Abstract
Objectives
Actionable, solid tumor activating neurotrophic receptor tyrosine kinase (NTRK) fusions are best detected via nucleic acid-based assays, while Pan-TRK immunohistochemistry (IHC) serves as a reasonable screening modality. We describe a practical and cost-effective approach to validate pan-TRK and discuss challenges that may be encountered.
Methods
Pan-TRK Clone EPR17341 was validated in accordance with the 2014 consensus statements set forth by the College of American Pathologists. Confirmation of IHC results were guided by the European Society of Medical Oncology recommendations for standard methods to detect NTRK fusions.
Results
Within 36 samples, ETV6-NTRK3 (n = 8) and TPM4-NTRK3 (n = 1) fusions were confirmed. ETV6-NTRK3 fusion positive cases revealed cytoplasmic and nuclear staining. A TPM4-NTRK3 fusion positive high grade malignant peripheral nerve sheath tumor revealed diffuse cytoplasmic staining. A high grade ovarian serous carcinoma revealed focal punctate staining and revealed a non-actionable NTRK1 truncation at intron 2. Diffuse cytoplasmic staining was observed in a case of fusion-negative polymorphous adenocarcinoma. Wild-type expression of TRK in pulmonary meningothelial-like nodules was discovered following a false-positive IHC interpretation.
Conclusion
Pan-TRK IHC shows some utility as a diagnostic and surrogate marker for NTRK screening however, physiologic or non-specific expression may lead to false-positive results.
Keywords: Pan-TRK, NTRK, Immunohistochemistry, Interpretation, Validation
Introduction
Oncogenic NTRK fusions are seen in many solid cancer types with a very low incidence and are typically mutually exclusive of other driver mutations. These fusions are particularly important to identify as they are the major determinants for the effectiveness of targeted molecular therapy against these alterations, which makes their routine detection a priority. Neurotrophic tyrosine receptor kinases (NTRK) are a family of transmembrane tyrosine kinases that are normally expressed in neural tissue and play a key role in the development and the function of the neural system [1–3]. Three members of this family, TRKA, TRKB, and TRKC, are encoded by the proto-oncogenes NTRK1, NTRK2, and NTRK3, respectively [4, 5]. Each of these proteins are structured with an extracellular ligand-binding domain, a transmembrane region, and an intracellular kinase [4]. In normal conditions, activation of the receptor through ligand binding activates the kinase domain, which leads to receptor homodimerization, phosphorylation and activation of downstream signaling pathways. Constitutive activation of the tropomyosin receptor kinase (TRK) receptors can occur through chromosomal inversions, deletions or translocations that result in an in-frame fusion of the C-terminal tyrosine kinase domain in any of the NTRK genes with an N-terminal fusion partner and constitute activation of several cascades or signaling pathways such as phospholipase C, Ras/MAPK/ERK, and PI3K/AKT pathways which promote tumorigenesis [2, 6–8].
Gene fusions involving NTRK genes are alterations with known oncogenic potential. Oncogenic fusions between the C-terminal kinase domain with the N-terminal fusion partner of the NTRK genes have been identified in high prevalence in rare subtypes of pediatric and adult tumors and are implicated in a small percentage of common adult patient cancers. The first TRK fusion protein was originally described more than 35 years ago in colorectal cancer [9]. In subsequent years, few other studies on NTRK gene alterations in colorectal carcinomas have been reported [10, 11]. ETV6-NTRK3 fusion involving a translocation of chromosome 12 and 15 was identified in infantile fibrosarcoma [12–14], and this same fusion was later also reported in both secretory carcinoma of the breast [15, 16] and secretory carcinoma of the salivary gland; previously known as mammary analog secretory carcinoma (MASC) [17]. NTRK fusions have also been identified in radiation-induced papillary thyroid carcinomas and in many other tumors, including carcinomas of the lung, gliomas, soft tissue sarcomas, inflammatory myofibroblastic tumors, certain hematologic malignancies, congenital mesoblastic nephroma, and melanocytic tumors, all of which collectively comprise less than 1% of all solid tumors [1, 18–29].
In the last two decades, promising preclinical data on the specific molecular mechanisms of NTRK gene fusions resulted in several clinical trials and accelerated the FDA approval of the selective, tumor agnostic NTRK inhibitors larotrectinib and entrectinib (pan-TRK, ROS1, and ALK inhibitor), both of which target a variety of solid tumors in the pediatric and adult populations harboring an NTRK fusion [2, 30–35].
These new therapeutic approvals for the treatment of cancers harboring NTRK fusions has prompted an urgent need for defining the routine testing criteria to help with clinical decision making and management. Screening for NTRK fusions may also be a prerequisite for inclusion into a clinical trial. Furthermore, in uncommon cancers such as infantile fibrosarcoma, congenital mesoblastic nephroma and secretory carcinoma of the breast and salivary gland; identification of NTRK fusions may be used to support the diagnosis. Although there are various available assays— such as fluorescence in situ hybridization (FISH), reverse transcription polymerase chain reaction (RT-PCR), DNA-based next-generation sequencing (NGS), and RNA-based NGS, which can detect these fusions at the DNA or RNA level; accurately detecting the variety of NTRK fusions with these modalities pose their own challenges and expense, given the limited number of cases that may harbor these alterations (Table 1). For example, DNA-based NGS, may miss fusion detection due to difficulty tiling introns and take several weeks [36]. RNA-based sequencing is advantageous in turnaround time (approximately 1 week) with fusion characterization dependent on RNA quality [36]. Further, the cost of implementation, longer turnaround time, and the need for molecular pathology expertise limits the wide applicable use of sequencing techniques.
Table 1.
NTRK1-3 Testing Methodologies
| Reverse transcriptase-polymerase chain reaction (RT-PCR) | ▪ Variable sensitivity ▪ High specificity |
▪ Fusion detection requires specific primers targeting suspected genes and exons ▪ Hindered by RNA degradation |
|---|---|---|
| RNA based NGS alone | ▪ Ability to access for unknown fusion partners, including other oncogenic alterations as well as splice variants | ▪ Hindered by RNA degradation |
| DNA-based NGS alone |
▪ Access for point mutations, fusions, and copy number changes ▪ Limited sensitivity to detect NTRK3 fusions |
▪ Reliant on decent tumor purity |
| Dual DNA/RNA based NGS | ▪ Superior analysis while likely being the most expensive methodology | ▪ Reliant on decent tumor purity |
| Fluorescent in situ hybridization (FISH) |
▪ Comparable turn-around time (approximately 1–3 days) compared to IHC ▪ Designed to detect specific breakpoints |
▪ Best utilized when there is high suspicion of ETV6-NTRK3 fusions |
| Pan-TRK IHC | ▪ Ability to screen for NTRK1-3 fusions while remaining cost effective |
▪ Limited specificity ▪ Detects physiologic wild-type TRK expression |
In contrast, immunohistochemistry (IHC) is a widely available methodology with several benefits, including quick turnaround time, lower cost and minimal tissue requirements, particularly from limited samples. Nevertheless, there is limited data available regarding the use of Pan-TRK IHC in detecting NTRK gene alterations in routine pathology practice [36–41]. Increasing requests for NTRK testing by treating physicians, often on limited specimens for advanced or metastatic disease in hopes of identifying an actionable alteration, prompted us to validate and use the pan-TRK diagnostic immunohistochemical assay Ventana pan-TRK (EPR17341) assay as a screening modality for actionable NTRK fusions in our laboratory.
The CAP 2014 evidence-based guidelines serve as a practical guide for ensuring accuracy and limiting variation among laboratories validating IHC assays. The CAP authorizes the use of orthogonal testing (non-IHC tests, such as FISH or NGS) to be employed in the validation of an IHC. As stated, “laboratories may use a combination of methods when appropriate” [48]. We present our experience with the validation of pan-TRK IHC in accordance with CAP recommendations with the key aim of making NTRK testing easily accessible for laboratories with limited resources. Molecular testing was not indicated on all cases as recommended by the European Society for Medical Oncology practice based guidelines on standard methods for NTRK fusion detection in daily practice and clinical research [39].
Methods
Design of the Validation Trial
The Ventana pan-TRK assay is a ready-to-use or pre-diluted Food and Drug Administration (FDA) Class I in-vitro diagnostic assay designed to detect the C-region of TRKA, TRKB and TRKC proteins which is conserved in NTRK fusions and wild-type proteins [1, 36, 38, 42]. As such, Class I IVD assays, are intended for use in the diagnosis of disease. The pan-TRK assay in this study is not authorized for predictive use or otherwise intended to influence therapeutic decision making alone. Nonetheless, the pan-TRK assay can be utilized as a screening method for NTRK fusions; an actionable, albeit uncommon biomarker for targeted therapy.
For validation, we adhered to the 2014 CAP based guidelines for validation and verification of a non-predictive IHC marker. For validation of a non-predictive IHC assay, the CAP recommends examination of at least 10 positive and 10 negative specimens for initial analytic validation of non-predictive IHC assays with a goal of achieving at least 90% overall concordance between the new test and the comparative test or expected result. Positive and negative controls are required for the validation of an IHC assay. We sought to correlate our assay results (positive or negative expression of pan-TRK IHC) with the expected result based on tumor morphology or tumor diagnosis [48]. We compared our assay result with testing of the same tissue at a reference laboratory with a validated or verified pan-TRK IHC assay; and finally we compared our expected results of the pan-TRK assay with a previously validated or verified non-IHC tests which included fluorescent in-situ hybridization (FISH) using break-apart probes for the detection of NTRK1, NTRK2 and NTRK3 genes or confirmation with a molecular assay using an RNA based next-generation sequencing which is limited to targeted regions involving NTRK1, NTRK3 and NTRK3 genes. Given the pan-TRK IHC is not authorized as a predictive biomarker, guidance on when confirmatory testing for NTRK fusions should be performed were adapted by recommendations set forth by the European Society of Medical Oncology (ESMO) on the standard methods to detect NTRK fusions [39].
This validation protocol is not designed to study the predictive nature of the pan-TRK IHC to identify NTRK fusions; therefore, based on ESMO recommendations, orthogonal testing for the presence of a pan-TRK fusion was not performed on each individual case. This protocol is specifically intended for the validation of pan-TRK IHC for clinical use while remaining cost-effective.
Pan-TRK IHC Staining Procedure
IHC staining for pan-TRK expression was performed with iVIEW DAB Detection Kit (Ventana Medical Systems, Tucson, AZ), using a commercially available pan-TRK assay (predilute rabbit monoclonal antibody, clone EPR17341, Assay, RTU, Roche, Ventana). Tissue slides were produced from surgical pathology and cytopathology formalin-fixed, paraffin-embedded (FFPE) neoplastic tissue samples. Tissue slides were pretreated with a high PH heat-induced epitope retrieval (CC1) for 88 min and endured a 16-min antibody incubation time. Immunohistochemical stains were performed on the automated Benchmark Ultra platform (Ventana Medical Systems, Tucson, AZ). All assays were performed in a CLIA certified- laboratory.
Normal appendix tissue was used as control which showed weak or strong membranous and cytoplasmic staining of nerves and ganglion cells and negative staining in mucosa, smooth muscle, lymphoid aggregates and glandular epithelium, all of which served as positive and negative internal controls.
Sample selection
Rare tumors which commonly harbor NTRK fusions were considered as the gold standard for the positive control group while tumors which uncommonly harbor NTRK fusions were considered for the negative control group. Relatively common tumors which infrequently harbor NTRK fusions and may be considered include non-small cell lung cancer, colorectal carcinoma, pancreatic adenocarcinoma, cutaneous melanoma, and invasive breast carcinoma [49].
We selected a total of 36 tumors diagnosed from 2015 to 2020. Clinicopathologic characteristics of the patients are represented on Table 2. Selected cases included 5 cytology and 31 surgical specimens. For the positive control group, we identified cases of secretory carcinoma of the breast, and secretory carcinoma of the salivary gland, including its historical synonym of mammary analogue secretory carcinoma. Secretory breast carcinoma and secretory carcinoma of the salivary gland harbor NTRK fusions in up to 92.78% and 79.86%, respectively [53].
Table 2.
Clinicopathological characteristics of 36 patients evaluated for Pan-TRK expression
| PATIENT CHARACTERISTICS | No | % | |
|---|---|---|---|
| Age range | 46–88 y | ||
| Median | 67.5 | ||
| Female | 19 | 52.8 | |
| Male | 17 | 47.2 | |
| HISTOLOGIC TYPE & SOURCE | SITE OF ORIGIN | ||
| Fusion positive cases (n = 9) | |||
| Secretory carcinoma of the salivary gland (MASC) | 6 | Parotid/salivary gland | 16.8 |
| Secretory carcinoma | 1 | Breast | 2.8 |
| Metastatic MEC, lung | 1 | Parotid | 2.8 |
| MPNST | 1 | Soft tissue | 2.8 |
| Fusion negative cases (n = 26) | |||
| Invasive carcinoma | 1 | Breast | 2.8 |
| SMARCA4-BRG-1-deficient neoplasm | 1 | Lung | 2.8 |
| Adenocarcinoma, lung | 9 | Lung | 25.2 |
| Adenocarcinoma, lung | 2 | Colorectal (n = 1), unknown origin (n = 1) | 5.6 |
| Polymorphous adenocarcinoma | 1 | Soft palate | 2.8 |
| Adenocarcinoma, lymph node | 5 | Lung (n = 4), urothelial (n = 1) | 14 |
| Adenocarcinoma, pericardial fluid | 1 | Lung | 2.8 |
| Adenocarcinoma, pleural fluid | 2 | Lung (n = 1), unknown origin (n = 1) | 5.6 |
| Adenocarcinoma, soft tissue | 1 | Lung | 2.8 |
| Adenocarcinoma, bone | 2 | Lung (n = 1), unknown origin (n = 1) | 5.6 |
| High grade serous carcinoma | 1 | Ovary | 2.8 |
| Fusion status N/A (n = 1) | |||
| Adenocarcinoma, pleura | 1 | Lung | 2.8 |
Abbreviations: MASC Mammary analog secretory carcinoma, MPNST Malignant peripheral nerve sheath tumor, HGSC High-grade serous carcinoma, N/A Not available
We did not identify cases of infantile fibrosarcoma or congenital mesoblastic nephroma which reportedly harbor ETV6-NTRK3 in approximately 90% of cases [51–53]. A various collection of tumors types which uncommonly harbor NTRK fusions were used for negative control specimens however, primarily consisted of lung adenocarcinoma given its exceedingly low incidence of NTRK fusions, which may be detected in less than 1% of cases [51]. Lastly, pathologic reports including addenda were searched for reported “NTRK” fusion results or other alterations. In general, neuroendocrine tumor and gliomas were not used given the cross-reactivity with normal or wild-type expression of TRKA, TRKB and TRKC proteins.
The tumor types and sites examined in our validation are summarized in Table 2. Hematoxylin and eosin (H&E)-stained sections were evaluated and the diagnoses were confirmed, including examination by a Head and Neck Pathologist for salivary gland tumors.
Pan-TRK scoring
Pan-TRK expression was considered positive if the following subcellular staining patterns of any intensity were observed in ≥ 1% of tumor cells [1, 36, 40, 42–44]. Staining intensity was graded as negative (0), weak (1 +), moderate (2 +) or strong (3 +). Cytoplasmic and nuclear reactivity was considered positive for expression. Punctate staining alone was considered non-specific or equivocal.
Pan-TRK IHC validation
For cases in our negative control group, internal interpretation of pan-TRK results were compared to an outside interpretation performed at a reference laboratory offering a pan-TRK assay which is validated for clinical use. As per ESMO recommendations, for tumors which uncommonly harbor NTRK fusions, we did not pursue RNA-based sequencing for negative IHC results [39]. Additionally, in accordance with CAP guidelines, results of our negative control group were compared with the expected results based on tumor morphology (i.e. the negative control group is expected to yield a negative result based on the tumor diagnosis and low incidence of pan-TRK fusions in the tested tumor type).
In keeping with ESMO recommendations, in tumors suspected to harbor NTRK fusions or in cases with positive pan-TRK IHC expression and no prior confirmatory testing, results were confirmed by RNA based next-generation sequencing performed at an outside reference laboratory for confirmation of the fusion transcript. However, RNA-based sequencing was also attempted on equivocal IHC results. Non-concordant IHC results were recorded as a discrepancy. Lastly, we referred to preexisting or reported RNA sequencing and FISH results, confirming the presence of an NTRK fusion in select cases as identified in the archives.
Pan-TRK IHC results and interpretation
All cases (n = 36) were tested by pan-TRK IHC which included 9 NTRK3-fusion positive cases (8 cases with ETV6-NTRK3 fusion and 1 case with TPM4-NTRK3 fusion).
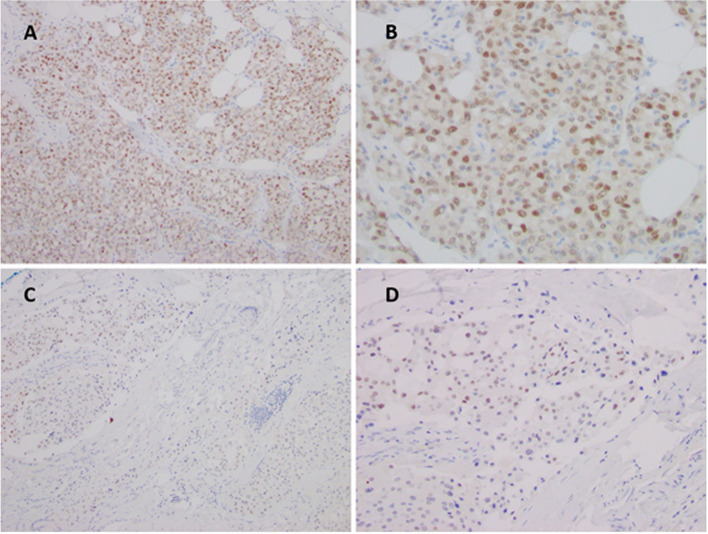
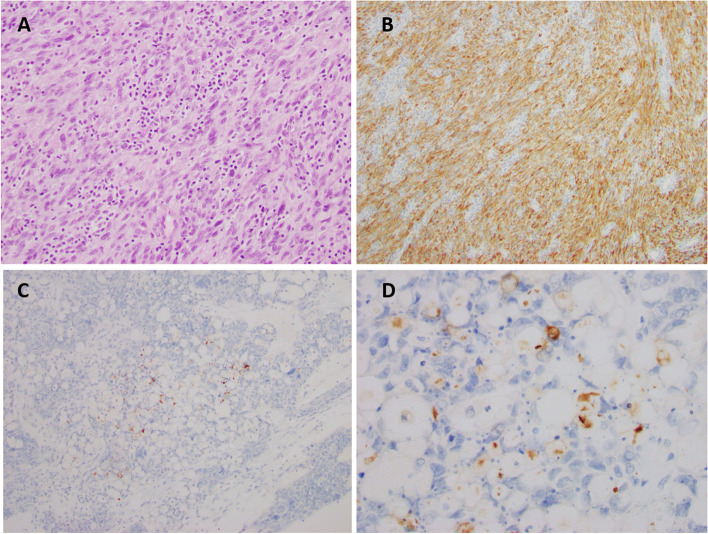
In the ETV6-NTRK3 fusion cohort, the pattern of Pan-TRK staining was both cytoplasmic and/or nuclear in all 8 out of 8 cases (Fig. 1A-B). The tissue distributions of cancers with ETV6-NTRK3 fusions included mammary analog secretory carcinoma (n = 6), metastatic mucoepidermoid carcinoma (n = 1), and breast secretory carcinoma (n = 1) and malignant peripheral nerve sheath tumor (n = 1) (Table 3). In two ETV6-NTRK3 fusion positive cases with Pan-TRK expression, staining was minimal (in approximately 5% of cells) (Fig. 1C-D). Of note, in three resection cases, nuclear staining was observed more at the periphery of the tumors and may be suggestive of a problem with adequate tissue fixation. One TPM4-NTRK3 fusion positive high grade malignant peripheral nerve sheath tumor (HGMPNST) revealed diffuse strong (3 +) cytoplasmic staining only. (Fig. 2A-B).
Fig. 1.
Different patterns of pan-TRK immunostaining with proven ETV6-NTRK3 fusion cases. A-B, Secretory carcinoma of the breast with diffuse nuclear and cytoplasmic staining. C-D, Focal and weak expression of Pan-TRK staining in secretory carcinoma of the parotid gland (Original magnification × 200, insets × 400)
Table 3.
Malignancies with Positive or equivocal (n=12) Internal (in-house) Pan-TRK IHC expression and NTRK NGS results
| Tumor location | Tumor diagnosis | NTRK NGS |
Pan-TRK IHC Reference lab |
Pan-TRK IHC In-house |
|---|---|---|---|---|
| Parotid gland | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (nuclear, 2+, 5%) |
| Parotid gland | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (cytoplasmic, 1+, 20%) |
| Parotid gland | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (cytoplasmic 1+, 100%; rare, focal nuclear staining, 2+, peripherally) |
| Parotid gland | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (cytoplasmic, peripherally, 1+, 5%) |
| Parotid gland | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (nuclear, peripherally, 2+, 15%) |
| Submaxillary gland | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (cytoplasmic 1+, 100%; scattered nuclear, 2+) |
| Lung | Metastatic MEC | ETV6-NTRK3 | Positive | Positive (cytoplasmic and nuclear staining, 1-2+, 100%) |
| Breast | Secretory carcinoma | ETV6-NTRK3 | Positive | Positive (nuclear and cytoplasmic, 2-3+, 70%) |
| Soft tissue-popliteal area | MPNST | TPM4-NTRK3 | Positive | Positive (cytoplasmic, 3+, 80%) |
| Soft palate | Polymorphous adenocarcinoma | No fusion detected | Positive | Positive (cytoplasmic, 2+, 90%) |
| Pleural fluid | Metastatic carcinoma, unknown origin | N/A | Positive, Focal | Equivocal (punctate, 1%) |
| Ovary | HGSC | NTRK1 truncation- intron 2 | Positive, Focal | Equivocal (punctate, 5%) |
Abbreviations: MPNST malignant peripheral nerve sheath tumor, HGSC high-grade serous carcinoma, MEC mucoepidermoid carcinoma, N/A not tested
Fig. 2.
Cases with various NTRK gene alterations. A, Representative H&E image of malignant peripheral nerve sheath tumor (MPNST). B, Strong membranous staining in a MPSNT harboring TPM4-NTRK3 fusion (Original magnification × 200, inset × 400). C-D, High grade ovarian serous carcinoma with focal punctate staining in areas with a microcystic pattern. This case revealed a NTRK1 truncation at intron 2 on NGS testing (Original magnification × 200, inset × 400)
Notably, focal or scattered nuclear staining was observed in 2 out of 8 ETV6-NTRK3 positive cases. Recently published studies showed that the staining patterns of the Pan-TRK antibody can vary in intensity and localization. Nuclear staining has been described in tumors harboring NTRK3 fusions, which can be used as a surrogate for the presence of NTRK3 fusions; however, the staining in these tumors can be weak and may cause false negative results [36, 37, 42, 43]. This weak focal staining may be responsible for a reduced sensitivity rate for NTRK3 fusions compared to fusions involving NTRK1 and NTRK2 [1, 42]. Other staining pattern described with ETV6-NTRK3 fusions include cellular membrane staining, cytoplasmic staining, and nuclear expression with accentuation of the nuclear envelope [28, 36, 40, 42]. All our cases with pan-TRK expression showed cytoplasmic staining, which was consistent with a prior assessment conducted by Hechtman et. al. using the same clone, EPR17341 [36] The staining patterns observed correlate with the subcellular localization of the most common NTRK fusion partners, ETV6 and TPM4.
Our analysis of polymorphous adenocarcinoma (n = 1) showed diffuse moderate cytoplasmic expression with pan-TRK IHC, both in-house and at the reference laboratory. While the IHC interpretation is concordant, orthogonal testing with RNA based NGS was negative for NTRK1-3 gene fusions.
We interpreted two cases as equivocal. One case of metastatic adenocarcinoma in the pleural fluid demonstrated granular cytoplasmic staining in scattered tumor cells and interpreted as equivocal staining internally and positive at the reference laboratory however; there was insufficient tissue for further orthogonal testing with RNA based NGS or FISH. Similarly, a discordant case of high grade ovarian serous carcinoma was interpreted as equivocal staining internally and interpreted as focally positive at a reference laboratory (Table 3). This case showed focal punctate staining in areas of tumor cells with a microcystic pattern (Fig. 2, C-D), which to our knowledge has not been described. Orthogonal testing revealed a NTRK1 truncation at intron 2 when reflexed to NGS NTRK gene fusion analysis at a reference laboratory. Although NTRK1 gene alterations in gynecologic tumors are rare, a recent case report presented a recurrent ovarian cancer with NTRK1-TPM3 fusion, in which entrectinib was ineffective. In this case report, NTRK1 gene fusion was detected by DNA-based NGS and IHC using Clone EPR17341 (Abcam, Cambridge, MA) was negative however; a scoring system for the interpretation of pan-TRK IHC was not described [47]. In contrast, cytoplasmic staining identified in tumors harboring NTRK1 fusions have been reported with different intensities.[1, 40, 41, 43].
Twenty-five tumors that uncommonly harbor NTRK fusions were interpreted as negative for pan-TRK expression internally and at the reference laboratory (Table 4). A discordant case of metastatic adenocarcinoma, favored to be of urothelial origin was interpreted as negative internally and focally positive at the reference laboratory. Similarly, a discordant case of pulmonary adenocarcinoma sampled by fine needle aspiration, was interpreted as negative for pan-TRK internally and as equivocal (focal) at the reference laboratory. Orthogonal testing was not pursued in these two cases given the tumor subtypes or unlikelihood of harboring an NTRK fusion and unequivocally negative internal result.
Table 4.
Malignancies with Negative (n = 24) Internal (in-house) results Pan-TRK IHC expression and NTRK results
| Tumor location | Tumor diagnosis | NTRK analysis | Pan-TRK IHC Reference lab |
Pan-TRK IHC In-house |
|---|---|---|---|---|
| Breast (n = 1) | Invasive carcinoma, exclude secretory carcinoma | ETV6-NTRK3 FISH negative | - | Negative |
| Lung (n = 1) | BRG-1 deficient tumor | - | Negative | Negative |
| Lung (n = 7) | Pulmonary adenocarcinoma | - | Negative | Negative |
| Pericardial fluid (n = 1) | Pulmonary adenocarcinoma | - | Negative | Negative |
| Lymph node (n = 1) | Adenocarcinoma with Neuroendocrine differentiation | - | Negative | Negative |
| Lymph node (n = 2) | Pulmonary adenocarcinoma | - | Negative | Negative |
| Lung (n = 1) | Colorectal adenocarcinoma | - | Negative | Negative |
| Bone (n = 1) | Poorly differentiated carcinoma | - | Negative | Negative |
| Bone (n = 1) | Pulmonary Adenocarcinoma | - | Negative | Negative |
| Soft tissue (n = 1) | Pulmonary mucinous adenocarcinoma | - | Negative | Negative |
| Lymph node (n = 1) | Pulmonary adenocarcinoma | - | Negative | Negative |
| Lymph node (n = 1) | Urothelial adenocarcinoma | - | Negative | Negative |
| Lung (n = 1) | Adenocarcinoma, unknown origin | - | Negative | Negative |
| Pleural fluid (n = 2) | Pulmonary adenocarcinoma | - | Negative | Negative |
| Lung, FNA (n = 1) | Pulmonary adenocarcinoma | - | Equivocal (focal) | Negative |
| Lung (n = 1) | Adenocarcinoma, Meningothelial-like nodule | Negative for NTRK1-3 fusion by NGS | Positive | Negative |
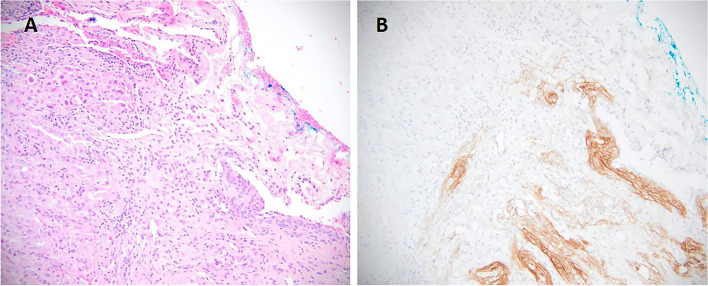
Retrospective review of H&E sections of a discordant case of lung adenocarcinoma which was interpreted as negative internally and positive at the reference laboratory, showed an adjacent entrapped meningothelial–like nodule (MLN) which exhibited strong, diffuse pan-TRK expression (Fig. 3, A-B). Co-occurrence of lesions with neural differentiation such as MLNs near or entrapped within the tumor bed, can make the interpretation of Pan-TRK IHC challenging. To distinguish between wild-type pan-TRK protein expression versus the presence of an NTRK fusion in MLNs, we expanded our analysis and performed RNA sequencing for NTRK fusions on 8 additional cases of MLNs identified from our archives. Out of 8 cases, 3 cases were excluded following microdissection due to insufficient lesional material being left for sequencing. 4 out of 5 of remaining cases had multiple MLNs on one section. All remaining 5 cases (100%) showed diffuse positive pan-TRK expression without confirmed NTRK fusions. Our findings therefore support wild-type expression of pan-TRK proteins in MLNs. Fusion analysis of MLNs was an important endeavor to distinguish between the potential identification of an NTRK fusion following orthogonal testing. Fortunately, a false positive pan-TRK result in the setting of MLNs will not lead to therapeutic implications as the presence of NTRK fusions in MLNs has not been identified.
Fig. 3.
Pan-TRK immunostaining in a meningothelial-like nodule (MLN). A, Representative H&E image of the meningothelial-like nodule located adjacent to the primary tumor in a lung resection specimen leading a false positive result. B, MLN shows strong cytoplasmic and membranous pan-TRK expression (original magnification × 200)
We were able to access the sensitivity and specificity based on any pan-TRK interpretation, and correlation with confirmatory testing results (Tables 3 and 4). Albeit a major limitation in accessing the predictability of this assay, NGS was not performed on all cases to make this a cost effective clinical validation. This was not intended to study the predictive correlation of pan-TRK expression and orthogonal testing results. As previously stated, per ESMO recommendations, orthogonal testing for the presence of NTRK fusion was not performed on negative cases given the exceedingly low probability of harboring a fusion based on specific tumor subtypes. In total, pan-TRK IHC was performed on 36 tumors and of those, 13 cases were interpreted as positive in-house or at the reference laboratory. One case with positive pan-TRK expression (pleural fluid) had insufficient tumor material for orthogonal testing. Therefore, NGS testing was performed on 12 of 13 cases.
Of these 12 pan-TRK positive cases, NTRK fusions were identified in 9 cases (true positives), while (following ESMO guidelines), testing for NTRK fusions was not performed on the presumed negative cohort (presumably, 0 false negatives). Therefore, the combined sensitivity of the pan-TRK IHC assay to identify NTRK fusions is 100%. Of the 26 expected and presumed fusion negative cases, pan/TRK IHC was positive in 3 cases (false positives). The combined specificity of the pan-TRK assay is 88%.
Among the tumors with pan-TRK expression and molecular testing, NTRK3 fusions (true positives) were identified with 100% sensitivity in 7/7 secretory carcinomas, 1/1 mucoepidermoid carcinoma and 1/1 malignant peripheral nerve sheath tumor.
Among the expected and presumed negative cohort (n = 26), false positive pan-TRK expression was observed in polymorphous adenocarcinoma (1/1), high grade ovarian serious carcinoma (1/1) and a single case of lung adenocarcinoma harboring a meningothelial-like nodule leading to false impression of epithelial cell staining.
Overall, there was 89% (32/36) concordance for the validation study between the pan-TRK IHC results conducted in-house and the reference laboratory including results for pan-TRK IHC, NTRK FISH, and RNA based NTRK 1–3 NGS fusion analysis used as a comparison (Tables 3 and 4). Interobserver agreement for pan-TRK IHC results between in the in-house pathologist and reference laboratory was observed; showing a Kappa coefficient of 0.63 (p < 0.001).
In conclusion, our overall concordance for our assay was slightly below the required parameters for the CAP recommendation of at least 90% overall concordance with expected results when compared to all comparator test i.e. outside reference laboratory and expected NTRK fusion positive results. Per CAP guidelines, if a laboratory director determines fewer validation cases are sufficient for a particular marker (such as for a rare antigen or tissue), the defendable rationale must be recorded in the validation summary. Given the rarity of tumors which harbor NTRK fusions, expected positive cases are difficult to come by, making meeting all CAP IHC validation parameters challenging, despite the relatively low number of minimum cases recommended for non-predictive markers (10 positive and 10 negative cases) and ease of use encountered with an in-vitro diagnostic assay. We therefore determined, the fewer number of positive controls were adequate for the validation, especially given our limited population showed positive pan-TRK expression in all (100%) NTRK rearranged tumors. Our results indicate that pan-TRK IHC is limited as a diagnostic marker however, has some utility as a marker to help screen for the identification of possible NTRK fusions; particularly ETV6-NTRK3 however, only if nuclear expression is observed and if an NTRK fusion is expected based on the tumor subtype.
Discussion
Pan-TRK IHC is a widely available, technically less challenging to perform and interpret, and may provide clinically actionable and time efficient results; especially if urgent therapeutic intervention is required, while awaiting orthogonal confirmation. Outside of major academic institutions with expanded fusion panels that may account for the detection of NTRK1-3 gene alterations, the vast majority of NTRK mutations may go undetected in advanced or metastatic disease. Detection of NTRK fusions in advanced or metastatic cases are recommended by National Comprehensive Cancer Network (NCCN) guidelines for cancers of lung, colon, breast, central nervous system cancers, pancreatic, gastrointestinal, hepatobiliary, thyroid and ovarian origin [50]. Validating Pan-TRK IHC may otherwise bridge this gap or delay in NTRK testing, especially if pan-TRK IHC is employed at the time of pathologic evaluation of advanced or metastatic cancers.
Evaluation of pan-TRK IHC is not without its challenges and may not be suitable for screening in all NCCN recommended cancers. A retrospective review of samples (2018–2021) to identify a correlation between positive pan-TRK (clone EPR17341) results and the presence of an NTRK gene fusion by Koehne de González et al., highlights the difficulty or lack of specificity when pan-TRK IHC is employed indiscriminately as well as the importance of NGS as the gold standard for detection of NTRK fusions. Koehne de González et al. reported a limited sensitivity to detect fusions however; 20/63 (31.7%) of cases with positive pan-TRK expression showed neuroendocrine or myogenic differentiation which are likely confounded by wild-type expression. Furthermore, weak or focal staining likely due to wild-type or non-specific expression was noted in 71.4% of cases, which did not include tumor types with a high incidence of NTRK fusions [50]. RNA fusion panel analysis of 127 cases identified 4 cases harboring NTRK fusions. Strong membranous and cytoplasmic pan-TRK IHC staining was reported in a case of TPM3-NTRK1 fusion positive colorectal carcinoma. Equivocal staining was noted in a pancreatic adenocarcinoma harboring a GOLGA4-NTRK3 fusion in which granular staining or “ISH-Like” staining was described. As an example of IHCs limited sensitivity, two cases (primary and metastatic tumors of lung) harboring ADAM19-NTRK3 fusions were negative for pan-TRK IHC. Per their institutional clinical practice, not all pan-TRK negative cases were accessed by RNA sequencing therefore the sensitivity could not be accessed [50]. As a general recommendation, the use of pan-TRK IHC as a screening modality should be limited in tumors with the potential of physiologic expression, such as gastrointestinal stromal tumors, neuroblastomas, glioblastomas and leiomyosarcomas [51].
A recent study by Mohamed et al. accessing the diagnostic value of pan-TRK (clone EPR17341, AbCam) in the detection of NTRK fusions in a subset (n = 23) of central nervous system (CNS) tumors, detected expression in 11/23 (47.8%) tumors. 2/11 tumors (liponeurocytoma and gliobastoma) with pan-TRK expression harbored AGBL4-NTRK, BEND5-NTRK2 fusions while no fusions were detected by NGS in all 12 tumors without pan-TRK expression (5). In the CNS, this study illustrates the inability of pan-TRK to distinguish between physiologic NTRK expression and the presence of an NTRK fusion however, the lack of staining can confidently exclude the latter.
The most common staining pattern encountered is cytoplasmic reactivity which can be observed in fusion positive and physiologic expression which can lead to a false-positive interpretation [51]. Given the conundrum of focal and weak staining which can be detected with pan-TRK IHC and the established lower limit of positivity (i.e. positivity in at least 1% of the tumor cell population), knowledge of the incidence of NTRK fusions in the tumor subtypes may influence whether such staining is interpreted as non-specific or equivocal. For example, NTRK fusions are detected in < 1% of NSCLCs, therefore, any increase in the frequency of staining in surgical or cytology specimens (after confirming a lack of background staining in the control tissue), should likely be regarded as equivocal and followed by reflexive testing with an orthogonal method. Since even NGS sequencing can be susceptible to a false negative result for NTRK fusion detection, your diagnostic consideration and insight into the incidence in NTRK fusions, such as the high incidence of NTRK fusions in secretory carcinoma of the salivary gland and breast, should prompt a conversation regarding the methodology or limitation of the NGS assay [51].
It’s important to note, false-positive staining has been described in approximately 8–10% of secretory carcinoma of the breast and salivary gland [51]. Among initial studies accessing pan-TRK IHC, Hechtman et al. reported a sensitivity and specificity of 95.2% and 100%, respectively and Rudzinski et al. reported a sensitivity and specificity or 97% and 98%, respectively, both of whom evaluated pan-TRK clone EPR17341 for the detection of NTRK rearrangements. Recently published studies additionally have shown sensitivities ranging from 75 to 92.5% and specificities between 81.1 and 100% [1, 36, 42, 45].
As a consideration, commercially available non-IVD pan-TRK IHC clones may be validated as a laboratory developed test. While the CAP guidelines for validation of an IHC antibody does serve to reduce variation amongst CAP accredited laboratories within the United States and its territories, a universal support plan for implementation of the pan-TRK assay does not exist, likely given the challenges posed, such as the implementation across various IHC platforms and validation as a laboratory developed test (for non-IVD antibodies).
In contrast, the Canadian CANTRK Ring Study, developed as a multicenter collaboration, underwent the endeavor of assisting laboratories across Canada to validate laboratory-developed IHC assays for pan-TRK screening, including the validation of NGS for the detection of NTRK fusions. For the adult population, the Canadian consensus group focused on methodologies in the detection of NTRK fusions in five specific neoplastic subgroups, which included thyroid carcinoma, colorectal carcinoma, NSCLC, soft tissue sarcomas and salivary gland carcinoma. In contrast to the ESMO guidelines which recommends a tumor agnostic approach based on the likelihood of harboring an NTRK fusions, the Canadian consensus group described recommendations for NGS testing dependent on the cancer subtype, extent of disease and provides a discussion on around available therapeutic options. While a separate publication on the Canadian consensus for NTRK biomarker testing in the pediatric populations exists, we intend to summarize key recommendations for the adult population by the Canadian consensus group on the use of pan-TRK IHC versus NGS analysis for NTRK1-3 genes, which may serve as a guide to direct subspecialty testing for NTRK alterations [53, 56].
TRK fusion positive thyroid carcinomas are clinically aggressive with high metastatic rates and multinodular growth [53]. For unresectable, metastatic or advanced thyroid cancer, pan-TRK IHC screening is recommended to be performed on all cases followed by NGS testing for NTRK to confirm IHC positive and equivocal cases [53].
For colorectal carcinoma, NTRK fusions are typically mutually exclusive of other mutations, thus the ideal scenario includes reflexive NTRK testing via NGS in all RAS/BRAF V600E wild-type, microsatellite instability (MSI)-high and mismatch repair deficient (dMMR) [53]. This strategy results in < 5% of metastatic patients requiring NTRK fusion testing; however, account for up to 90% of detected NTRK fusion positive colorectal carcinomas [53]. Further, since 11–23% of TRK fusion positive colorectal carcinomas have been identified in microsatellite stable/ MMR proficient (pMMR) tumors; it reasonable to consider NTRK gene fusion testing in MSI stable/ pMMR, RAS/ BRAF V600E wild-type patients however; testing this population accounts for 40% of cases tested [53]. If resources do not allow NTRK1-3 gene fusion testing, pan-TRK IHC is a suitable alternative with NGS testing performed to confirm a positive result.
The incidence of NTRK fusions in NSCLC is extremely low (0.23%), with a predilection to occur in younger patients (Median age of 47.6 years) with a 0–5-year smoking history. This specific demographic accounts for 73% of NTRK gene fusions in NSCLC with the vast majority of fusions detected in pulmonary adenocarcinoma (82%) [21, 53]. Since NTRK fusions are considered to be mutually exclusive, NTRK gene fusion testing should be ideally be considered as part of a larger molecular panel testing for ALK, ROS1, EGFR, KRAS, RET, BRAF, ERBB2 exon 20, NRG1 and MET. The Canadian consensus recommends integration of routine testing for NTRK1-3 genes in Stage III/IV patients with locally advanced or metastatic disease, in patients with non-squamous or adenocarcinoma components. NTRK testing can be considered for squamous cell carcinoma with mixed histology (example adenosquamous carcinoma) or in patients with a light smoking history. For limited molecular panels, the use of pan-TRK IHC is permissible, with or without NTRK fusion testing to confirm wild-type patients in non-squamous NSCLC. If pan-TRK IHC is performed after sequential molecular testing to first exclude the possibility of more common driver mutations, pan-TRK IHC testing would then be limited to 65% of NSCLC cases.
Within the vast soft tissue tumor differentials, inflammatory myofibroblastic tumor-like, fibrosarcoma/ malignant peripheral nerve sheath tumor-like or lipofibromatosis-like morphologic patterns show a higher incidence of harboring NTRK fusions [53]. Currently, NTRK testing is not recommended in leiomyosarcoma or liposarcoma since NTRK fusions have not been identified to date. NTRK fusions are mutually exclusive and are uncommonly identified in a subset of KIT/PDGFRA wild-type gastrointestinal stromal tumors (GIST). In this setting, NTRK1-3 gene fusion and BRAF mutation testing via NGS is recommended [53]. Otherwise, pan-TRK IHC may be utilized on other locally advanced or metastatic non-GIST soft tissue tumors with are negative for known or diagnostic molecular alterations to screen for NTRK fusions; followed by NGS for positive of IHC equivocal results [53].
Secretory carcinoma of the salivary gland accounts for 4–5% of salivary gland carcinomas, with the vast majority of them harboring NTRK3-ETV6 gene fusions. NGS is recommended for actionable NTRK and RET gene fusions [53]. A broad molecular panel is recommended if there is unclear histology. However, for all other salivary gland carcinomas, NGS is recommended in only locally advanced or metastatic disease [53]. NTRK testing may also be pursed following androgen receptor and HER2 IHC analysis [53]. Given the low incidence of salivary gland carcinomas, pan-TRK IHC is not recommended by the Canadian consensus group however, it may be considered to conserve resources and recommends confirmatory NGS or other molecular test following a positive or equivocal IHC result.
In summary, the Canadian consensus for biomarker testing and treatment of TRK fusion cancers offers a practical approach to testing for NTRK fusions, including the implementation of pan-TRK IHC; and to our knowledge remains the only expert consensus in North America to guide testing in the adult population to-date. While ultimately, the optimal testing strategy may be guided by the availability of institutional assays and expertise, comprehensive NGS remains the gold standard for detection of NTRK fusions and other targetable alterations across multiple tumor types.
At our institution, clinically advanced or metastatic cases are typically sent for RNA NGS to test for various alterations for targeted therapy or clinical trial inclusion while pan-TRK IHC serves to supplement initial analysis of a limited molecular panel. If NGS is not routinely performed in advanced malignant tumors, most algorithms support using pan-TRK IHC as screening method following by confirmation of positive cases with an orthogonal method, while acknowledging feasibility and cost should be taken into consideration [51].
There are benefits and limitations to consider with other testing methodologies utilized for the confirmation of NTRK fusions. Reverse transcriptase-polymerase chain reaction (RT-PCR) for example, shows variable sensitivity and high specificity however, detection of fusions requires specific primers targeting the involved or suspected genes and exons. RNA based NGS, like RT-PCR, may be hindered by RNA degradation however, provides the benefit of assessing for unknown fusion partners across multiple gene types, including other oncogenic alterations as well as splice variants [54]. While DNA-based NGS alone may access for point mutations, fusions, and copy number changes, the sensitivity for NTRK3 is limited (76.9%) and is reliant on decent tumor purity. Dual DNA/RNA based NGS offers superior analysis while likely being the most expensive methodology. FISH results offer a comparable turn-around time (approximately 1–3 days) compared to IHC however, FISH is designed to detect specific breakpoints and is likely best utilized when there is high suspicion of ETV6-NTRK3 fusions [54]. IHC, while limited in specificity, displays the ability to detect NTRK1-3 fusions, and wild-type TRK expression (54).
To date, only NTRK1-3 gene fusions have been identified as targetable alterations responsive to TRK inhibitors [54]. In-vitro analysis of tumor cells harboring known NTRK point mutations show no tumor driver potential but demonstrated impaired receptor activation and downstream signaling of no functional difference from wild-type receptors [54]. TRKB overexpression in neuroblastoma was associated with higher grade tumors with preclinical studies demonstrating responsiveness to TRK inhibitors in cells lines. Furthermore, high expression of full-length TRKC (NTRK3) in a EWSR1-WT1 fusion positive desmoplastic small round cell tumor showed sensitivity to TRK inhibitor therapy [54]. Diffuse membranous and cytoplasmic staining with NTRK3 IHC in DSRCT was identified confirming wild-type protein expression. In this study, two NTRK3-null DSRCT cell lines showed reduced tumor viability, supporting inhibition of NTRK3 by entrectinib and reprotrectinib. This study remarkably shows the transcriptional activation of NTRK3 by EWSR1-WT1 plays a significant role in the proliferation of DSRCT. This example highlights that further studies are needed to elucidate the functional significance of wild-type NTRK expression in tumor subtypes with known alterations if identified with pan-TRK IHC [53, 54].
We hope our experience and discussion will provide guidance into the validation, interpretation and implementation of pan-TRK IHC while creating awareness of potential pitfalls and challenges with interpretation. While further literature contributions and expert consensus from the Association for Molecular Pathology (AMP), may later define a standardized role for pan-TRK-IHC, for the foreseeable future it seems, pan-TRK IHC will continue to have a role as a screening method for actionable NTRK fusions until other testing methodologies continue to become widely available. Establishing an optimal algorithm for the detection of NTRK fusions may be dependent on the tumor subtype and perhaps best established by subspecialty consensus when possible. The goal of screening with immunohistochemistry or other methodology is intended to prolong patient survival by identifying patients who may benefit from TRK inhibitors, in a population that would otherwise go undetected.
Acknowledgements
Not applicable.
Disclaimers
No conflicts of interest to declare. Dr. Velez, received a one-time honorarium (Roche) to discuss his validation method of pan-TRK at the CAP annual meeting 2021.
Author’s contributions
MV: Conceptualization, Supervision, Writing, Editing, Data analysis; CK: Conceptualization, Writing, Editing, Data Analysis; TL: Data analysis; EG: Review, Editing; DH: Review, Editing.
Funding
Not applicable.
Availability of data and materials
All data supporting the findings and conclusions of this study is included within the article.
Declarations
Ethics approval and consent to participte
The study was exempt by the Institutional Review Board of University of Rochester.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
All authors consent for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019;32(1):147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 2.Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58–66. doi: 10.1016/j.pharmthera.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169(2):107–114. doi: 10.1016/S0304-3835(01)00530-4. [DOI] [PubMed] [Google Scholar]
- 4.Amatu A, Sartore-Bianchi A, Bencardino K, Pizzutilo EG, Tosi F, Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol. 2019;30. [DOI] [PMC free article] [PubMed]
- 5.Belli C, Penault-Llorca F, Ladanyi M, Normanno N, Scoazec JY, Lacroix L, et al. ESMO recommendations on the standard methods to detect RET fusions and mutations in daily practice and clinical research. Ann Oncol. 2021;32(3):337–350. doi: 10.1016/j.annonc.2020.11.021. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19(11):1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5(1):25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tognon C, Garnett M, Kenward E, Kay R, Morrison K, Sorensen PH. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 2001;61(24):8909–8916. [PubMed] [Google Scholar]
- 9.Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319(6056):743–748. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 10.Lasota J, Chłopek M, Lamoureux J, Christiansen J, Kowalik A, Wasąg B, et al. Colonic Adenocarcinomas Harboring NTRK Fusion Genes: A Clinicopathologic and Molecular Genetic Study of 16 Cases and Review of the Literature. Am J Surg Pathol. 2020;44(2):162–173. doi: 10.1097/PAS.0000000000001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Li ZW, Ou Q, Wu X, Nagasaka M, Shao Y, Ou SI, Yang Y. NTRK fusion positive colorectal cancer is a unique subset of CRC with high TMB and microsatellite instability. Cancer Med. 2022;11(13):2541–2549. doi: 10.1002/cam4.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knezevich SR, Garnett MJ, Pysher TJ, Beckwith JB, Grundy PE, Sorensen PH. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58(22):5046–5048. [PubMed] [Google Scholar]
- 13.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18(2):184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 14.Bourgeois JM, Knezevich SR, Mathers JA, Sorensen PH. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol. 2000;24(7):937–946. doi: 10.1097/00000478-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/S1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 16.Vasudev P, Onuma K. Secretory breast carcinoma: unique, triple-negative carcinoma with a favorable prognosis and characteristic molecular expression. Arch Pathol Lab Med. 2011;135(12):1606–1610. doi: 10.5858/arpa.2010-0351-RS. [DOI] [PubMed] [Google Scholar]
- 17.Skálová A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34(5):599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 18.Anderson J, Gibson S, Sebire NJ. Expression of ETV6-NTRK in classical, cellular and mixed subtypes of congenital mesoblastic nephroma. Histopathology. 2006;48(6):748–753. doi: 10.1111/j.1365-2559.2006.02400.x. [DOI] [PubMed] [Google Scholar]
- 19.Brzeziańska E, Karbownik M, Migdalska-Sek M, Pastuszak-Lewandoska D, Włoch J, Lewiński A. Molecular analysis of the RET and NTRK1 gene rearrangements in papillary thyroid carcinoma in the Polish population. Mutat Res. 2006;599(1–2):26–35. doi: 10.1016/j.mrfmmm.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, et al. ALK, ROS1, and NTRK Rearrangements in Metastatic Colorectal Cancer. JNCI: Journal of the National Cancer Institute. 2017;109(12). [DOI] [PubMed]
- 21.Farago AF, Taylor MS, Doebele RC, Zhu VW, Kummar S, Spira AI, et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis Oncol. 2018;2018. [DOI] [PMC free article] [PubMed]
- 22.Xu T, Wang H, Huang X, Li W, Huang Q, Yan Y, et al. Gene Fusion in Malignant Glioma: An Emerging Target for Next-Generation Personalized Treatment. Transl Oncol. 2018;11(3):609–618. doi: 10.1016/j.tranon.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alassiri AH, Ali RH, Shen Y, Lum A, Strahlendorf C, Deyell R, et al. ETV6-NTRK3 Is Expressed in a Subset of ALK-Negative Inflammatory Myofibroblastic Tumors. Am J Surg Pathol. 2016;40(8):1051–1061. doi: 10.1097/PAS.0000000000000677. [DOI] [PubMed] [Google Scholar]
- 24.Lezcano C, Shoushtari AN, Ariyan C, Hollmann TJ, Busam KJ. Primary and Metastatic Melanoma With NTRK Fusions. Am J Surg Pathol. 2018;42(8):1052–1058. doi: 10.1097/PAS.0000000000001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Busam KJ, Benayed R, Cimera R, Wang J, Denley R, et al. Identification of NTRK3 Fusions in Childhood Melanocytic Neoplasms. J Mol Diagn. 2017;19(3):387–396. doi: 10.1016/j.jmoldx.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol. 2010;321(1):44–49. doi: 10.1016/j.mce.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Ricarte-Filho JC, Li S, Garcia-Rendueles ME, Montero-Conde C, Voza F, Knauf JA, et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest. 2013;123(11):4935–4944. doi: 10.1172/JCI69766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang S, Cotzia P, Hyman DM, Drilon A, Tap WD, Zhang L, et al. NTRK Fusions Define a Novel Uterine Sarcoma Subtype With Features of Fibrosarcoma. Am J Surg Pathol. 2018;42(6):791–798. doi: 10.1097/PAS.0000000000001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferguson SD, Zhou S, Huse JT, de Groot JF, Xiu J, Subramaniam DS, et al. Targetable Gene Fusions Associate With the IDH Wild-Type Astrocytic Lineage in Adult Gliomas. J Neuropathol Exp Neurol. 2018;77(6):437–442. doi: 10.1093/jnen/nly022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7(4):400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidaway P. Targeted therapy: Larotrectinib effective against TRK-fusion-positive cancers. Nat Rev Clin Oncol. 2018;15(5):264. doi: 10.1038/nrclinonc.2018.40. [DOI] [PubMed] [Google Scholar]
- 33.Menichincheri M, Ardini E, Magnaghi P, Avanzi N, Banfi P, Bossi R, et al. Discovery of Entrectinib: A New 3-Aminoindazole As a Potent Anaplastic Lymphoma Kinase (ALK), c-ros Oncogene 1 Kinase (ROS1), and Pan-Tropomyosin Receptor Kinases (Pan-TRKs) inhibitor. J Med Chem. 2016;59(7):3392–3408. doi: 10.1021/acs.jmedchem.6b00064. [DOI] [PubMed] [Google Scholar]
- 34.Ardini E, Menichincheri M, Banfi P, Bosotti R, De Ponti C, Pulci R, et al. Entrectinib, a Pan-TRK, ROS1, and ALK Inhibitor with Activity in Multiple Molecularly Defined Cancer Indications. Mol Cancer Ther. 2016;15(4):628–639. doi: 10.1158/1535-7163.MCT-15-0758. [DOI] [PubMed] [Google Scholar]
- 35.Lange AM, Lo HW. Inhibiting TRK Proteins in Clinical Cancer Therapy. Cancers (Basel). 2018;10(4). [DOI] [PMC free article] [PubMed]
- 36.Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am J Surg Pathol. 2017;41(11):1547–1551. doi: 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung YP, Fletcher CDM, Hornick JL. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology. 2018;73(4):634–644. doi: 10.1111/his.13666. [DOI] [PubMed] [Google Scholar]
- 38.Rudzinski ER, Lockwood CM, Stohr BA, Vargas SO, Sheridan R, Black JO, et al. Pan-Trk Immunohistochemistry Identifies NTRK Rearrangements in Pediatric Mesenchymal Tumors. Am J Surg Pathol. 2018;42(7):927–935. doi: 10.1097/PAS.0000000000001062. [DOI] [PubMed] [Google Scholar]
- 39.Marchiò C, Scaltriti M, Ladanyi M, Iafrate AJ, Bibeau F, Dietel M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol. 2019;30(9):1417–1427. doi: 10.1093/annonc/mdz204. [DOI] [PubMed] [Google Scholar]
- 40.Brčić I, Godschachner TM, Bergovec M, Igrec J, Till H, Lackner H, et al. Broadening the spectrum of NTRK rearranged mesenchymal tumors and usefulness of pan-TRK immunohistochemistry for identification of NTRK fusions. Mod Pathol. 2021;34(2):396–407. doi: 10.1038/s41379-020-00657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi Y, Won YJ, Lee S, Kim A, Kim Y, Park WY, et al. Cytoplasmic TrkA Expression as a Screen for Detecting NTRK1 Fusions in Colorectal Cancer. Transl Oncol. 2018;11(3):764–770. doi: 10.1016/j.tranon.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020;33(1):38–46. doi: 10.1038/s41379-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Winne K, Sorber L, Lambin S, Siozopoulou V, Beniuga G, Dedeurwaerdere F, et al. Immunohistochemistry as a screening tool for NTRK gene fusions: results of a first Belgian ring trial. Virchows Arch. 2021;478(2):283–291. doi: 10.1007/s00428-020-02921-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strohmeier S, Brcic I, Popper H, Liegl-Atzwanger B, Lindenmann J, Brcic L. Applicability of pan-TRK immunohistochemistry for identification of NTRK fusions in lung carcinoma. Sci Rep. 2021;11(1):9785. doi: 10.1038/s41598-021-89373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell D, Ferrarotto R, Liang L, Goepfert RP, Li J, Ning J, et al. Pan-Trk immunohistochemistry reliably identifies ETV6-NTRK3 fusion in secretory carcinoma of the salivary gland. Virchows Arch. 2020;476(2):295–305. doi: 10.1007/s00428-019-02640-7. [DOI] [PubMed] [Google Scholar]
- 46.Ziegler YS, Moresco JJ, Tu PG, Yates JR, 3rd, Nardulli AM. Plasma membrane proteomics of human breast cancer cell lines identifies potential targets for breast cancer diagnosis and treatment. PLoS ONE. 2014;9(7):e102341. doi: 10.1371/journal.pone.0102341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endo Y, Watanabe T, Saito M, Saito K, Suzuki R, Sano H, et al. A rare case of recurrent ovarian cancer with TPM3-NTRK1 gene rearrangement: A case report. Mol Clin Oncol. 2022;16(4):90. doi: 10.3892/mco.2022.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitzgibbons PL, Bradley LA, Fatheree LA, et al. Principles of analytic validation of immunohistochemical assays: guideline from the college of american pathologists pathology and laboratory quality center. Arch Pathol Lab Med. 2014;138(11):1432–1443. doi: 10.5858/arpa.2013-0610-CP. [DOI] [PubMed] [Google Scholar]
- 49.Forsythe A, Zhang W, Phillip Strauss U, Fellous M, Korei M, Keating K. A systematic review and meta-analysis of neurotrophic tyrosine receptor kinase gene fusion frequencies in solid tumors. Ther Adv Med Oncol. 2020;21(12):1758835920975613. doi: 10.1177/1758835920975613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koehne de González A, Mansukhani MM, Fernandes H, Hsiao SJ. Pan-tumor screening for NTRK gene fusions using pan-TRK immunohistochemistry and RNA NGS fusion panel testing. Cancer Genet. 2022;262–263:47–52. [DOI] [PubMed]
- 51.Conde E, Hernandez S, Sanchez E, et al. Pan-TRK Immunohistochemistry: An Example-Based Practical Approach to Efficiently Identify Patients With NTRK Fusion Cancer. Arch Pathol Lab Med. 2021;145(8):1031–1040. doi: 10.5858/arpa.2020-0400-RA. [DOI] [PubMed] [Google Scholar]
- 52.Zhao X, Kotch C, Fox E, et al. NTRK Fusions Identified in Pediatric Tumors: The Frequency, Fusion Partners, and Clinical Outcome. JCO Precis Oncol. 2021;1:PO.20.00250. Published 2021 Jan 14. [DOI] [PMC free article] [PubMed]
- 53.Bebb DG, Banerji S, Blais N, et al. Canadian Consensus for Biomarker Testing and Treatment of TRK Fusion Cancer in Adults. Curr Oncol. 2021;28(1):523–548. Published 2021 Jan 15. [DOI] [PMC free article] [PubMed]
- 54.Hechtman JF. NTRK insights: best practices for pathologists. Mod Pathol. 2022;35(3):298–305. doi: 10.1038/s41379-021-00913-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohamed F, Kurdi M, Baeesa S, Sabbagh AJ, Hakamy S, Maghrabi Y, Alshedokhi M, Dallol A, Halawa TF, Najjar AA, Fdl-Elmula I. The Diagnostic Value of Pan-Trk Expression to Detect Neurotrophic Tyrosine Receptor Kinase (NTRK) Gene Fusion in CNS Tumours: A Study Using Next-Generation Sequencing Platform. Pathol Oncol Res. 2022;28(28):1610233. doi: 10.3389/pore.2022.1610233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perreault S, Chami R, Deyell RJ, El Demellawy D, Ellezam B, Jabado N, Morgenstern DA, Narendran A, Sorensen PHB, Wasserman JD, Yip S. Canadian Consensus for Biomarker Testing and Treatment of TRK Fusion Cancer in Pediatric Patients. Curr Oncol. 2021;28(1):346–366. doi: 10.3390/curroncol28010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings and conclusions of this study is included within the article.