogranin A by IHC performed on tumors collected at experimental end point for each of the treatment arms revealed an increased NE cell subpopulation in enzalutamide-treated versus control tumors in both models (Fig. 4, B to F). This was consistent with the previously described ability of targeted therapies to induce or select for the outgrowth of histologically transdifferentiated cells (4, 10). We observed reduced representation of this cell subset in the selinexor- and combination-treated arms (Fig. 4, B to F), suggesting that exportin 1 inhibition can inhibit NE transformation. To further dissect this phenotype, we performed transcriptome sequencing in tumors from all treatment arms collected at control arm end points for the DKO 22PC model (day 31; Fig. 4A). In line with the immunohistochemical data, we observed increased expression of the NE markers ASCL1 (P = 0.046), CHGA (P = 0.039), and SYP (P = 0.040) in the enzalutamide-treated but not selinexor- or combination-treated cohorts (Fig. 4G). In addition to classical NE marker expression, we observed a transcriptomic profile compatible with NE transformation in the enzalutamide-treated tumors, characterized by increased expression of (i) HES6 and DLL3, markers of Notch signaling down-regulation; (ii) EZH2, the catalytic component of the PRC2 complex, an epigenetic remodeling complex involved in lineage plasticity and histological transformation (4, 9); and (iii) transcription factors previously involved in NE transformation including ONECUT2, FOXN4, and POU3F2 (fig. S2B) (4, 19, 20). This characteristic transcriptomic signature was suppressed by selinexor in the combination-treated tumors, which also exhibited higher maintained expression of AR, AR target genes such as FKBP5 and NDRG1, and luminal markers including KRT8 and KRT18 (Fig. 4G and fig. S2B). In addition, we observed that selinexor prevented the acquisition of a basal-like phenotype, previously described to be potentiated in TP53/RB1-deficient PRAD after targeted therapy treatment, in parallel with NE features (fig. S2C) (8), further supporting the capacity of exportin 1 inhibition to constrain lineage plasticity in this setting.
Fig. 4. Exportin 1 inhibition interferes with NE transformation.
In vivo treatment of cell line xenografts for TP53/RB1 DKO 22PC (A) and LnCap/AR (B) PRAD cells with enzalutamide, selinexor, or their combination. Five to 10 female (22PC) or male (LnCap/AR) 6-week-old athymic nude mice were subcutaneously engrafted per treatment arm and grown until tumors reached 100 to 150 mm3. At that point, mice were randomized into groups and treated with either vehicle (n = 7 for 22PC and n = 4 for LnCap/AR), selinexor (10 mg/kg, p.o., QDx3, n = 7 for 22PC and n = 4 for LnCap/AR), enzalutamide (10 mg/kg, p.o., QDx5, n = 7 for 22PC and n = 5 for LnCap/AR), or the combinations of enzalutamide + selinexor at the previously mentioned doses (n = 9 for 22PC and n = 5 for LnCap/AR). Mice weights and tumor volumes were measured twice a week, and mice were euthanized when tumors reached a humane end point (volume, 1000 mm3). Tumor volumes are shown as normalized volume in arbitrary units (au). Each tumor was normalized to its volume at day 0 of treatment. Representative IHC images for synaptophysin (SYP) and chromogranin A (CHGA) staining in DKO 22PC (C) and LnCap/AR (D) tumors. Quantification of SYP- or CHGA-positive cells, normalized to tissue area, in immunohistochemical tissue stains in DKO 22PC (n = 6, 5, 4, and 4 tumors for control, selinexor-, enzalutamide, and combo-treated arms, respectively) (E) and LnCap/AR (n = 5, 5, 6, and 6 randomly selected tissue pieces for control, selinexor-, enzalutamide, and combo-treated arms, respectively) (F) tumors. Positive cells were counted, tissue area (viable tumor area) was estimated using the SketchAndCalc online app (https://sketchandcalc.com/), and positive-stained cells were normalized by estimated area. (G) RNA-seq data from tumors from (A) collected at control arm experimental end point (day 31), showing mRNA expression for genes of interest, involved in NE transformation, divided by treatment arm (n = 4, 3, 3, and 3 tumors for the control, enzalutamide-, selinexor-, and combo-treated tumors). mRNA expression values are shown as TPM. (H) H&E and IHC staining for markers of interest for the EGFR-mutant combined NSCLC/SCLC PDX tumor MSK_Lx151. (I) In vivo treatment of the MSK_Lx151 PDX with vehicle (n = 5), osimertinib (n = 5), selinexor (n = 5), or their combination (n = 5). Five to 10 female 6-week-old NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice were subcutaneously engrafted per treatment arm and grown until tumors reached 100 to 150 mm3. At that point, mice were randomized into groups and treated with either vehicle, selinexor (10 mg/kg, p.o. QDx3), enzalutamide (10 mg/kg, p.o. QDx5), osimertinib (25 mg/kg, p.o. QDx5), or the combination of osimertinib + selinexor at the previously mentioned doses. Mice weights and tumor volumes were measured twice a week, and mice were euthanized when tumors reached a humane end point (volume, 1000 mm3). Tumor volumes are shown as normalized volume in arbitrary units (au). Each tumor was normalized to its volume at day 0 of treatment. Pathway enrichment analysis on DEGs from enzalutamide versus control (J) and combination versus enzalutamide (K) conditions in the transcriptomic data from TP53/RB1 DKO 22PC xenografts treated in vivo and collected at control arm experimental end point (day 31). Categorized pathways of interest, previously involved in NE transformation (3, 4), are shown. For (A), (C), (D), and (E), P values were calculated using the Student’s t test (unpaired, heterogeneous variances, and two-tailed). *P < 0.05, **P < 0.01, and ***P < 0.001.
A parallel model of in vivo LUAD-to-SCLC transformation has not been described. However, we identified a PDX (MSK_Lx151) derived from an EGFR-mutant mixed histology tumor, which retained both LUAD and SCLC components in the mouse (Fig. 4H), potentially representative of an intermediate state of transformation. In this PDX model, both osimertinib and selinexor demonstrated limited efficacy as single agents, with T/C values of 73.30 and 57.67%, respectively, at control arm end point (Fig. 4I). Again, the combination of osimertinib and selinexor demonstrated greater efficacy, with a T/C value of 34.15% at control arm end point and no additional toxicity compared to osimertinib monotherapy (fig. S2D). No significant differences were observed in the tumors collected at the end point of each of the treatment arms under study in terms of expression of the LUAD marker TTF-1 and the NE markers synaptophysin and chromogranin A (fig. S2E), suggesting that the combination of osimertinib and selinexor may not be able to revert NE transformation after it has occurred. Together, these results suggest that exportin 1 inhibition suppresses the acquisition of an NE phenotype in models of NE transformation and that its combination with targeted therapy might prevent or delay NE relapse in LUADs and PRADs prone to transformation.
Exportin 1 inhibition blocks SOX2 induction in models of NE transformation
To investigate the mechanisms by which exportin 1 inhibition interferes with NE relapse, we performed differential gene expression (DEG) and pathway enrichment analyses on the transcriptomic data from the DKO 22PC transformation model (Fig. 4, A, C, E, and G). As expected, in the comparison of enzalutamide-treated versus control tumors, we observed up-regulation of genes in several pathways previously implicated in NE transformation (3, 4, 21), including those related to epithelial-to-mesenchymal transition, stemness, PRC2 complex, AKT, and Wnt signaling (Fig. 4J). In comparing combination-versus enzalutamide-treated tumors, we observed down-regulation of all these pathways when selinexor was added (Fig. 4K). Leveraging previously published (12) transcriptomic data on selinexor-treated de novo SCLC cell lines, we observed down-regulation of these pathways as well (fig. S2F), further supporting the ability of exportin 1 inhibition to interfere with these NE transformation-related pathways.
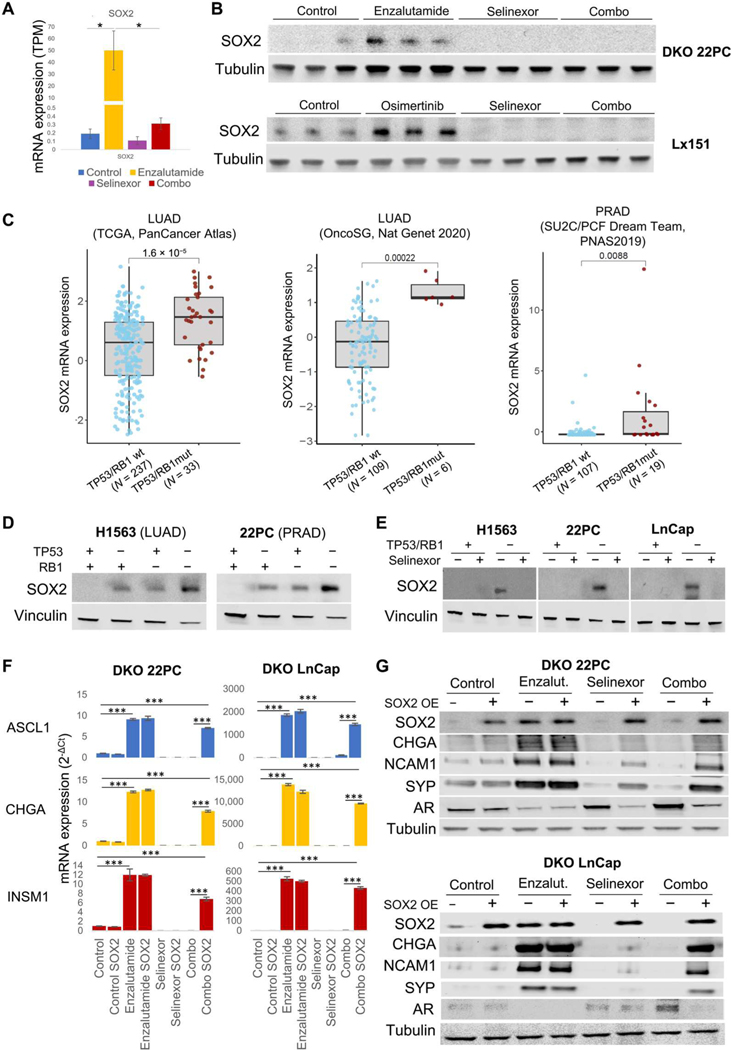
SOX2, a transcription factor implicated in maintenance of stem cell capacity, was found to be essential for NE transformation in a prostate cancer model (8) and is highly overexpressed in SCLC (5). SOX2 expression was induced by enzalutamide in the TP53/RB1-inactivated (DKO) 22PC model, and this induction was inhibited by selinexor in the combination-treated group (Fig. 5A). We observed SOX2 protein up-regulation induced by single-agent targeted therapy in both the DKO 22PC and Lx151 models, again suppressed by the addition of selinexor (Fig. 5B). SOX2 expression was also down-regulated by selinexor in de novo SCLC cell lines (fig. S2G) (12). Up-regulation of SOX2 expression has been reported as an early event in NE transformation of prostate cancer models after inactivation of TP53 and RB1 (8). Consistent with these preclinical observations, we also observed up-regulated SOX2 expression in double TP53/RB1-mutant LUAD and PRAD clinical specimens relative to their wild-type counterparts (Fig. 5C and fig. S3A). SOX2 mRNA and protein expression was also elevated in isogenic TP53/RB1-inactivated LUAD and PRAD cell lines (Fig. 5D and fig. S3B). This induction was prevented by selinexor treatment at both mRNA and protein levels (Fig. 5E and fig. S3B). Previous reports have implicated miR-145 in the down-regulation of SOX2 mediated by XPO1 inhibition (22). We were not able to detect miR-145 expression in any of the lung or PRAD samples under study. Together, these data supported the hypothesis that exportin 1 inhibition might interfere with NE transformation in TP53/RB1-deficient adenocarcinomas by preventing SOX2 up-regulation in early steps of transformation, consistent with a prior report that ectopic SOX2 suppression prevented induction of NE features in PRAD models and maintained enzalutamide sensitivity (8). To test this hypothesis, we exogenously overexpressed SOX2 in the DKO PC22 and LnCap/AR NE transformation models and treated it as previously with enzalutamide, selinexor, or their combination, with the aim of characterizing changes in NE marker and AR expression in vitro (Fig. 5, F and G). In the untreated condition, SOX2 ectopic overexpression did not alter expression of either, suggesting that even if SOX2 is required for NE transformation (8), its sole overexpression in a TP53/RB1-deficient background may not be enough to induce an NE phenotype. As observed in vivo, enzalutamide-treated cells exhibited high SOX2 expression, comparable to the expression induced by ectopic overexpression (Fig. 5G), together with induction of NE markers and suppression of AR. In the absence of exogenous SOX2 expression, selinexor was able to prevent enzalutamide-induced NE marker expression and AR suppression, but these effects were abrogated by expression of exogenous SOX2 (Fig. 5, F and G). However, SOX2 ectopic overexpression was not able to revert the increased sensitivity to selinexor reported after inactivation of TP53 and RB1 (Fig. 3F and fig. S3B). We tested whether overexpression of factors other than SOX2, which our data implicate in NE transformation, would rescue the NE phenotype inhibited by selinexor in these models. Overexpression of POU3F2, ONECUT2, or a constitutively active isoform of AKT (myrAKT) did not revert selinexor-induced NE marker expression (fig. S3C), suggesting that SOX2 might regulate NE transformation upstream of any of these factors. These results implicate prevention of SOX2 induction as a mechanism by which exportin 1 inhibition interferes with NE transformation.
Fig. 5. Exportin 1 inhibition down-regulates SOX2 expression, hindering the acquisition of NE features.
(A) SOX2 mRNA expression in tumors divided by treatment condition. mRNA expression values are shown as TPM (n = 4, 3, 3, and 3 tumors for the control, enzalutamide-, selinexor-, and combo-treated tumors, respectively). (B) Western blot showing SOX2 protein expression of TP53/RB1 DKO 22PC and Lx151 models treated in vivo (n = 3 per treatment condition). Five to 10 female 6-week-old NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice (Lx151) or female 6-week-old athymic nude mice (22PC xenografts) were subcutaneously engrafted per treatment arm and grown until tumors reached 100 to 150 mm3. At that point, mice were randomized into groups and treated with either vehicle (n = 7 for 22PC and n = 5 for Lx151) selinexor (10 mg/kg, p.o., QDx3, n = 7 for 22PC, and n = 5 for Lx151), enzalutamide (for 22PC, 10 mg/kg, p.o., QDx5, n = 7), osimertinib (for Lx151, 25 mg/kg, p.o., QDx5, n = 5), or the combinations of enzalutamide + selinexor (22PC, n = 9) or osimertinib + selinexor (Lx151, n = 5) at the previously mentioned doses. Mice weights and tumor volumes were measured twice a week, and mice were euthanized when tumors reached a humane end point (volume, 1000 mm3). (C) SOX2 mRNA expression in LUAD clinical specimens, categorized by their TP53/RB1 status. P values are shown. Data obtained from LUAD TCGA (PanCancer, n = 237 wt and 33 mutated), LUAD OncoSG (OncoSG, Nat Genetics 2020, n = 109 wt and mutated), and PRAD TCGA (PanCancer, n = 107 wt and 19 mutated) (15, 16). See also fig. S3. (D) Western blot showing SOX2 protein abundance in isogenic control and TP53-and/or RB1-inactivated H1563 (LUAD) and 22PC (PRAD) cell lines. (E) SOX2 protein abundance in control and TP53/RB1-inactivated H1563 (LUAD), 22PC, and LnCap/AR (PRAD) cell lines treated with selinexor (5 nM) for 4 days. ASCL1, CHGA, and INSM1 mRNA expression (F) and SOX2, CHGA, NCAM1, SYP, and AR protein abundance (G) in DKO 22PC cells treated with enzalutamide (150 nM), selinexor (5 nM), or their combination for 4 days. For Western blots and mRNA plots, representative images are shown. P values were calculated using the Student’s t test (unpaired, heterogeneous variances, and two-tailed). *P < 0.05, and ***P < 0.001.
DISCUSSION
Lineage plasticity facilitating histologic transdifferentiation is increasingly recognized as a mechanism of tumor evolution across multiple solid tumors—in part because of research efforts including repeated biopsy after treatment failure to define mechanisms of acquired resistance. Beyond EGFR- and AR-targeted therapy in lung and PRADs (2, 3), respectively, NE transformation has been identified in multiple other oncogene-driven settings (4, 23). The development of highly potent and specific inhibitors of oncogenic drivers across the spectrum of cancer types may increase the frequency of occurrence of such histological transdifferentiation. The poor prognoses of patients after NE transformation across tumor types make defining strategies to constrain or prevent plasticity and to treat NE transformation more effectively key areas of need. This identification of pharmacologically tractable targets in these contexts could have a substantial clinical impact.
One of the major hurdles to identify drivers of histologic transdifferentiation has been a paucity of pre- and posttransformation clinical specimens amenable for molecular analysis. A continuing limitation is the minimal number of viable, manipulable in vitro or in vivo models with which to interrogate this phenomenon. Leveraging microdissected LUAD and SCLC subdomains from combined histology tumors as closely associated putative intermediate states of transformation, we nominated several genes and pathways as possible candidates contributing to NE transformation in the lung (4). These included transcription factors ONECUT2 and POU3F2, members of the AKT signaling pathway, and members of the PRC2 epigenetic remodeling complex. In addition, all of these were also identified as putative factors contributing to NE transformation in the prostate (3, 21, 24, 25). These parallel results suggested to us that shared mechanisms promote lineage plasticity across disease types, increasing the likelihood that discoveries in one tumor type may have broader implications.
We have previously studied the exportin 1 dependence of de novo SCLC tumors, demonstrating in multiple models that exportin 1 inhibition can increase the durability of chemotherapy response (12). Here, we first sought to assess whether the effects of exportin 1 inhibition observed in de novo SCLCs could be extended to T-SCLC and NEPC. Similar to de novo SCLC, we observed exportin 1 up-regulation in NE transdifferentiating clinical specimens from patients with both lung and prostate cancer, highlighting the parallelisms between NE tumors of different origin (26). Although de novo and T-SCLCs do show some divergent molecular profiles (4, 17), with T-SCLCs retaining features of their parental adenocarcinoma state, such as decreased neuronal differentiation and increased Notch signaling (4, 11), we observed consistent combinatorial efficacy of selinexor and cisplatin in both T-SCLC and NEPC PDX models. This combination outperformed that of cisplatin and etoposide, a standard first-line clinical regimen for these tumors.
Defining more durably effective treatments for transformed NE carcinomas is of immediate clinical relevance, but preventing emergence of these aggressive cancer derivatives could have even greater clinical impact. Exportin 1 up-regulation was observed in pretransformation LUAD carrying concomitant TP53 and RB1 inactivation (fig. S4). Loss of these two key tumor suppressors is not sufficient to drive full NE transdifferentiation (6, 11) but defines patient subsets in both lung and prostate cancer at high risk of transformation (6) and with reduced durations of response to targeted therapy (6, 27). Linking the mutational context to the target, TP53 and RB1 inactivation drove exportin 1 up-regulation and induced increased sensitivity to the exportin 1 inhibitor selinexor.
The mechanisms contributing to the apparent increased dependence of NE tumors on nuclear transport by exportin 1 have not been determined. Exportin 1 might promote efficient transport of transcripts controlling cell cycle regulatory and DNA damage repair pathways, on which tumor cells are increasingly dependent after loss of TP53 and RB1 (13). In line with this hypothesis, our transcriptomic data in TP53/RB1-inactivated adenocarcinoma models show down-regulation of cell cycle and DNA damage repair genes upon treatment with selinexor. However, given the broad relevance of nuclear transport, other essential pathways may also be affected. Further investigation will be required to elucidate mechanisms by which TP53/RB1-inactivated tumors become more sensitive to exportin 1 inhibition.
Selinexor was able to suppress enzalutamide-triggered NE transformation in vivo in an adenocarcinoma-to-NE transformation prostate cancer model. In our prostate and lung models, selinexor down-regulates key transcription factors linked to the NE phenotype, including POU3F2 and ONECUT2 (4), and gene expression for pathways implicated in NE transdifferentiation, including AKT signaling and the PRC2 complex (3, 21, 24). The relative importance of these several effects is likely to vary in different contexts. Nonetheless, mechanistically, the capacity of selinexor to interfere with NE transformation was associated with its ability to down-regulate expression of SOX2, as seen in different models in both lung and prostate contexts (fig. S4), and ectopic SOX2 overexpression restored the NE phenotype suppressed by selinexor in a prostate model of transformation. These results support exportin 1 inhibition leading to SOX2 down-regulation as a mechanism constraining lineage plasticity and abrogating NE transformation in these models.
Our study has some limitations. The potential benefits of selinexor in constraining lineage plasticity need to be weighed against known toxicities of this agent. In a prior phase 2 study of selinexor in patients with metastatic prostate cancer, selinexor dosed at 65 mg/m2 twice weekly was associated with significant anorexia, nausea, and fatigue, requiring dose reduction to 60 mg of flat dosing twice weekly (28). However, the latter regimen was tolerable and demonstrated improved progression-free survival over placebo in a subsequent randomized phase 3 study of 285 patients with dedifferentiated liposarcoma (29). Neither trial involved patient selection by predictive biomarkers: Our study may help to inform future targeted clinical trials of selinexor. Another limitation of the current work is that the transformed small cell carcinoma models used in the present study show limited sensitivity to chemotherapy, even at maximally tolerated chemotherapy doses. These results do not fully model clinical observations, where such tumors typically show at least transient response to treatment (3, 7). Furthermore, the number of PDXs tested was limited because of the scarcity of such models. Although our data are in line with the results from our group on de novo SCLC PDXs (12), these results should be interpreted with caution.
In summary, the results presented here nominate exportin 1 as a therapeutic target to inhibit NE transformation in patients with adenocarcinomas at high risk of NE transformation and to augment the efficacy of standard chemotherapy against NE tumors after transformation. The clinical availability of potent and safe exportin 1 inhibitors could facilitate clinical translation of these findings in an increasingly relevant disease context in which the currently available therapeutic options are generally few and ineffective.
MATERIALS AND METHODS
Study design
The purpose of this study was to evaluate the potential efficacy of therapeutic targeting of exportin 1 (XPO1) at preventing NE transformation and to investigate the underlying mechanism of action. Exportin 1 expression was analyzed in clinical specimens analyzed at the mRNA and protein level, and isogenic cell lines were generated to study how early molecular alterations in the transformation process affect exportin 1 expression. Different preclinical xenograft models of NE transformation were treated in vivo with exportin 1 inhibitors in combination with targeted therapy or chemotherapy to assess efficacy at controlling tumor growth. End points for each treatment group were defined as the time when group average size would reach 1000 mm3 of volume. Where specified, tumors were harvested, and the expression of NE and other markers of interest was assessed. All experiments were randomized and blinded where possible. Sample sizes were determined on the basis of expected effect sizes from pilot experiments. In general, group sizes of five or more mice were used. Differences in tumor growth were tested using Student’s t test (two-tailed), correcting for multiple measurements. All in vitro experiments were run at least in biological triplicates each including technical triplicates.
Cell lines
H1563 (CRL-5875) and H660 (CRL-5813) were purchased from the American Type Culture Collection (ATCC). LnCap/AR and 22PC cell lines were shared by the Sawyers laboratory at Memorial Sloan Kettering Cancer Center (MSKCC) and maintained as previously described [RPMI 1640 + 10% fetal bovine serum + 1% L-glutamine (Corning, no. 10–041-CM) + 1% sodium pyruvate (Sigma-Aldrich, no. S8636–100ML), with the addition of 0.1 nM dihydrotestosterone (Sigma-Aldrich, no. 521–18-6) for 22PC] (8). Cell lines were authenticated through the short tandem repeats (STR) characterization method and regularly tested for mycoplasma (Universal Mycoplasma Detection Kit, no. 30–1012K, ATCC). All experiments were performed in low-passage cells. All cell lines were cultured according to ATCC guidelines or as previously described (8). TP53/RB1-deficient PRAD cell lines were generated as previously described (8), with CRISPR-Cas9 technology. The CRISPR-Cas9 lentiviral vectors used to inactivate TP53 and RB1 genes were pLKO5.sgR-NA.EFS.tRFP (Addgene, no. 57823) and lentiCRISPR v2 (Addgene, no. 52961), which were transduced into Cas9-expressing cell lines. TP53/RB1-deficient LUAD cell lines were generated by lentiviral transduction of a construct expressing a dominant negative TP53 isoform and a short hairpin RNA against RB1 produced from the FU-CYW vector that was previously described (21) and shared by O. Witte. Other lentiviral overexpression plasmids used in this work included SOX2 (EX-T2547-Lv105-B), POU3F2 (EX-A3238-Lv151), and ONECUT2 (EX-Z4476-Lv151)—all purchased from Genecopoeia.
Cell cycle assays
Cell cycle was studied by flow cytometry. Cells were seeded in six-well plates and treated for 4 days with selinexor at the indicated doses. At day 4, cells were washed with phosphate-buffered saline (PBS, Lonza) and fixed in 70% ethanol for a maximum period of 1 week at 4°C. Next, cells were washed twice with PBS (Lonza) and incubated with FxCycle propidium iodide/ribonuclease (RNase) staining solution (Invitrogen) for 1 hour at room temperature. Cell cycle was determined using a BD LSRFortessa cell analyzer (BD Bioscience), and cell cycle phases were determined using FlowJo software v10.
Monotherapy cytotoxicity assay and in vitro treatments
Cytotoxic assays were performed as described in (30), with a total of 1500 cells per well seeded in 96-well plates and treated with the drugs/doses described for 96 hours. Viability was assessed with the CellTiter-Glo 2.0 Assay (Promega, G9242) as indicated by the manufacturer and normalized to the untreated control wells.
Synergy assays
Cells were seeded in 96-well plates (1500 cells per well) and treated with the interval of concentrations of cisplatin or selinexor for 5 days. Then, cell viability was assessed with CellTiter-Glo 2.0 Assay (Promega, G9242) and normalized to the untreated wells. Synergy was calculated using the zero interaction potency (ZIP) method using the SynergyFinder web application (2.0) (31).
Promoter reporter assays
A promoter reporter clone for the human XPO1 gene (HPRM44900-LvPG04, Genecopoeia) was used in combination with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)–positive control clone (GAPDH-LvPG04, Genecopoeia) and a negative control clone (NEG-LvPG04, Genecopoeia). Such clones were purchased in a lentiviral vector (LvPG04). Lentiviral particles were produced and used to infect isogenic cell lines of interest, as described previously (12), through concurrent transfection of human embryonic kidney 293T cells (ATCC, no. CRL-1573) with a 3:2:1 ratio of lentiviral plasmid:psPAX2:pMD2.G and JetPrime transfection reagent (Polyplus, no. 114–15) at a 2:1 JetPrime:DNA ratio. Medium was changed 24 hours after transfection, and viral supernatants were collected 72 hours after transfection. Viral supernatants were syringe-filtered with a 0.45-μm polyvinylidene difluoride filter (Millipore, no. SLHVM33RS) and concentrated approximately 20-fold with a Lenti-X concentrator (Takara Bio, no. 631232) according to the manufacturer’s protocol. Promoter reporter assays were performed as specified by the manufacturer using the Secrete-Pair Gaussia luciferase dual and single luminescence assay kits (LF032, Genecopoeia), where signal from constitutively secreted alkaline phosphatase activity was used to normalized XPO1 promoter–dependent Gaussia luciferase activity.
Immunoblot
Protein extraction and Western blot were performed as previously described (32) from frozen cell pellets or flash-frozen tumor samples using radioimmunoprecipitation assay lysis buffer with 1× HALT protease inhibitor cocktail (Thermo Fisher Scientific, no. 78446). Cell pellets were resuspended in five volumes of cold lysis buffer and incubated on ice for 30 min. Lysates were clarified by centrifugation at 20,000g for 10 min at 4°C. Antibodies for Western blotting included XPO1 (no. 46249, Cell Signaling Technology), pAKT (no. 4060, Cell Signaling Technology), pPRAS40 (no. 13175, Cell Signaling Technology), chromogranin A (no. ab85554, Abcam), synaptophysin (no. 36406, Cell Signaling Technology), CD56 (no. 99746, Cell Signaling Technology), AR (no. 5153, Cell Signaling Technology), POU3F2 (no. 12137, Cell Signaling Technology), ONECUT2 (no. ab28466, Abcam), vinculin (no. 13901, Cell Signaling Technology), and tubulin (no. 3873, Cell Signaling Technology). Quantifications were performed with Image Studio software (version 3.1, Li-Cor). Antibodies used for IHC included exportin 1 (no. 611833 from BD), synaptophysin (Dako, no. A0010), and chromogranin A (Dako, no. A0430). IHC was performed on formalin-fixed paraffin-embedded (FFPE) tissue from resected tumor samples obtained from patients with de novo and transforming LUAD, SCLC, PRAD, and NEPC. For immunohistochemical staining, slides were deparaffinized and steamed for 45 min in Target Retrieval Solution (Dako). Immunocomplexes were detected using PV Poly-horseradish peroxidase anti-mouse immunoglobulin G (Leica Microsystems, no. PV6114) followed by a Tyr-amide Signal Amplification (TSA) biotin amplification step (PerkinElmer) with 3,3’-diaminobenzidine (DAB) as the chromogen. Tissue sections were counterstained with hematoxylin, and slides were digitized on a Ventana DP 200 slide scanner (Roche). Expression was scored in a blinded manner by pathologists, whereby the optical density level (“0” for no brown color, “1” for faint and fine brown chromogen deposition, “2” for intermediate chromogen deposition, and “3” for prominent chromogen deposition) was multiplied by the percentage of cells at each staining level, resulting in a total H-score range of 0 to 300. All study participants had provided signed informed consent for biospecimen analyses under Institutional Review Board–approved protocols (nos. 14–209 and 14–091).
In vivo treatments
Four to ten female (22PC, PDXs) or male (LnCap/AR) 6-week-old NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice (PDXs) or female 6-week-old athymic nude mice (cell line xenografts) were subcutaneously engrafted per treatment arm and until tumors reached 100 to 150 mm3. At that point, mice were randomized into groups and treated with either vehicle, cisplatin [2 mg/kg intraperitoneally (i.p.) once per week], etoposide [3 mg/kg, i.p., quaque die (QD) x3], selinexor [10 mg/kg per os (p.o.), QDx3], enzalutamide (10 mg/kg, p.o., QDx5), osimertinib (25 mg/kg, p.o., QDx5), or the combinations of cisplatin + etoposide, cisplatin + selinexor, enzalutamide + selinexor, or osimertinib + selinexor at the previously mentioned doses. Mice weights and tumor volumes were measured twice a week, and mice were euthanized when tumors reached a humane end point (volume, 1000 mm3). The number of mice per treatment arm was selected according to previous experience with the models and response to treatments. Blinding was not performed. All animal experiments were approved by the MSKCC Animal Care and Use Committee (no. 13–07-007).
RNA extraction
Frozen tissues or cell pellets were weighed and homogenized in RLT, and nucleic acids were extracted using the AllPrep DNA/RNA Mini Kit (QIAGEN, no. 80204) according to the manufacturer’s instructions. RNA was eluted in nuclease-free water.
RNA-seq alignment and quantification
Transcript abundances were quantified using RNA-seq reads by Salmon v1.1.0 (33). Raw reads of RNA-seq were mapped to 25-mer indexed hg38 genome. In addition to default settings, mapping validation (--validatemappings), bootstrapping with 30 resamplings (--numBootstraps), sequence-specific bias correction (--seqBias), coverage bias correction (--posBias), and GC bias correction (--gcBias) were also enabled. Transcripts were mapped to genes based on Ensembl 92 (34), normalized by size factor at gene level. Subsequently, the differential gene expression was evaluated on Salmon output files using Sleuth v0.30.0 (35) in gene mode. A Wald test was performed on differential gene expressions. Genes were marked as significantly differentially expressed if the false discovery rates, q, calculated using the Benjamini-Hochberg method, were less than 0.05, and β (Sleuth-based estimation of log2 fold change) > 0.58, which is approximately equivalent to a log2 fold change of 1.5.
Publicly available RNA-seq dataset analyses
Public datasets leveraged in the present manuscript accessible through cbioportal.com include Abida et al. (16), LUAD the Cancer Genome Atlas (TCGA) PanCancer, LUAD OncoSG, Nat Gen 2020, and PRAD TCGA PanCancer. The dataset from (36) can be accessed at the Gene Expression Omnibus portal (GSE104786). The public sets were divided into four groups according to their mutation status of TP53 and RB1: TP53WT/RB1WT, TP53MT/RB1WT, TP53WT/RB1MT, and TP53MT/RB1MT. RNA-seq expression distribution of XPO1, SOX2, and CDC7 was presented in box plots for the above four groups of samples. RNA-seq expression values were downloaded through cBioPortal.
Data type 1, RSEM: The expression quantification for LUAD (OncoSG, Nat Genet 2020) is in RSEM (RNA-seq by expectation-maximization), which has been normalized using DESeq2 v.1.16.1 followed by log transformation, whereas that for PRAD (TCGA, PanCancer) is in batch-normalized RSEM then followed by log transformation.
Data type 2, RSEM z score: Log-transformed mRNA expression z scores compared with the expression distribution of all samples were downloaded for both LUAD [OncoSG, Nat Genet 2020, (37)] and PRAD (TCGA, PanCancer) (38). The pairwise comparisons of mean expressions were conducted among the four previously mentioned groups and evaluated by Wilcoxon test. [Using traditional RNA-seq DEG approaches to evaluate DE P value by the limma pipeline, we applied linear modeling on the normalized and log-transformed RSEM values, which are assumed to be normally distributed using limma (v3.28.14) (39). The coefficients and SEs were then estimated for each pair of contrast from the linear model. Empirical Bayes statistics for differential expressions was carried out to evaluate the significance value.]
The expression values of XPO1 and SOX2 were correlated in scatter plots for the seven previously mentioned cohorts. RNA-seq expression values were downloaded through cBioPortal.
Data type 1, RSEM: The expression values are in RSEM, which has been using DESeq2 v.1.16.1 normalization, LUAD [OncoSG, Nat Genet 2020, (37)], or batch-normalized followed by log transformation.
Data type 2, RSEM z score: Log-transformed mRNA expression z scores compared with the expression distribution of all samples were downloaded. The expression correlations were evaluated by Pearson (Spearman).
Pathway enrichment analyses
Gene set enrichment analysis (40) was conducted on the full sets of differential gene expression output from the previously mentioned comparisons. Genes were ranked by P value scores computed as −log10 (P value) * (β). The annotations of gene set taken from the Molecular Signatures Database [MSigDB v7.0.1, (40, 41)] of gene set enrichment were evaluated using permutation test, and the P value was adjusted by the Benjamini-Hochberg procedure. Any enriched gene sets with adjusted P value ≤ 0.1 were regarded as significant. This analysis was conducted using ClusterProfiler R package v3.18.1 (42). Some enriched gene sets of interests were selected, and their pathway annotations were concatenated manually to remove redundancy and achieve high generality. When the pathway terms were merged, median enrichment score was taken as the new group enrichment score, P values were aggregated using Fisher’s method from the Aggregation R package (43), and core enrichment of genes was collapsed. The consolidated gene set enrichments were then presented in dot plots.
ATAC sequencing
The reads were trimmed for both quality and Illumina adaptor sequences using trim_galore v0.4.4 (https://github.com/FelixKrueger/TrimGalore) in the pair-end mode. Then, the raw reads were aligned to human assembly hg38 using bowtie2 v2.3.4 (44) using the default parameters. Aligned reads with the same start site and orientation were removed using the Picard tool (https://broadinstitute.github.io/picard/). Enriched regions in individual samples were called using MACS2 (45) and then filtered against genomic “blacklisted” regions (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/hg38-human/hg38.blacklist.bed.gz ). The filtered peaks within 500 base pairs were merged to create a union of peak atlas. Raw read counts were tabulated over this peak atlas using featureCounts v1.6.0 (46). The read counts were then normalized with DESeq2. The read density profile in the format of bigwig file for each sample was created using the BED-Tools suite (https://bedtools.readthedocs.io) with the normalization factor from DESeq2 (47). All bigwig genome tracks on XPO1 gene region were generated using pyGenomeTracks v3.5 (48).
Reverse transcription qPCR
Retrotranscription was performed with the Superscript IV VILO kit (Thermo Fisher Scientific, no. 11756050) following the manufacturer’s instructions. The following TaqMan (Thermo Fisher Scientific) probes were used: CHGA (Hs00900370_m1), INSM1 (Hs00357871_s1) and ASCL1 (Hs00269932_m1), XPO1 (Hs00185645_m1), SOX2 (Hs04234836_s1), and UBC (Hs00824723_m1). Data were analyzed as previously described (30).
Clinical samples
All study participants had provided signed informed consent for biospecimen analyses under an Institutional Review Board–approved protocol. Metastatic prostate cancer samples were collected as part of the Prostate Cancer Donor Program at the University of Washington. Tissue microarrays sampling PRAD and NEPC FFPE tissues were used in this study.
Statistical analyses
Comparisons between two groups were performed using paired or unpaired two-tailed Student’s t test, as indicated in the figure legends. A P value < 0.05 was considered statistically significant (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001). N indicates the number of biological replicates; all bars within the graphs represent mean values; and the error bars represent SEMs or SD, as indicated in the figure legends. All in vitro experiments were replicated a minimum of three times (biological replicates). All Western blots have been replicated a minimum of two times (biological replicates). Please refer to previous sections for detailed statistical analyses of the bioinformatic data.
Supplementary Material
Acknowledgments:
We acknowledge the use of the PPBC Biobank, Pathology Core Facility, and Integrated Genomics Operation Core, funded by the NCI Cancer Center support grant (P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We thank the patients and their families, C. Higano, E. Yu, E. Mostaghel, H. Cheng, P. Nelson, B. Montgomery, M. Schweizer, A. Hsieh, J. Wright, D. Lin, F. Vakar-Lopez, X. Zhang, M. Roudier, L. True, E. Corey, and the rapid autopsy teams for contributions to the University of Washington Prostate Cancer Donor Rapid Autopsy Program. We thank E. Corey from the University of Washington for providing the LuCAP49 model.
Funding:
This study was supported by PO1 NIH PO1CA163227 (Prostate Cancer Donor Program), NIH T32 CA1600001 (to A.Q.-V.), K08 CA-248723 (to A.C.), R01CA264078 (to C.M.R. and H.A.Y.), the Doris Duke Foundation (grant 2021184) (to M.C.H.), P50CA97186 (to M.C.H. and C.M.), R35 CA263816 (to C.M.R.), U24 CA213274 (to C.M.R.), P30 CA008748, Yasuda Medical Foundation (to K.K.), the American Lung Association (to A.Q.-V.), the Druckenmiller Center for Lung Cancer Research (to A.Q.-V., K.K., and C.M.R.), and the Howard Hughes Medical Institute (to C.L.S.).
Footnotes
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. Datasets generated in the present manuscript are available at the Gene Expression Omnibus (GEO) database with reference numbers GSE237071 (ATAC-seq) and GSE237072 (RNA-seq). Lung cancer patient–derived xenograft models will be provided under a material transfer agreement by contacting the corresponding author.
Competing interests: A.Q.-V. has received honoraria from AstraZeneca. W.R.K. is an inventor of organoid technology. C.M.R. has consulted regarding oncology drug development with AbbVie, Amgen, Astra Zeneca, D2G, Daiichi Sankyo, Epizyme, Genentech/Roche, Ipsen, Jazz, Kowa, Lilly, Merck, and Syros. He serves on the scientific advisory boards of Auron, Bridge Medicines, DISCO, Earli, and Harpoon Therapeutics. C.L.S. serves on the board of directors of Novartis, is a cofounder of ORIC Pharmaceuticals, and is coinventor of enzalutamide and apalutamide. He is a science adviser to Arsenal, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ, and PMV. A.Q.-V. and C.M.R. have filed two patent applications entitled “Methods for treating or preventing neuroendocrine tumor formation using XPO1 inhbitors” (no.115872–2576) and “Methods for predicting and treating chemoresistance in small cell lung cancer patients pending” (WO2022256371A1).
REFERENCES AND NOTES
- 1.Marcoux N, Gettinger SN, O’Kane G, Arbour KC, Neal JW, Husain H, Evans TL, Brahmer JR, Muzikansky A, Bonomi PD, del Prete S, Wurtz A, Farago AF, DiasSantagata D, Mino-Kenudson M, Reckamp KL, Yu HA, Wakelee HA, Shepherd FA, Piotrowska Z, Sequist LV, EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: Clinical outcomes. J. Clin. Oncol. 37, 278–285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, Weinstein AS, Friedl V, Zhang C, Witte ON, Lloyd P, Gleave M, Evans CP, Youngren J, Beer TM, Rettig M, Wong CK, True L, Foye A, Playdle D, Ryan CJ, Lara P, Chi KN, Uzunangelov V, Sokolov A, Newton Y, Beltran H, Demichelis F, Rubin MA, Stuart JM, Small EJ, Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J. Clin. Oncol. 36, 2492–2503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintanal-Villalonga Á, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, Rudin CM, Lineage plasticity in cancer: A shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 17, 360–371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, Uddin F, Manoj P, Donoghue MTA, Won HH, Chan JM, Ciampricotti M, Chow A, Offin M, Chang JC, Ray-Kirton J, Tischfield SE, Egger J, Bhanot UK, Linkov I, Asher M, Sinha S, Silber J, Iacobuzio-Donahue CA, Roehrl MH, Hollmann TJ, Yu HA, Qiu J, de Stanchina E, Baine MK, Rekhtman N, Poirier JT, Loomis B, Koche RP, Rudin CM, Sen T, Multi-omic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discov. 11, 3028–3047 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, Rivers CS, Foo CK, Bhatt D, Stinson J, Gnad F, Haverty PM, Gentleman R, Chaudhuri S, Janakiraman V, Jaiswal BS, Parikh C, Yuan W, Zhang Z, Koeppen H, Wu TD, Stern HM, Yauch RL, Huffman KE, Paskulin DD, Illei PB, Varella-Garcia M, Gazdar AF, De Sauvage FJ, Bourgon R, Minna JD, Brock MV, Seshagiri S, Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, Rekhtman N, Daneshbod Y, Quintanal-Villalonga A, Penson A, Hellmann MD, Arcila ME, Ladanyi M, Péer D, Kris MG, Rudin CM, Yu HA, Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J. Thorac. Oncol. 14, 1784–1793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudin CM, Brambilla E, Faivre-Finn C, Sage J, Small-cell lung cancer. Nat. Rev. Dis. Primers. 7, 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, Wongvipat J, Ku SY, Gao D, Cao Z, Shah N, Adams EJ, Abida W, Watson PA, Prandi D, Huang CH, De Stanchina E, Lowe SW, Ellis L, Beltran H, Rubin MA, Goodrich DW, Demichelis F, Sawyers CL, SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Zheng D, Zhou T, Song H, Hulsurkar M, Su N, Liu Y, Wang Z, Shao L, Ittmann M, Gleave M, Han H, Xu F, Liao W, Wang H, Li W, Androgen deprivation pro- motes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, Uddin F, Allaj V, Manoj P, Shah NS, Chan JM, Ciampricotti M, Chow A, Offin M, Ray-Kirton J, Egger JD, Bhanot UK, Linkov I, Asher M, Roehrl MH, Ventura K, Qiu J, de Stanchina E, Chang JC, Rekhtman N, Houck-Loomis B, Koche RP, Yu HA, Sen T, Rudin CM, Comprehensive molecular characterization of lung tumors implicates AKT and MYC signaling in adenocarcinoma to squamous cell transdifferentiation. J. Hematol. Oncol. 14, 170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T, Howe E, Fulton LE, Mulvey HE, Bernardo LA, Mohamoud F, Miyoshi N, VanderLaan PA, Costa DB, Jänne PA, Borger DR, Ramaswamy S, Shioda T, Iafrate AJ, Getz G, Rudin CM, Mino-Kenudson M, Engelman JA, RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 6, 6377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quintanal-Villalong A, Taniguchi H, Hao Y, Chow A, Zhan YA, Chavan SS, Uddin F, Allaj V, Manoj P, Shah NS, Chan JM, Offin M, Ciampricotti M, Ray-Kirto J, Egger J, Bhanot U, Linkov I, Asher M, Roehrl MH, Qiu J, De Stanchina E, Hollmann TJ, Koche RP, Sen T, Poirier JT, Rudin CM, Inhibition of XPO1 sensitizes small cell lung cancer to first- and second-line chemotherapy. Cancer Res. 82, 472–483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang AY, Liu H, The past, present, and future of CRM1/XPO1 inhibitors. Stem Cell Investig. 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nachmias B, Schimmer AD, Targeting nuclear import and export in hematological malignancies. Leukemia 34, 2875–2886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzelepi V, Zhang J, Lu J-F, Kleb B, Wu G, Wan X, Hoang A, Efstathiou E, Sircar K, Navone NM, Troncoso P, Liang S, Logothetis CJ, Maity SN, Aparicio AM, Modeling a lethal prostate cancer variant with small-cell carcinoma features. Clin. Cancer Res. 18, 666–677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, Cieslik M, Benelli M, Robinson D, Van Allen EM, Sboner A, Fedrizzi T, Mosquera JM, Robinson BD, De Sarkar N, Kunju LP, Tomlins S, Wu YM, Rodrigues DN, Loda M, Gopalan A, Reuter VE, Pritchard CC, Mateo J, Bianchini D, Miranda S, Carreira S, Rescigno P, Filipenko J, Vinson J, Montgomery RB, Beltran H, Heath EI, Scher HI, Kantoff PW, Taplin M-E, Schultz N, Debono JS, Demichelis F, Nelson PS, Rubin MA, Chinnaiyan AM, Sawyers CL, Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 166, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park C-K, Oh I-J, Kim Y-C, Is transformed small cell lung cancer (SCLC) different from de novo SCLC? Transl. Cancer Res. 8, 346–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.True LD, Buhler K, Quinn J, Williams E, Nelson PS, Clegg N, Macoska JA, Norwood T, Liu A, Ellis W, Lange P, Vessella R, A neuroendocrine/small cell prostate carcinoma xenograft—LuCaP 49. Am. J. Pathol. 161, 705–715 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, Ci X, Ahmed M, Hua JT, Soares F, Lin D, Puca L, Vosoughi A, Xue H, Li E, Su P, Chen S, Nguyen T, Liang Y, Zhang Y, Xu X, Xu J, Sheahan AV, Ba-Alawi W, Zhang S, Mahamud O, Vellanki RN, Gleave M, Bristow RG, Haibe-Kains B, Poirier JT, Rudin CM, Tsao M-S, Wouters BG, Fazli L, Feng FY, Ellis L, van der Kwast T, Berlin A, Koritzinsky M, Boutros PC, Zoubeidi A, Beltran H, Wang Y, He HH, ONECUT2 is a driver of neuroendocrine prostate cancer. Nat. Commun. 10, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, Jama R, Nip KM, Angeles A, Johnson F, Wyatt AW, Fazli L, Gleave ME, Lin D, Rubin MA, Collins CC, Wang Y, Beltran H, Zoubeidi A, The master neural transcription factor BRN2 is an androgen receptor–suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 7, 54–71 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, Smith BA, Cheng C, Tsai BL, Cheng D, Huang J, Kurdistani SK, Graeber TG, Witte ON, Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 362, 91–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azmi AS, Li Y, Muqbil I, Aboukameel A, Senapedis W, Baloglu E, Landesman Y, Shacham S, Kauffman MG, Philip PA, Mohammad RM, Exportin 1 (XPO1) inhibition leads to restoration of tumor suppressor miR-145 and consequent suppression of pancreatic cancer cell proliferation and migration. Oncotarget 8, 82144–82155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fares AF, Lok BH, Zhang T, Cabanero M, Lau SCM, Stockley T, Patel D, Bradbury PA, Sacher A, Yasufuku K, Morash BA, Sabatini PJB, Nguyen LN, Leighl NB, Tsao M-S, Shepherd FA, Liu G, Martins-Filho SN, Pal P, ALK-rearranged lung adenocarcinoma transformation into high-grade large cell neuroendocrine carcinoma: Clinical and molecular description of two cases. Lung Cancer 146, 350–354 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, Cyrta J, Sboner A, Noorzad Z, MacDonald T, Cheung C, Yuen KS, Gao D, Chen Y, Eilers M, Mosquera J-M, Robinson BD, Elemento O, Rubin MA, Demichelis F, Rickman DS, N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30, 563–577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JM, Quintanal-Villalonga Á, Gao VR, Xie Y, Allaj V, Chaudhary O, Masilionis I, Egger J, Chow A, Walle T, Mattar M, Yarlagadda DVK, Wang JL, Uddin F, Offin M, Ciampricotti M, Qeriqi B, Bahr A, de Stanchina E, Bhanot UK, Lai WV, Bott MJ, Jones DR, Ruiz A, Baine MK, Li Y, Rekhtman N, Poirier JT, Nawy T, Sen T, Mazutis L, Hollmann TJ, Pe’er D, Rudin CM, Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 39, 1479–1496.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawasaki K, Rekhtman N, Quintanal-Villalonga Á, Rudin CM, Neuroendocrine neoplasms of the lung and gastrointestinal system: Convergent biology and a path to better therapies. Nat. Rev. Clin. Oncol 20, 16–32 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbé DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW, Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei XX, Siegel AP, Aggarwal R, Lin AM, Friedlander TW, Fong L, Kim W, Louttit M, Chang E, Zhang L, Ryan CJ, A phase II trial of selinexor, an oral selective inhibitor of nuclear export compound, in abiraterone- and/or enzalutamide-refractory metastatic castration-resistant prostate cancer. Oncologist 23, 656–e64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gounder MM, Razak AA, Somaiah N, Chawla S, Martin-Broto J, Grignani G, Schuetze SM, Vincenzi B, Wagner AJ, Chmielowski B, Jones RL, Riedel RF, Stacchiotti S, Loggers ET, Ganjoo KN, Cesne AL, Italiano A, Del Muro XG, Burgess M, Ryan SP-NC, Mulcahy MF, Forscher C, Penel N, Okuno S, Elias A, Hartner L, Philip T, Alcindor T, Kasper B, Reichardt P, Lapeire L, Blay JY, Chevreau C, Morales CMV, Schwartz GK, Chen JL, Deshpande H, Davis EJ, Nicholas G, Gröschel S, Hatcher H, Duffaud F, Herráez AC, Beveridge RD, Badalamenti G, Eriksson M, Meyer C, Mehren MV, Van Tine BA, Götze K, Mazzeo F, Yakobson A, Zick A, Lee A, Gonzalez AE, Napolitano A, Dickson MA, Michel D, Meng C, Li L, Liu J, Ben-Shahar O, Van Domelen DR, Walker CJ, Chang H, Landesman Y, Shah JJ, Shacham S, Kauffman MG, Attia S, Selinexor in advanced, metastatic dedifferentiated liposarcoma: A multinational, randomized, double-blind, placebo-controlled trial. J. Clin. Oncol. 40, 2479–2490 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintanal-Villalonga Á, Ferrer I, Guruceaga E, Cirauqui C, Marrugal Á, Ojeda L, García S, Zugazagoitia J, Muñoz-Galván S, Lopez-Rios F, Montuenga L, Vicent S, Molina-Pinelo S, Carnero A, Paz-Ares L, FGFR1 and FGFR4 oncogenicity depends on n-cadherin and their co-expression may predict FGFR-targeted therapy efficacy. EBioMedicine 53, 102683 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ianevski A, Giri AK, Aittokallio T, SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 48, W488–W493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, Ni A, Khodos I, de Stanchina E, Nguyen T, Sage J, Campbell JE, Ribich S, Rekhtman N, Dowlati A, Massion PP, Rudin CM, Poirier JT, Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell 31, 286–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T; To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P, Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L, Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Tsai HK, Lehrer J, Alshalalfa M, Erho N, Davicioni E, Lotan TL, Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 17, 759–NaN (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, Alvarez JJS, Lu B, Lim JQ, Takano A, Nahar R, Lee YY, Phua CZJ, Chua KP, Suteja L, Chen PJ, Chang MM, Koh TPT, Ong BH, Anantham D, Hsu AAL, Gogna A, Too CW, Aung ZW, Lee YF, Wang L, Lim TKH, Wilm A, Choi PS, Ng PY, Toh CK, Lim WT, Ma S, Lim B, Liu J, Tam WL, Skanderup AJ, Yeong JPS, Tan EH, Creasy CL, Tan DSW, Hillmer AM, Zhai W, Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 52, 177–186 (2020). [DOI] [PubMed] [Google Scholar]
- 38.The Cancer Genome Atlas Research Network, The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP, Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang L-G, Han Y, He Q-Y, ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi L, Pimentel H, Bray NL, Pachter L, Gene-level differential analysis at transcript-level resolution. Genome Biol. 19, 53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Shirley XS, Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W, FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Grüning B, Ramírez F, Manke T, pyGenomeTracks: Reproducible plots for multivariate genomic datasets. Bioinformatics 37, 422–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.